Tentative Peptide‒Lipid Bilayer Models Elucidating Molecular Behaviors and Interactions Driving Passive Cellular Uptake of Collagen-Derived Small Peptides
Abstract
:1. Introduction
2. Results
2.1. Molecular Docking Model
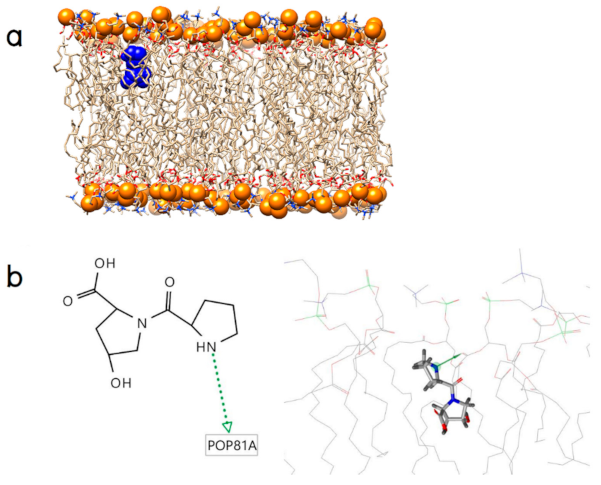
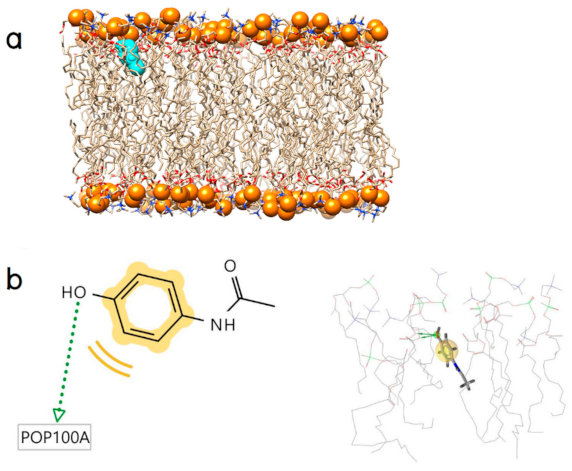
2.2. Initial Binding Interaction
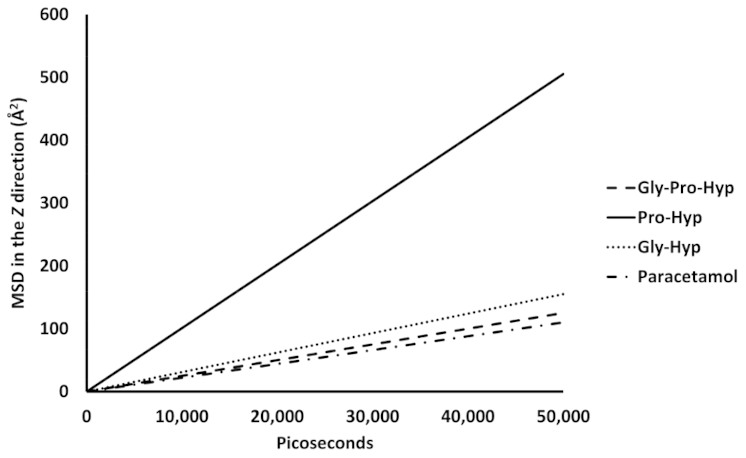
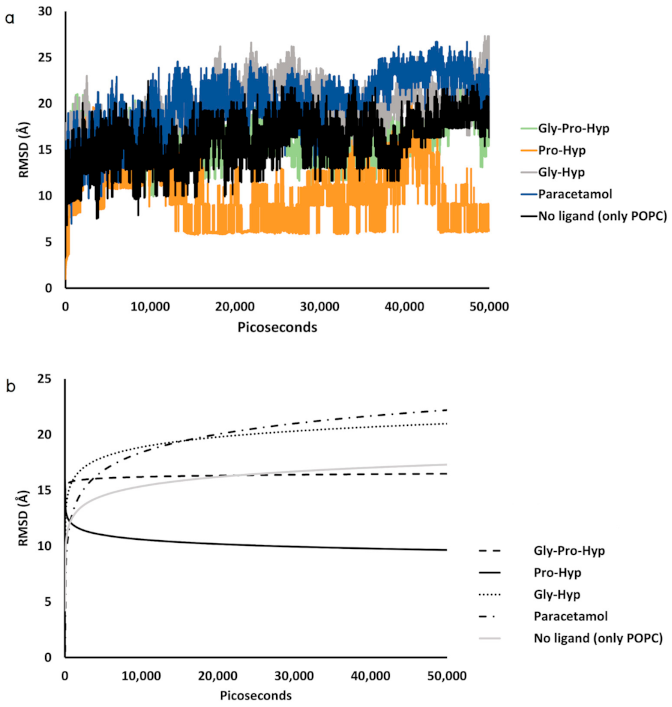
2.3. Dynamic Insertion Interaction and Mobility of the Small Collagen Peptides
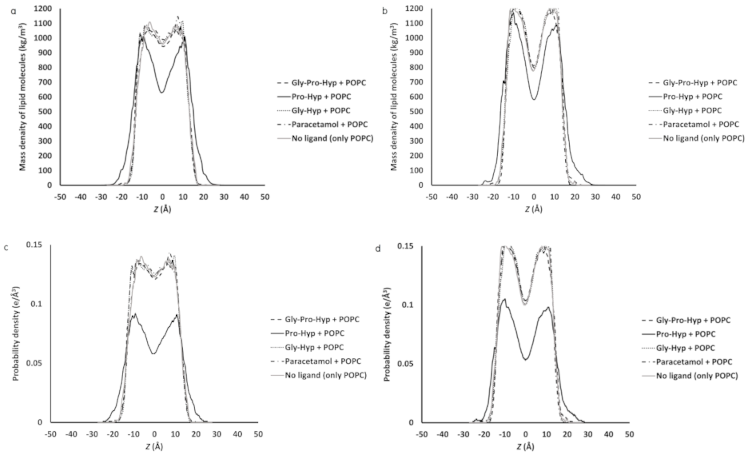
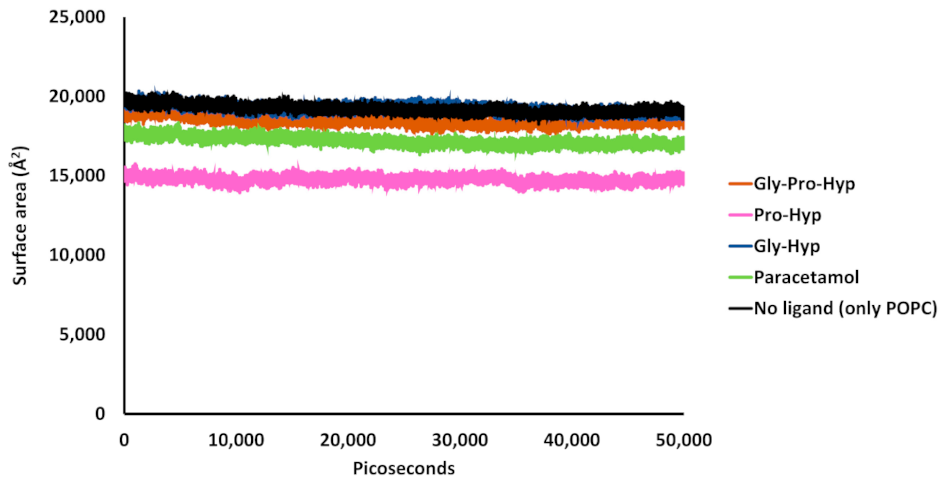
2.4. Effect of the Small Collagen Peptide on the Membrane Structure
3. Discussion
4. Materials and Methods
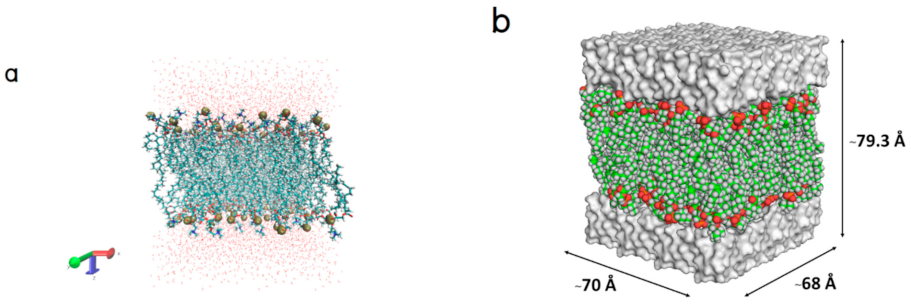
4.1. Lipid Bilayer Structure Preparation
4.2. Ligand Structure Preparation
4.3. Molecular Docking
4.4. MD Simulations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| AMBER | Assisted model building with energy refinement |
| CHARMM | Chemistry at Harvard Macromolecular Mechanics |
| DMPC | 1,2-Dimyristoyl-sn-glycero-3-phosphocholine |
| DPPC | Dipalmitoylphosphatidylcholine |
| GA | Genetic algorithm |
| Gly | Glycine |
| Gly-Hyp | Glycyl-hydroxyproline |
| Gly-Pro-Hyp | Glycylprolylhydroxyproline |
| GPU | Graphics processing unit |
| GUI | Graphical user interface |
| Hyp | Hydroxyproline |
| K | Lysine |
| kcal/mol | Kilocalorie per mole |
| LGA | Lamarckian genetic algorithm |
| MD | Molecular dynamics |
| MSD | Mean square displacement |
| PDB | Protein data bank |
| PEPT | Peptide transporter |
| PHT | Peptide histidine transporter |
| PMEMD | Particle mesh Ewald molecular dynamics |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| Pro | Proline |
| Pro-Hyp | Prolyl-hydroxyproline |
| Ps | Picoseconds |
| Q | Glutamine |
| RMSD | Root mean square deviation |
| TIP3P | Transferable intermolecular potential with 3 points |
| UCSF | The University of California, San Francisco |
| VMD | Visual molecular dynamics |
| Å | Angstrom |
References
