A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study
Abstract
1. Introduction
2. Results
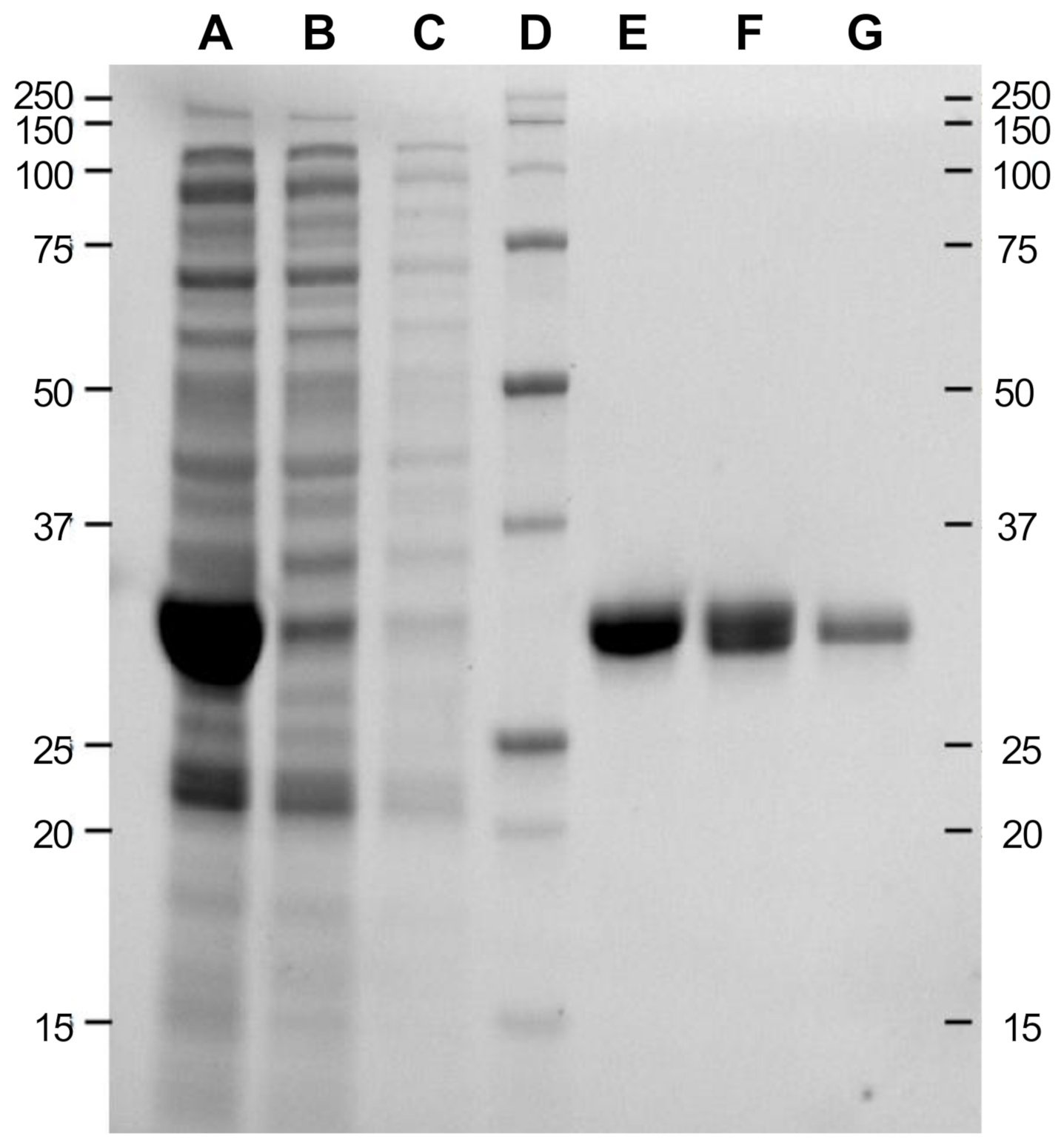
2.1. Recombinant Expression and Purification of TtEst2
2.2. Enzymatic Characterization
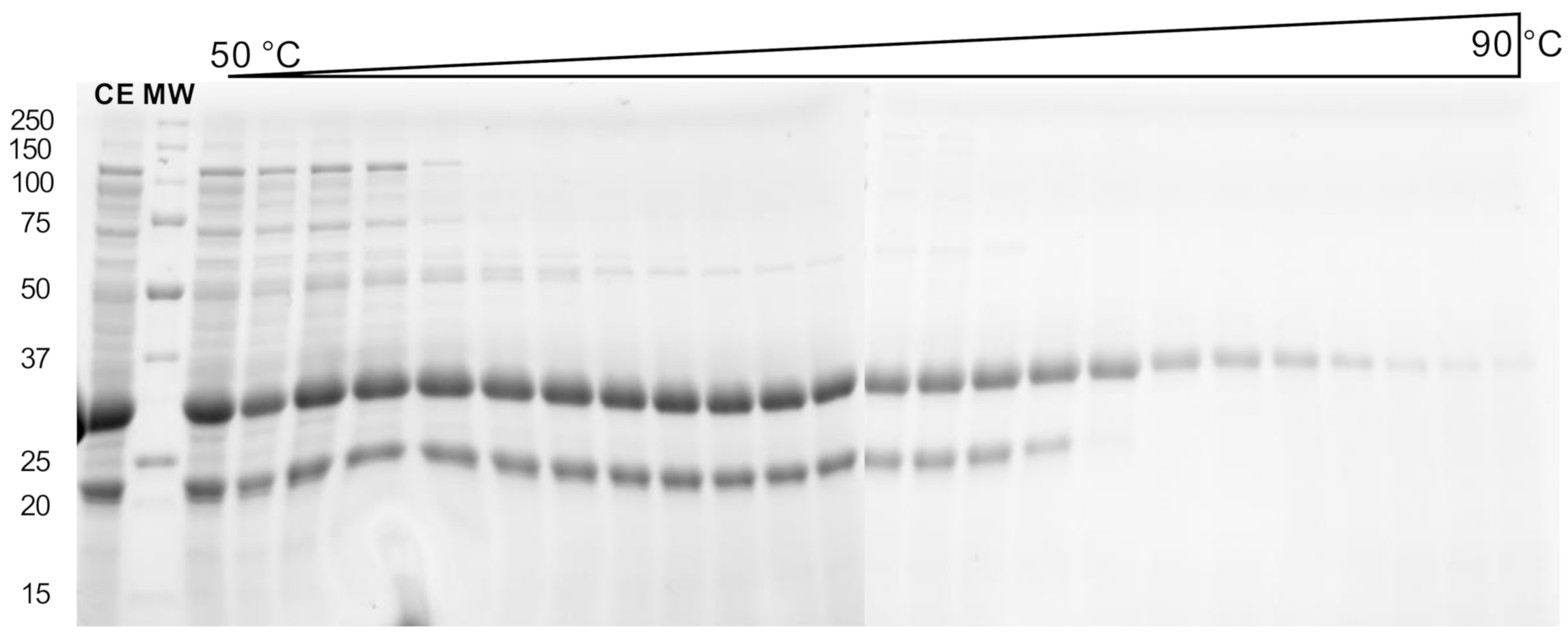
2.3. Biophysical Characterization
2.4. Crystallization of TtEst2
2.5. X-ray Structures of TtEst2 and OPNA Conjugates
2.5.1. Structure of Apo-TtEst2
2.5.2. Structures of VX- and Sarin-TtEst2 Conjugates
2.5.3. Structure of Tabun-TtEst2, Cyclosarin, and Paraoxon Conjugates
3. Discussion and Conclusions
4. Materials and Methods
4.1. Recombinant Expression and Purification of TtEst2
4.2. Acrylamide Gel Electrophoresis under Denaturing Condition (SDS-PAGE)
4.3. Enzymatic Assay and Inhibition Measurements
4.4. Size Exclusion Chromatography Coupled to Multi-Angle Light Scattering (SEC-MALS)
4.5. Thermal Shift Assay
4.6. Protein Crystallization and Crystal Inhibitions
4.7. Structure Determination
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Watson, A.; Opresko, D.; Young, R.A.; Hauschild, V.; King, J.; Bakshi, K. Organophosphate Nerve Agents. In Handbook of Toxicology of Chemical Warfare Agents, 2nd ed.; Gupta, R.C., Ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 87–110. [Google Scholar]
- Mew, E.J.; Padmanathan, P.; Konradsen, F.; Eddleston, M.; Chang, S.S.; Phillips, M.R.; Gunnell, D. The global burden of fatal self-poisoning with pesticides 2006-15: Systematic review. J. Affect. Disord. 2017, 219, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Mercey, G.; Verdelet, T.; Renou, J.; Kliachyna, M.; Baati, R.; Nachon, F.; Jean, L.; Renard, P.Y. Reactivators of acetylcholinesterase inhibited by organophosphorus nerve agents. Acc. Chem. Res. 2012, 45, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Gorecki, L.; Gerlits, O.; Kong, X.; Cheng, X.; Blumenthal, D.K.; Taylor, P.; Ballatore, C.; Kovalevsky, A.; Radic, Z. Rational design, synthesis, and evaluation of uncharged, “smart” bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase. J. Biol. Chem. 2020, 295, 4079–4092. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Brazzolotto, X.; Trovaslet, M.; Masson, P. Progress in the development of enzyme-based nerve agent bioscavengers. Chem. Biol. Interact. 