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Ann. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogura, Y.; Tanaka, Y.; Hase, E.; Yamashita, T.; Yasui, T. Texture analysis of second-harmonic-generation images for quantitative analysis of reticular dermal collagen fibre in vivo in human facial cheek skin. Exp. Dermatol. 2019, 28, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Hayasaka, F.; Deguchi, K.; Okudera, T.; Furusawa, T.; Sakai, Y. Absorption and plasma kinetics of collagen tripeptide after peroral or intraperitoneal administration in rats. Biosci. Biotechnol. Biochem. 2015, 79, 2026–2033. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, S.; Deguchi, K.; Onuma, M.; Numata, N.; Sakai, Y. Absorption and Urinary Excretion of Peptides after Collagen Tripeptide Ingestion in Humans. Biol. Pharm. Bull. 2016, 39, 428–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Sugita, K.; Nihei, K.-I.; Yoneyama, K.; Tanaka, H. Absorption of Hydroxyproline-Containing Peptides in Vascularly Perfused Rat Small Intestine In Situ. Biosci. Biotechnol. Biochem. 2009, 73, 1741–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sontakke, S.B.; Jung, J.H.; Piao, Z.; Chung, H.J. Orally Available Collagen Tripeptide: Enzymatic Stability, Intestinal Permeability, and Absorption of Gly-Pro-Hyp and Pro-Hyp. J. Agric. Food Chem. 2016, 64, 7127–7133. [Google Scholar] [CrossRef]
- Ruaro, B.; Soldano, S.; Smith, V.; Paolino, S.; Contini, P.; Montagna, P.; Pizzorni, C.; Casabella, A.; Tardito, S.; Sulli, A.; et al. Correlation between circulating fibrocytes and dermal thickness in limited cutaneous systemic sclerosis patients: A pilot study. Rheumatol. Int. 2019, 39, 1369–1376. [Google Scholar] [CrossRef]
- Soldano, S.; Trombetta, A.C.; Contini, P.; Tomatis, V.; Ruaro, B.; Brizzolara, R.; Montagna, P.; Sulli, A.; Paolino, S.; Pizzorni, C.; et al. Increase in circulating cells coexpressing M1 and M2 macrophage surface markers in patients with systemic sclerosis. Ann. Rheum. Dis. 2018, 77, 1842–1845. [Google Scholar] [CrossRef]
- Maquart, F.X.; Monboisse, J.C. Extracellular matrix and wound healing. Pathol. Biol. 2014, 62, 91–95. [Google Scholar] [CrossRef]
- Surazynski, A.; Miltyk, W.; Palka, J.; Phang, J.M. Prolidase-dependent regulation of collagen biosynthesis. Amino Acids 2008, 35, 731–738. [Google Scholar] [CrossRef]
- Sugihara, F.; Inoue, N.; Kuwamori, M.; Taniguchi, M. Quantification of hydroxyprolyl-glycine (Hyp-Gly) in human blood after ingestion of collagen hydrolysate. J. Biosci. Bioeng. 2012, 113, 202–203. [Google Scholar] [CrossRef] [PubMed]
- Aito-Inoue, M.; Lackeyram, D.; Fan, M.Z.; Sato, K.; Mine, Y. Transport of a tripeptide, Gly-Pro-Hyp, across the porcine intestinal brush-border membrane. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2007, 13, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Nishikimi, A.; Koyama, Y.I.; Ishihara, S.; Kobayashi, S.; Tometsuka, C.; Kusubata, M.; Kuwaba, K.; Hayashida, O.; Hattori, S.; Katagiri, K. Collagen-derived peptides modulate CD4(+) T-cell differentiation and suppress allergic responses in mice. Immun. Inflamm. Dis. 2018, 6, 245–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostofian, B.; Johnson, Q.R.; Smith, J.C.; Cheng, X. Carotenoids promote lateral packing and condensation of lipid membranes. Phys. Chem. Chem. Phys. PCCP 2020, 22, 12281–12293. [Google Scholar] [CrossRef] [PubMed]
- Khoshakhlagh, P.; Johnson, R.; Nawroth, T.; Langguth, P.; Schmueser, L.; Hellmann, N.; Decker, H.; Szekely, N.K. Nanoparticle structure development in the gastro-intestinal model fluid FaSSIFmod6.5 from several phospholipids at various water content relevant for oral drug administration. Eur. J. Lipid Sci. Technol. 2014, 116, 1155–1166. [Google Scholar] [CrossRef]
- Moradi, S.; Nowroozi, A.; Shahlaei, M. Shedding light on the structural properties of lipid bilayers using molecular dynamics simulation: A review study. RSC Adv. 2019, 9, 4644–4658. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Ren, X.; Meng, F. Molecular dynamics simulation of six β-blocker drugs passing across POPC bilayer. Mol. Simul. 2016, 42, 56–63. [Google Scholar] [CrossRef]
- Kokate, A.; Li, X.; Williams, P.J.; Singh, P.; Jasti, B.R. In silico prediction of drug permeability across buccal mucosa. Pharmaceut. Res. 2009, 26, 1130–1139. [Google Scholar] [CrossRef]
- Seelig, J. Thermodynamics of lipid-peptide interactions. Biochim. Biophys. Acta 2004, 1666, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Porasso, R.D.; Ale, N.M.; Ciocco Aloia, F.; Masone, D.; Del Pópolo, M.G.; Ben Altabef, A.; Gomez-Zavaglia, A.; Diaz, S.B.; Vila, J.A. Interaction of glycine, lysine, proline and histidine with dipalmitoylphosphatidylcholine lipid bilayers: A theoretical and experimental study. RSC Adv. 2015, 5, 43537–43546. [Google Scholar] [CrossRef]