2013, 206, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Mann, T.M.; Price, M.E.; Whitmore, C.L.; Perrott, R.L.; Laws, T.R.; McColm, R.R.; Emery, E.R.; Tattersall, J.E.H.; Green, A.C.; Rice, H. Bioscavenger is effective as a delayed therapeutic intervention following percutaneous VX poisoning in the guinea-pig. Toxicol. Lett. 2018, 293, 198–206. [Google Scholar] [CrossRef]
- Mumford, H.; Price, M.E.; Cerasoli, D.M.; Teschner, W.; Ehrlich, H.; Schwarz, H.P.; Lenz, D.E. Efficacy and physiological effects of human butyrylcholinesterase as a post-exposure therapy against percutaneous poisoning by VX in the guinea-pig. Chem. Biol. Interact. 2010, 187, 304–308. [Google Scholar] [CrossRef]
- Saxena, A.; Hastings, N.B.; Sun, W.; Dabisch, P.A.; Hulet, S.W.; Jakubowski, E.M.; Mioduszewski, R.J.; Doctor, B.P. Prophylaxis with human serum butyrylcholinesterase protects Gottingen minipigs exposed to a lethal high-dose of sarin vapor. Chem. Biol. Interact. 2015, 238, 161–169. [Google Scholar] [CrossRef]
- Saxena, A.; Sun, W.; Dabisch, P.A.; Hulet, S.W.; Hastings, N.B.; Jakubowski, E.M.; Mioduszewski, R.J.; Doctor, B.P. Pretreatment with human serum butyrylcholinesterase alone prevents cardiac abnormalities, seizures, and death in Gottingen minipigs exposed to sarin vapor. Biochem. Pharmacol. 2011, 82, 1984–1993. [Google Scholar] [CrossRef]
- Ashani, Y.; Pistinner, S. Estimation of the upper limit of human butyrylcholinesterase dose required for protection against organophosphates toxicity: A mathematically based toxicokinetic model. Toxicol. Sci. 2004, 77, 358–367. [Google Scholar] [CrossRef]
- Geyer, B.C.; Kannan, L.; Cherni, I.; Woods, R.R.; Soreq, H.; Mor, T.S. Transgenic plants as a source for the bioscavenging enzyme, human butyrylcholinesterase. Plant Biotechnol. J. 2010, 8, 873–886. [Google Scholar] [CrossRef]
- Huang, Y.J.; Huang, Y.; Baldassarre, H.; Wang, B.; Lazaris, A.; Leduc, M.; Bilodeau, A.S.; Bellemare, A.; Cote, M.; Herskovits, P.; et al. Recombinant human butyrylcholinesterase from milk of transgenic animals to protect against organophosphate poisoning. Proc. Natl. Acad. Sci. USA 2007, 104, 13603–13608. [Google Scholar] [CrossRef] [PubMed]