- Chen, Z.; Mao, Y.; Yang, J.; Zhang, T.; Zhao, L.; Yu, K.; Zheng, M.; Jiang, H.; Yang, H. Characterizing the binding of annexin V to a lipid bilayer using molecular dynamics simulations. Proteins 2014, 82, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.E.; White, S.H. The nature of the hydrophobic binding of small peptides at the bilayer interface: Implications for the insertion of transbilayer helices. Biochemistry 1989, 28, 3421–3437. [Google Scholar] [CrossRef]
- Babakhani, A.; Gorfe, A.A.; Gullingsrud, J.; Kim, J.E.; Andrew McCammon, J. Peptide insertion, positioning, and stabilization in a membrane: Insight from an all-atom molecular dynamics simulation. Biopolymers 2007, 85, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Phadke, R.S.; Govil, G.; Rao, C.N.R. Fluidity, permeability and antioxidant behaviour of model membranes incorporated with α-tocopherol and vitamin E acetate. Biochim. Biophys. Acta Biomembr. 1983, 734, 353–362. [Google Scholar] [CrossRef]
- Deepak, R.N.; Sankararamakrishnan, R. Unconventional N-H…N Hydrogen Bonds Involving Proline Backbone Nitrogen in Protein Structures. Biophys. J. 2016, 110, 1967–1979. [Google Scholar] [CrossRef] [Green Version]
- Diehl, R.C.; Guinn, E.J.; Capp, M.W.; Tsodikov, O.V.; Record, M.T., Jr. Quantifying additive interactions of the osmolyte proline with individual functional groups of proteins: Comparisons with urea and glycine betaine, interpretation of m-values. Biochemistry 2013, 52, 5997–6010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateo Marti, E.; Barlow, S.M.; Haq, S.; Raval, R. Bonding and assembly of the chiral amino acid S-proline on Cu(110): The influence of structural rigidity. Surf. Sci. 2002, 501, 191–202. [Google Scholar] [CrossRef]
- Scheidt, H.A.; Meyer, T.; Nikolaus, J.; Baek, D.J.; Haralampiev, I.; Thomas, L.; Bittman, R.; Müller, P.; Herrmann, A.; Huster, D. Cholesterol’s Aliphatic Side Chain Modulates Membrane Properties. Angew. Chem. Int. Ed. Engl. 2013, 52, 12848–12851. [Google Scholar] [CrossRef] [Green Version]
- Olsen, B.N.; Schlesinger, P.H.; Ory, D.S.; Baker, N.A. Side-chain oxysterols: From cells to membranes to molecules. Biochim. Biophys. Acta 2012, 1818, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Mangiarotti, A.; Genovese, D.M.; Naumann, C.A.; Monti, M.R.; Wilke, N. Hopanoids, like sterols, modulate dynamics, compaction, phase segregation and permeability of membranes. Biochim. Biophys. Biomembr. 2019, 1861, 183060. [Google Scholar] [CrossRef]
- Ho, C.S.; Khadka, N.K.; She, F.; Cai, J.; Pan, J. Polyglutamine aggregates impair lipid membrane integrity and enhance lipid membrane rigidity. Biochim. Biophys. Acta 2016, 1858, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Baumber, J.; Ball, B.A.; Gravance, C.G.; Medina, V.; Davies-Morel, M.C. The effect of reactive oxygen species on equine sperm motility, viability, acrosomal integrity, mitochondrial membrane potential, and membrane lipid peroxidation. J. Androl. 2000, 21, 895–902. [Google Scholar] [PubMed]
- Devi, K.P.; Nisha, S.A.; Sakthivel, R.; Pandian, S.K. Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J. Ethnopharmacol. 2010, 130, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009, 97, 50–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, C.J.; Madej, B.D.; Skjevik, A.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef]
- Rubbens, J.; Mols, R.; Brouwers, J.; Augustijns, P. Exploring gastric drug absorption in fasted and fed state rats. Int. J. Pharm. 2018, 548, 636–641. [Google Scholar] [CrossRef]
- Li, Q.; Cheng, T.; Wang, Y.; Bryant, S.H. PubChem as a public resource for drug discovery. Drug Discov. Today 2010, 15, 1052–1057. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhou, X.; Wang, D.; Yin, D.; Lin, Z. Using molecular docking between organic chemicals and lipid membrane to revise the well known octanol-water partition coefficient of the mixture. Environ. Toxicol. Pharmacol. 2012, 34, 59–66. [Google Scholar] [CrossRef]
- Raffa, R.B.; Pawasauskas, J.; Pergolizzi, J.V., Jr.; Lu, L.; Chen, Y.; Wu, S.; Jarrett, B.; Fain, R.; Hill, L.; Devarakonda, K. Pharmacokinetics of Oral and Intravenous Paracetamol (Acetaminophen) When Co-Administered with Intravenous Morphine in Healthy Adult Subjects. Clin. Drug Investig. 2018, 38, 259–268. [Google Scholar] [CrossRef] [Green Version]