- Brazzolotto, X.; Wandhammer, M.; Ronco, C.; Trovaslet, M.; Jean, L.; Lockridge, O.; Renard, P.Y.; Nachon, F. Human butyrylcholinesterase produced in insect cells: Huprine-based affinity purification and crystal structure. FEBS J. 2012, 279, 2905–2916. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, L.M.; Lockridge, O.; David, E.; Hinrichs, S.H. Purification of human butyrylcholinesterase from frozen Cohn fraction IV-4 by ion exchange and Hupresin affinity chromatography. PLoS ONE 2019, 14, e0209795. [Google Scholar] [CrossRef] [PubMed]
- Bigley, A.N.; Raushel, F.M. The evolution of phosphotriesterase for decontamination and detoxification of organophosphorus chemical warfare agents. Chem. Biol. Interact. 2019, 308, 80–88. [Google Scholar] [CrossRef]
- Jacquet, P.; Daude, D.; Bzdrenga, J.; Masson, P.; Elias, M.; Chabriere, E. Current and emerging strategies for organophosphate decontamination: Special focus on hyperstable enzymes. Environ. Sci. Pollut. Res. Int. 2016, 23, 8200–8218. [Google Scholar] [CrossRef]
- Masson, P.; Nachon, F. Cholinesterase reactivators and bioscavengers for pre- and post-exposure treatments of organophosphorus poisoning. J. Neurochem. 2017, 142 (Suppl. 2), 26–40. [Google Scholar] [CrossRef]
- Lenfant, N.; Hotelier, T.; Velluet, E.; Bourne, Y.; Marchot, P.; Chatonnet, A. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: Tools to explore diversity of functions. Nucleic Acids Res. 2013, 41, D423–D429. [Google Scholar] [CrossRef]
- Sayer, C.; Szabo, Z.; Isupov, M.N.; Ingham, C.; Littlechild, J.A. The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta terrifontis Reveals an Open Active Site Due to a Minimal ‘Cap’ Domain. Front. Microbiol. 2015, 6, 1294. [Google Scholar] [CrossRef]
- Worek, F.; Thiermann, H.; Szinicz, L.; Eyer, P. Kinetic analysis of interactions between human acetylcholinesterase, structurally different organophosphorus compounds and oximes. Biochem. Pharmacol. 2004, 68, 2237–2248. [Google Scholar] [CrossRef]
- Afonine, P.V.; Moriarty, N.W.; Mustyakimov, M.; Sobolev, O.V.; Terwilliger, T.C.; Turk, D.; Urzhumtsev, A.; Adams, P.D. FEM: Feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 646–666. [Google Scholar] [CrossRef]