- Holloway, A.C. Nicotine. In Encyclopedia of Toxicology, 3rd ed.; Wexler, P., Ed.; Academic Press: Oxford, UK, 2014; pp. 514–516. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Wongrattanakamon, P.; Nimmanpipug, P.; Sirithunyalug, B.; Chaiyana, W.; Jiranusornkul, S. Investigation of the Skin Anti-photoaging Potential of Swertia chirayita Secoiridoids Through the AP-1/Matrix Metalloproteinase Pathway by Molecular Modeling. Int. J. Pept. Res. Ther. 2019, 25, 517–533. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Computat. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miguel, V.; Defonsi Lestard, M.E.; Tuttolomondo, M.E.; Díaz, S.B.; Altabef, A.B.; Puiatti, M.; Pierini, A.B. Molecular view of the interaction of S-methyl methanethiosulfonate with DPPC bilayer. Biochim. Biophys. Acta Biomembr. 2016, 1858, 38–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villalaín, J. Epigallocatechin-3-gallate location and interaction with late endosomal and plasma membrane model membranes by molecular dynamics. J. Biomol. Struct. Dyn. 2019, 37, 3122–3134. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wongrattanakamon, P.; Yooin, W.; Sirithunyalug, B.; Nimmanpipug, P.; Jiranusornkul, S. Tentative Peptide‒Lipid Bilayer Models Elucidating Molecular Behaviors and Interactions Driving Passive Cellular Uptake of Collagen-Derived Small Peptides. Molecules 2021, 26, 710. https://doi.org/10.3390/molecules26030710
Wongrattanakamon P, Yooin W, Sirithunyalug B, Nimmanpipug P, Jiranusornkul S. Tentative Peptide‒Lipid Bilayer Models Elucidating Molecular Behaviors and Interactions Driving Passive Cellular Uptake of Collagen-Derived Small Peptides. Molecules. 2021; 26(3):710. https://doi.org/10.3390/molecules26030710
Chicago/Turabian StyleWongrattanakamon, Pathomwat, Wipawadee Yooin, Busaban Sirithunyalug, Piyarat Nimmanpipug, and Supat Jiranusornkul. 2021. "Tentative Peptide‒Lipid Bilayer Models Elucidating Molecular Behaviors and Interactions Driving Passive Cellular Uptake of Collagen-Derived Small Peptides" Molecules 26, no. 3: 710. https://doi.org/10.3390/molecules26030710
APA StyleWongrattanakamon, P., Yooin, W., Sirithunyalug, B., Nimmanpipug, P., & Jiranusornkul, S. (2021). Tentative Peptide‒Lipid Bilayer Models Elucidating Molecular Behaviors and Interactions Driving Passive Cellular Uptake of Collagen-Derived Small Peptides. Molecules, 26(3), 710. https://doi.org/10.3390/molecules26030710