- Masson, P.; Nachon, F.; Lockridge, O. Structural approach to the aging of phosphylated cholinesterases. Chem. Biol. Interact. 2010, 187, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Mandrich, L.; Merone, L.; Manco, G. Hyperthermophilic phosphotriesterases/lactonases for the environment and human health. Environ. Technol. 2010, 31, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Slobodkina, G.B.; Kovaleva, O.L.; Miroshnichenko, M.L.; Slobodkin, A.I.; Kolganova, T.V.; Novikov, A.A.; van Heerden, E.; Bonch-Osmolovskaya, E.A. Thermogutta terrifontis gen. nov., sp. nov. and Thermogutta hypogea sp. nov., thermophilic anaerobic representatives of the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 2015, 65, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Sayer, C.; Isupov, M.N.; Bonch-Osmolovskaya, E.; Littlechild, J.A. Structural studies of a thermophilic esterase from a new Planctomycetes species, Thermogutta terrifontis. FEBS J. 2015, 282, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.; Radic, Z. The cholinesterases: From genes to proteins. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 281–320. [Google Scholar] [CrossRef]
- Ordentlich, A.; Barak, D.; Sod-Moriah, G.; Kaplan, D.; Mizrahi, D.; Segall, Y.; Kronman, C.; Karton, Y.; Lazar, A.; Marcus, D.; et al. Stereoselectivity toward VX is determined by interactions with residues of the acyl pocket as well as of the peripheral anionic site of AChE. Biochemistry 2004, 43, 11255–11265. [Google Scholar] [CrossRef]
- Benschop, H.P.; De Jong, L.P.A. Nerve agent stereoisomers: Analysis, isolation and toxicology. Acc. Chem. Res. 1988, 21, 368–374. [Google Scholar] [CrossRef]
- Zhao, H.; Brown, P.H.; Schuck, P. On the distribution of protein refractive index increments. Biophys. J. 2011, 100, 2309–2317. [Google Scholar] [CrossRef]
- Boivin, S.; Kozak, S.; Meijers, R. Optimization of protein purification and characterization using Thermofluor screens. Protein Expr. Purif. 2013, 91, 192–206. [Google Scholar] [CrossRef]
- Brazzolotto, X.; Igert, A.; Guillon, V.; Santoni, G.; Nachon, F. Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development. Molecules 2017, 22, 1828. [Google Scholar] [CrossRef]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A.; Diederichs, K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 2015, 34, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Legrand, P. XDSME: XDS Made Easier, GitHub Repository. 2017. Available online: https://github.com/legrandp/xdsme (accessed on 26 July 2020).
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Casanal, A.; Lohkamp, B.; Emsley, P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2020, 29, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Moriarty, N.W.; Grosse-Kunstleve, R.W.; Adams, P.D. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
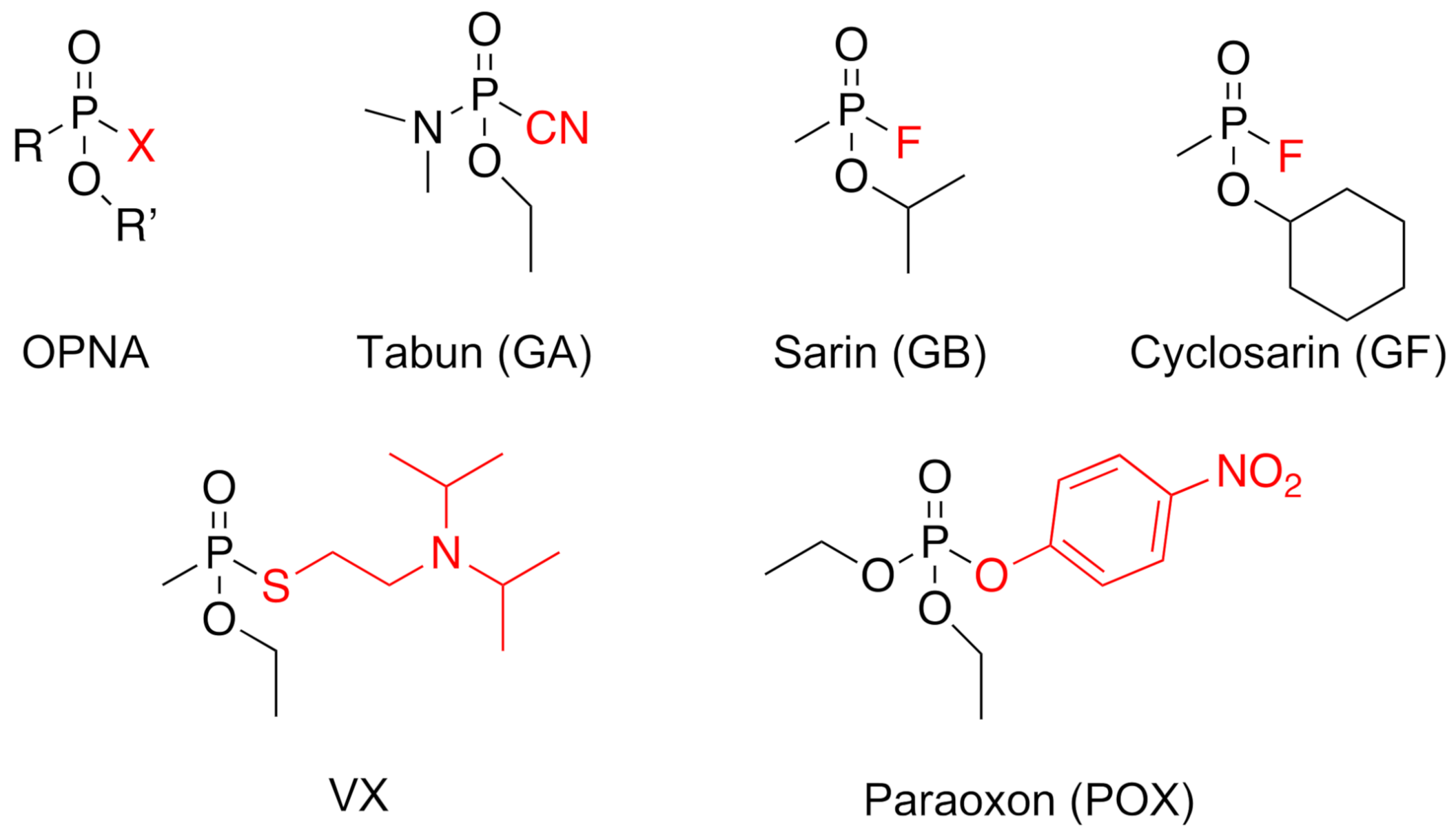

| ki (M−1·min−1) | |||
|---|---|---|---|
| OPNA | TtEst2 | hAChE from [20] | hAChE/TtEst2 |
| POX | 2.41 ± 0.12 × 104 | 2.2 × 106 | 91 |
| GA | 1.6 ± 0.4 × 106/5.1 ± 0.8 × 104 | 7.4 × 106 | 5/145 |
| GB | 1.51 ± 0.12 × 106 | 2.7 × 107 | 18 |
| GF | 1.29 ± 0.11 × 106 | 4.9 × 108 | 230 |
| VX | 540 ± 15 | 1.2 × 108 | 220,000 |
| Ligand | Apo | VX | Tabun | Sarin | Cyclosarin | Paraoxon |
|---|---|---|---|---|---|---|
| Pdb Code | 7bfn | 7bfo | 7bft | 7bfu | 7bfv | 7bfr |
| Data Collection | ||||||
| wavelength (Å) | 0.9786 | 0.9801 | 0.9801 | 0.9789 | 0.9786 | 0.9801 |
| Resolution range (Å) (highest-resolution shell) | 33.85–1.7 (1.77–1.7) | 42.11–1.994 (2.065–1.994) | 37.47–1.993 (2.064–1.993) | 36.5–1.65 (1.709–1.65) | 33.87–1.84 (1.906–1.84) | 36.74–1.99 (2.061–1.99) |
| space group, mol/AU | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| unit cell parameters (Å) | 56.21 67.7 74.58 90 90 90 | 53.7 67.84 75.76 90 90 90 | 53.74 67.87 74.94 90 90 90 | 52.86 67.71 75.71 90 90 90 | 55.83 67.74 74.52 90 90 90 | 53.52 67.64 75.97 90 90 90 |
| Total reflections | 405,998 (28,562) | 253,233 (23,090) | 247,497 (21,226) | 375,032 (31,442) | 325,398 (31,661) | 249,281 (19,651) |
| Unique reflections | 31,810 (2990) | 19,424 (1872) | 19,260 (1851) | 32,804 (3175) | 25,095 (2435) | 19,251 (1664) |
| Multiplicity | 12.8 (9.5) | 13.0 (12.3) | 12.9 (11.5) | 11.4 (9.8) | 13.0 (13.0) | 12.9 (11.8) |
| Completeness (%) | 98.66 (94.94) | 99.75 (98.37) | 99.54 (96.90) | 98.07 (96.59) | 99.26 (97.41) | 98.59 (86.79) |
| Mean I/σ (I) | 13.71 (1.07) | 29.15 (9.54) | 17.14 (5.33) | 17.40 (1.14) | 22.16 (4.76) | 27.16 (8.19) |
| Wilson B-factor | 35.45 | 26.79 | 25.37 | 26.44 | 33.92 | 25.44 |
| R-merge | 0.0860 (1.603) | 0.0597 (0.2776) | 0.1300 (0.5887) | 0.08227 (1.503) | 0.06329 (0.397) | 0.0801 (0.3037) |
| R-meas | 0.0897 (1.694) | 0.0622 (0.2896) | 0.1356 (0.6157) | 0.08608 (1.584) | 0.06611 (0.414) | 0.0835 (0.3169) |
| R-pim | 0.0251 (0.5321) | 0.0174 (0.0810) | 0.0380 (0.1758) | 0.0248 (0.4871) | 0.01879 (0.115) | 0.0232 (0.0890) |
| CC1/2 | 0.998 (0.776) | 0.999 (0.99) | 0.998 (0.978) | 0.999 (0.641) | 0.999 (0.99) | 0.999 (0.991) |
| CC * | 0.999 (0.935) | 1 (0.997) | 0.999 (0.995) | 1 (0.884) | 1 (0.998) | 1 (0.998) |
| Refinement Statistics | ||||||
| Reflections used | 31,554 (2986) | 19,410 (1871) | 19,232 (1844) | 32,727 (3175) | 24,979 (2408) | 19,232 (1662) |
| Reflections for R-free | 1264 (119) | 972 (94) | 962 (93) | 1310 (127) | 1249 (122) | 961 (83) |
| R-work | 0.2316 (0.4505) | 0.1601 (0.1708) | 0.1595 (0.1859) | 0.1785 (0.4862) | 0.1802 (0.2521) | 0.1611 (0.1760) |
| R-free | 0.2622 (0.4827) | 0.2034 (0.2237) | 0.1928 (0.2294) | 0.2204 (0.4904) | 0.2273 (0.2804) | 0.2073 (0.2858) |
| CC(work) | 0.954 (0.783) | 0.970 (0.956) | 0.966 (0.953) | 0.970 (0.442) | 0.962 (0.960) | 0.965 (0.962) |
| CC(free) | 0.954 (0.780) | 0.849 (0.827) | 0.966 (0.920) | 0.978 (0.269) | 0.925 (0.914) | 0.968 (0.844) |
| Number of non-H atoms | 2187 | 2435 | 2315 | 2461 | 2266 | 2261 |
| macromolecule | 2092 | 2191 | 2079 | 2187 | 2064 | 2037 |
| ligands | 5 | 3 | 5 | 3 | 6 | 10 |
| solvent | 90 | 241 | 231 | 271 | 196 | 214 |
| Protein residues | 268 | 277 | 264 | 276 | 264 | 260 |
| RMSD (bonds; Å) | 0.008 | 0.004 | 0.008 | 0.008 | 0.011 | 0.009 |
| RMSD (angles; deg) | 1.02 | 0.75 | 1.06 | 1.02 | 1.13 | 1.05 |
| Ramachandran favored (%) | 94.70 | 96.69 | 97.67 | 96.31 | 96.89 | 96.88 |
| Ramachandran allowed (%) | 5.30 | 2.57 | 2.33 | 2.95 | 3.11 | 3.12 |
| Ramachandran outliers (%) | 0.00 | 0.74 | 0.00 | 0.74 | 0.00 | 0.00 |
| Rotamer outliers (%) | 0.00 | 0.45 | 0.47 | 2.25 | 0.48 | 0.48 |
| Clashscore | 7.19 | 6.17 | 6.26 | 7.79 | 5.11 | 4.67 |
| Average B-factor (Å2) | 64.89 | 36.62 | 32.09 | 35.57 | 52.22 | 30.33 |
| macromolecules (Å2) | 65.30 | 36.14 | 31.27 | 34.95 | 52.18 | 29.53 |
| ligands (Å2) | 64.92 | 25.64 | 28.22 | 26.64 | 47.00 | 29.53 |
| solvent (Å2) | 55.28 | 41.16 | 39.61 | 40.67 | 52.82 | 37.91 |
| Number of TLS groups | 1 | 1 | 1 | 1 | 1 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bzdrenga, J.; Trenet, E.; Chantegreil, F.; Bernal, K.; Nachon, F.; Brazzolotto, X. A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study. Molecules 2021, 26, 657. https://doi.org/10.3390/molecules26030657
Bzdrenga J, Trenet E, Chantegreil F, Bernal K, Nachon F, Brazzolotto X. A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study. Molecules. 2021; 26(3):657. https://doi.org/10.3390/molecules26030657
Chicago/Turabian StyleBzdrenga, Janek, Elodie Trenet, Fabien Chantegreil, Kevin Bernal, Florian Nachon, and Xavier Brazzolotto. 2021. "A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study" Molecules 26, no. 3: 657. https://doi.org/10.3390/molecules26030657
APA StyleBzdrenga, J., Trenet, E., Chantegreil, F., Bernal, K., Nachon, F., & Brazzolotto, X. (2021). A Thermophilic Bacterial Esterase for Scavenging Nerve Agents: A Kinetic, Biophysical and Structural Study. Molecules, 26(3), 657. https://doi.org/10.3390/molecules26030657






