NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax
Abstract
1. Introduction
2. Results and Discussion
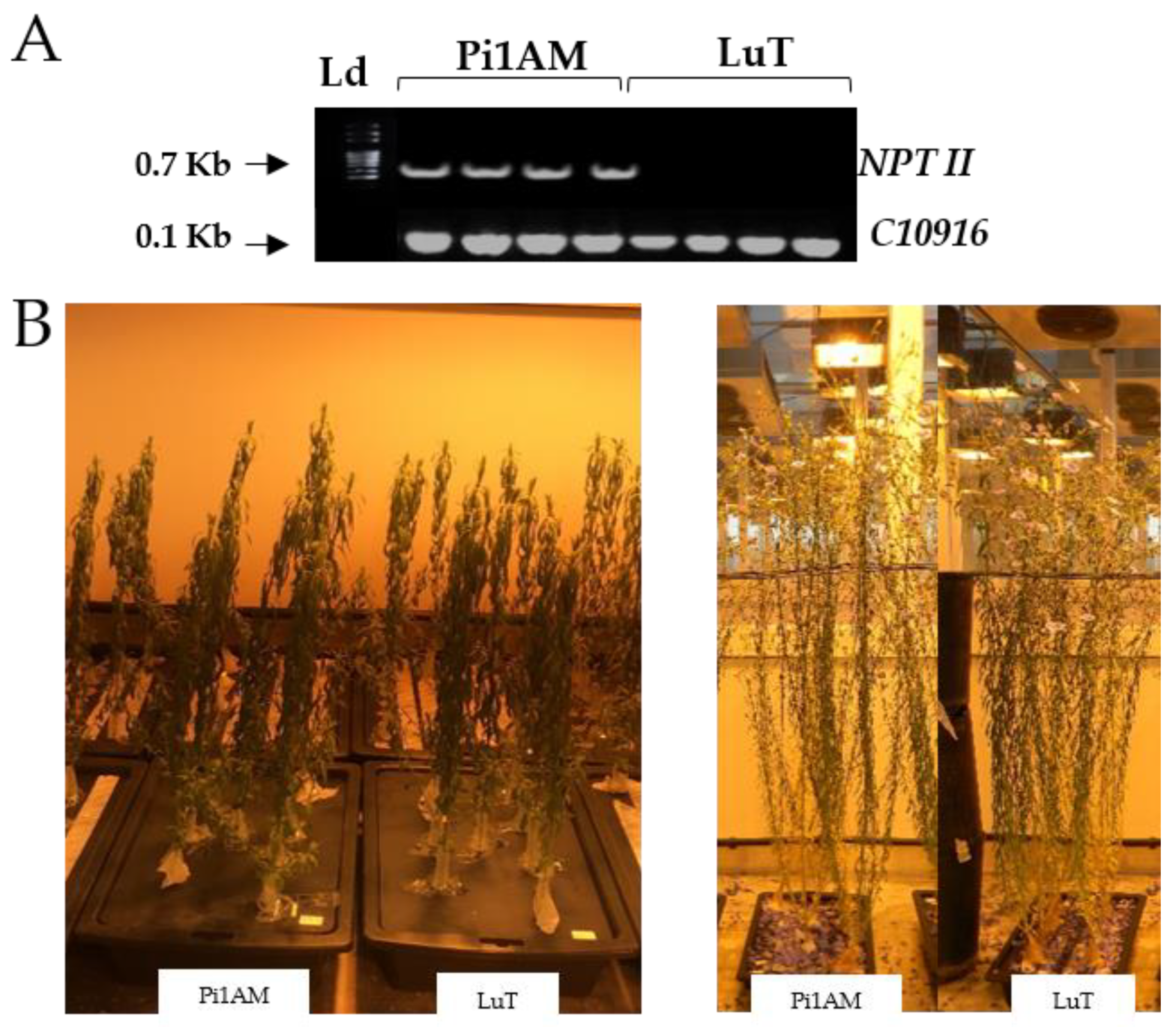
2.1. Phenotype Analysis
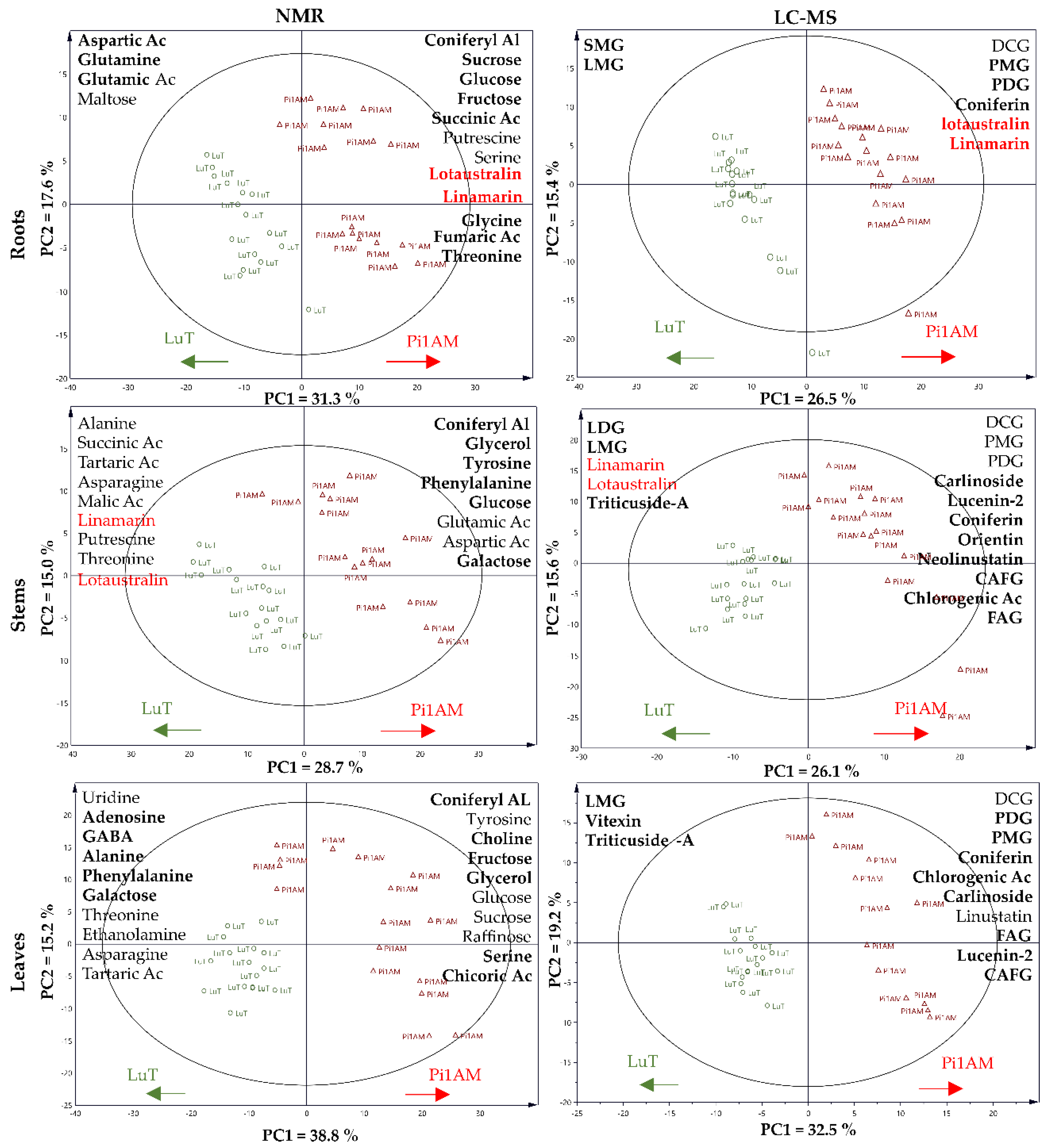
2.2. Metabolite Profiling
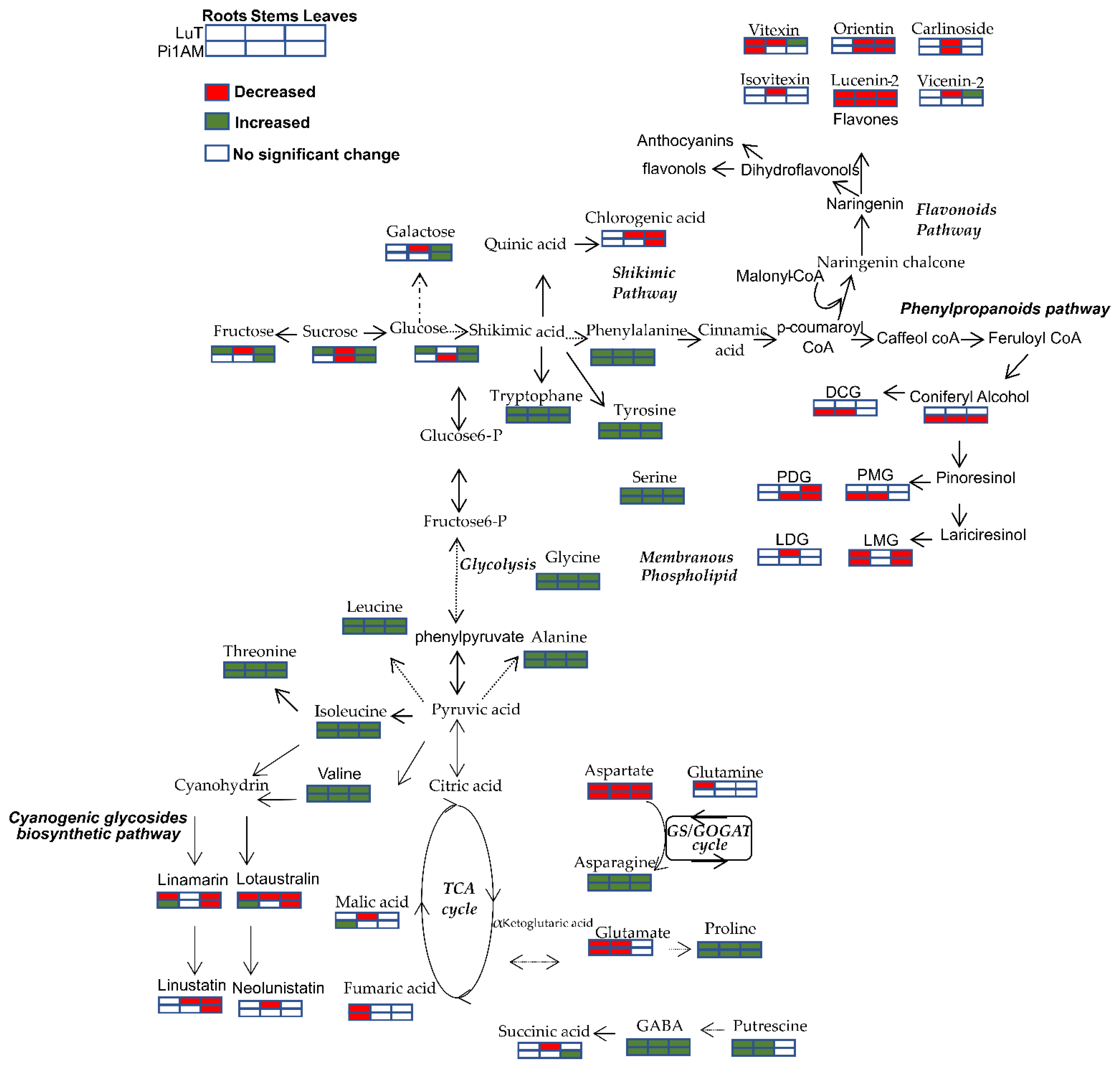
2.2.1. Difference of Metabolic Profiles between Wild-Type and PLR1-RNAi Transgenic Lines under Control Conditions
Changes in Primary Metabolism
Changes in Secondary Metabolism
2.2.2. Metabolites Change in Response to Osmotic Stress in Wild-Type and PLR1-RNAi Transgenic Line
Metabolic Profiles in Response to Osmotic Stress in Roots
Metabolic Profiles in Response to Osmotic Stress in Stems
Metabolic Profiles in Response to Osmotic Stress in Leaves
2.2.3. Comparison of the Metabolites in Wild-Type and PLR1-RNAi Transgenic Line under Osmotic Stress Conditions
3. Materials and Methods
3.1. Plant Material
3.2. PCR Analysis of Transgenic Plants
3.3. Metabolite Wxtractions
3.4. Metabolite Analysis by NMR
3.4.1. Sample Preparation
3.4.2. NMR Data Acquisition
3.4.3. NMR Data Treatment
3.4.4. Metabolite Analysis by LC/MS
Sample Preparation
LC-MS Data Acquisition
LC-MS/MS Data Processing
Statistical Treatment of Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Imran, M.; Ahmad, N.; Anjum, F.M.; Khan, M.K.; Mushtaq, Z.; Nadeem, M.; Hussain, S. Potential Protective Properties of Flax Lignan Secoisolariciresinol Diglucoside. Nutr. J. 2015, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T. Diversity in Lignan Biosynthesis. Phytochem. Rev. 2003, 2, 371–390. [Google Scholar] [CrossRef]
- Schmidt, T.; Hemmati, S.; Fuss, E.; Alfermann, A. A Combined HPLC-UV and HPLC-MS Method for the Identification of Lignans and Its Application to the Lignans of Linum Usitatissimum L. and L. Bienne Mill. Phytochem. Anal. Phytochem Anal. 2006, 17, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Hano, C.; Martin, I.; Fliniaux, O.; Legrand, B.; Gutierrez, L.; Arroo, R.R.J.; Mesnard, F.; Lamblin, F.; Lainé, E. Pinoresinol–Lariciresinol Reductase Gene Expression and Secoisolariciresinol Diglucoside Accumulation in Developing Flax (Linum Usitatissimum) Seeds. Planta 2006, 224, 1291–1301. [Google Scholar] [CrossRef]
- Hemmati, S.; Heimendahl, C.; Klaes, M.; Alfermann, A.; Schmidt, T.; Fuss, E. Pinoresinol-Lariciresinol Reductases with Opposite Enantiospecificity Determine the Enantiomeric Composition of Lignans in the Different Organs of Linum Usitatissimum L. Planta Med. 2010, 76, 928–934. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Mateljak, I.; Markulin, L.; Decourtil, C.; Renouard, S.; Lopez, T.; Doussot, J.; Lamblin, F.; Auguin, D.; et al. Functional Characterization of the Pinoresinol-Lariciresinol Reductase-2 Gene Reveals Its Roles in Yatein Biosynthesis and Flax Defense Response. Planta 2017, 246, 405–420. [Google Scholar] [CrossRef]
- Lamblin, F.; Hano, C.; Fliniaux, O.; Mesnard, F.; Fliniaux, M.-A.; Lainé, E. [Interest of lignans in prevention and treatment of cancers]. Med. Sci. MS 2008, 24, 511–519. [Google Scholar] [CrossRef]
- Dikshit, A.; Gao, C.; Small, C.; Hales, K.; Hales, D.B. Flaxseed and Its Components Differentially Affect Estrogen Targets in Pre-Neoplastic Hen Ovaries. J. Steroid Biochem. Mol. Biol. 2016, 159, 73–85. [Google Scholar] [CrossRef][Green Version]
- Hano, C.; Corbin, C.; Drouet, S.; Quéro, A.; Rombaut, N.; Savoire, R.; Molinié, R.; Thomasset, B.; Mesnard, F.; Lainé, E. The Lignan (+)-Secoisolariciresinol Extracted from Flax Hulls Is an Effective Protectant of Linseed Oil and Its Emulsion against Oxidative Damage. Eur. J. Lipid Sci. Technol. 2017, 119, 1600219. [Google Scholar] [CrossRef]
- Kezimana, P.; Dmitriev, A.A.; Kudryavtseva, A.V.; Romanova, E.V.; Melnikova, N.V. Secoisolariciresinol Diglucoside of Flaxseed and Its Metabolites: Biosynthesis and Potential for Nutraceuticals. Front. Genet. 2018, 9. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Liu, Y.; Tian, H.; Flickinger, B.; Empie, M.W.; Sun, S.Z. Dietary Flaxseed Lignan Extract Lowers Plasma Cholesterol and Glucose Concentrations in Hypercholesterolaemic Subjects. Br. J. Nutr. 2008, 99, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-García, C.; Sánchez-Quesada, C.; Toledo, E.; Delgado-Rodríguez, M.; Gaforio, J. Naturally Lignan-Rich Foods: A Dietary Tool for Health Promotion? Molecules 2019, 24, 917. [Google Scholar] [CrossRef] [PubMed]
- Pan, A.; Sun, J.; Chen, Y.; Ye, X.; Li, H.; Yu, Z.; Wang, Y.; Gu, W.; Zhang, X.; Chen, X.; et al. Effects of a Flaxseed-Derived Lignan Supplement in Type 2 Diabetic Patients: A Randomized, Double-Blind, Cross-Over Trial. PLoS ONE 2007, 2, e1148. [Google Scholar] [CrossRef] [PubMed]
- Senizza, A.; Rocchetti, G.; Mosele, J.I.; Patrone, V.; Callegari, M.L.; Morelli, L.; Lucini, L. Lignans and Gut Microbiota: An Interplay Revealing Potential Health Implications. Molecules 2020, 25, 5709. [Google Scholar] [CrossRef]
- Chang, C.W.; Lin, M.T.; Lee, S.S.; Liu, K.C.; Hsu, F.L.; Lin, J.Y. Differential Inhibition of Reverse Transcriptase and Cellular DNA Polymerase-Alpha Activities by Lignans Isolated from Chinese Herbs, Phyllanthus Myrtifolius Moon, and Tannins from Lonicera Japonica Thunb and Castanopsis Hystrix. Antivir. Res. 1995, 27, 367–374. [Google Scholar] [CrossRef]
- Ekalu, A.; Ayo, R.G.-O.; Habila, J.D.; Hamisu, I. In Vitro Antimicrobial Activity of Lignan from the Stem Bark of Strombosia Grandifolia Hook.f. Ex Benth. Bull. Natl. Res. Cent. 2019, 43, 115. [Google Scholar] [CrossRef]
- Akiyama, K.; Yamauchi, S.; Nakato, T.; Maruyama, M.; Sugahara, T.; Kishida, T. Antifungal Activity of Tetra-Substituted Tetrahydrofuran Lignan, (−)-Virgatusin, and Its Structure-Activity Relationship. Biosci. Biotechnol. Biochem. 2007, 71, 1028–1035. [Google Scholar] [CrossRef][Green Version]
- Cho, J.Y.; Choi, G.J.; Son, S.W.; Jang, K.S.; Lim, H.K.; Lee, S.O.; Sung, N.D.; Cho, K.Y.; Kim, J.-C. Isolation and Antifungal Activity of Lignans from Myristica Fragrans against Various Plant Pathogenic Fungi. Pest. Manag. Sci. 2007, 63, 935–940. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Geng, Z.; Guo, S.; Cao, J.; Zhang, Z.; Pang, X.; Chen, Z.; Du, S.; Deng, Z. Antifeedant Activities of Lignans from Stem Bark of Zanthoxylum Armatum DC. against Tribolium Castaneum. Molecules 2018, 23, 617. [Google Scholar] [CrossRef]
- Gang, D.R.; Kasahara, H.; Xia, Z.Q.; Vander Mijnsbrugge, K.; Bauw, G.; Boerjan, W.; Van Montagu, M.; Davin, L.B.; Lewis, N.G. Evolution of Plant Defense Mechanisms. Relationships of Phenylcoumaran Benzylic Ether Reductases to Pinoresinol-Lariciresinol and Isoflavone Reductases. J. Biol. Chem. 1999, 274, 7516–7527. [Google Scholar] [CrossRef]
- Gang, D.R.; Fujita, M.; Davin, L.B.; Lewis, N.G. The “Abnormal Lignins”: Mapping Heartwood Formation through the Lignan Biosynthetic Pathway. In Lignin and Lignan Biosynthesis; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1998; Volume 697, pp. 389–421. ISBN 978-0-8412-3566-3. [Google Scholar]
- Thongphasuk, P.; Suttisri, R.; Bavovada, R.; Verpoorte, R. Antioxidant Lignan Glucosides from Strychnos Vanprukii. Fitoterapia 2004, 75, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Mahendra Kumar, C.; Singh, S.A. Bioactive Lignans from Sesame (Sesamum Indicum L.): Evaluation of Their Antioxidant and Antibacterial Effects for Food Applications. J. Food Sci. Technol. 2015, 52, 2934–2941. [Google Scholar] [CrossRef]
- Li, Y.; Wei, J.; Fang, J.; Lv, W.; Ji, Y.; Aioub, A.A.A.; Zhang, J.; Hu, Z. Insecticidal Activity of Four Lignans Isolated from Phryma Leptostachya. Molecules 2019, 24, 1976. [Google Scholar] [CrossRef] [PubMed]
- Niculaes, C.; Morreel, K.; Kim, H.; Lu, F.; McKee, L.S.; Ivens, B.; Haustraete, J.; Vanholme, B.; Rycke, R.D.; Hertzberg, M.; et al. Phenylcoumaran Benzylic Ether Reductase Prevents Accumulation of Compounds Formed under Oxidative Conditions in Poplar Xylem. Plant. Cell 2014, 26, 3775–3791. [Google Scholar] [CrossRef]
- Corbin, C.; Decourtil, C.; Marosevic, D.; Bailly, M.; Lopez, T.; Renouard, S.; Doussot, J.; Dutilleul, C.; Auguin, D.; Giglioli-Guivarc’h, N.; et al. Role of Protein Farnesylation Events in the ABA-Mediated Regulation of the Pinoresinol-Lariciresinol Reductase 1 (LuPLR1) Gene Expression and Lignan Biosynthesis in Flax (Linum Usitatissimum L.). Plant Physiol. Biochem. PPB 2013, 72, 96–111. [Google Scholar] [CrossRef]
- Renouard, S.; Tribalatc, M.-A.; Lamblin, F.; Mongelard, G.; Fliniaux, O.; Corbin, C.; Marosevic, D.; Pilard, S.; Demailly, H.; Gutierrez, L.; et al. RNAi-Mediated Pinoresinol Lariciresinol Reductase Gene Silencing in Flax (Linum Usitatissimum L.) Seed Coat: Consequences on Lignans and Neolignans Accumulation. J. Plant. Physiol. 2014, 171, 1372–1377. [Google Scholar] [CrossRef]
- Barros-Rios, J.; Serrani-Yarce, J.; Chen, F.; Baxter, D.; Venables, B.; Dixon, R. Role of Bifunctional Ammonia-Lyase in Grass Cell Wall Biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Bernard, S.M.; Habash, D.Z. The Importance of Cytosolic Glutamine Synthetase in Nitrogen Assimilation and Recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef]
- Corbin, C.; Renouard, S.; Lopez, T.; Lamblin, F.; Lainé, E.; Hano, C. Identification and Characterization of Cis-Acting Elements Involved in the Regulation of ABA- and/or GA-Mediated LuPLR1 Gene Expression and Lignan Biosynthesis in Flax (Linum Usitatissimum L.) Cell Cultures. J. Plant. Physiol. 2013, 170, 516–522. [Google Scholar] [CrossRef]
- Wu, R.; Wang, L.; Wang, Z.; Shang, H.; Liu, X.; Zhu, Y.; Qi, D.; Deng, X. Cloning and Expression Analysis of a Dirigent Protein Gene from the Resurrection Plant Boea Hygrometrica. Prog. Nat. Sci. 2009, 19, 347–352. [Google Scholar] [CrossRef]
- Ramsay, A.; Fliniaux, O.; Fang, J.; Molinié, R.; Roscher, A.; Grand, E.; Guillot, X.; Kovensky, J.; Fliniaux, M.-A.; Schneider, B.; et al. Development of an NMR Metabolomics-Based Tool for Selection of Flaxseed Varieties. Metabolomics 2014, 10. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef]
- Jez, J.; Ferrer, J.-L.; Bowman, M.; Austin, M.; Schröder, J.; Dixon, R.; Noel, J. Structure and Mechanism of Chalcone Synthase-like Polyketide Synthases. J. Ind. Microbiol. Biotechnol. 2002, 27, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Marchiosi, R.; dos Santos, W.D.; Constantin, R.P.; de Lima, R.B.; Soares, A.R.; Finger-Teixeira, A.; Mota, T.R.; de Oliveira, D.M.; de Pavia Foletto-Felipe, M.; Abrahão, J.; et al. Biosynthesis and Metabolic Actions of Simple Phenolic Acids in Plants. Phytochem. Rev. 2020, 19, 865–906. [Google Scholar] [CrossRef]
- Shahidi, F.; Chandrasekara, A. Hydroxycinnamates and Their in Vitro and in Vivo Antioxidant Activities. Phytochem. Rev. 2009, 9, 147–170. [Google Scholar] [CrossRef]
- Quéro, A.; Molinié, R.; Elboutachfaiti, R.; Petit, E.; Pau-Roblot, C.; Guillot, X.; Mesnard, F.; Courtois, J. Osmotic Stress Alters the Balance between Organic and Inorganic Solutes in Flax (Linum Usitatissimum). J. Plant. Physiol. 2014, 171, 55–64. [Google Scholar] [CrossRef]
- Delauney, A.J.; Verma, D.P.S. Proline Biosynthesis and Osmoregulation in Plants. Plant. J. 1993, 4, 215–223. [Google Scholar] [CrossRef]
- Hare, P.D.; Natal, P.U.; Cress, W.A. Metabolic Implications of Stress-Induced Proline Accumulation in Plants. Plant Growth Regul. Neth. 1997, 21, 79–102. [Google Scholar] [CrossRef]
- Munns, R. Comparative Physiology of Salt and Water Stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Kovács, Z.; Simon-Sarkadi, L.; Vashegyi, I.; Kocsy, G. Different Accumulation of Free Amino Acids during Short- and Long-Term Osmotic Stress in Wheat. Available online: https://www.hindawi.com/journals/tswj/2012/216521/ (accessed on 25 November 2020).
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Sharma, S.; Verslues, P.E. Mechanisms Independent of Abscisic Acid (ABA) or Proline Feedback Have a Predominant Role in Transcriptional Regulation of Proline Metabolism during Low Water Potential and Stress Recovery. Plant Cell Environ. 2010, 33, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Abbas, S.R.; Ahmad, S.D.; Sabir, S.M.; Shah, A.H. Detection of Drought Tolerant Sugarcane Genotypes (Saccharum Officinarum) Using Lipid Peroxidation, Antioxidant Activity, Glycine-Betaine and Proline Contents. J. Soil Sci. Plant Nutr. 2014, 14, 233–243. [Google Scholar] [CrossRef]
- Vetter, J. Plant Cyanogenic Glycosides. Toxicon Off. J. Int. Soc. Toxinol. 2000, 38, 11–36. [Google Scholar] [CrossRef]
- Bartels, D. Desiccation Tolerance Studied in the Resurrection Plant Craterostigma Plantagineum. Integr. Comp. Biol. 2005, 45, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Koster, K.L.; Leopold, A.C. Sugars and Desiccation Tolerance in Seeds. Plant. Physiol. 1988, 88, 829–832. [Google Scholar] [CrossRef]
- Liu, F.; Jensen, C.R.; Andersen, M.N. Drought Stress Effect on Carbohydrate Concentration in Soybean Leaves and Pods during Early Reproductive Development: Its Implication in Altering Pod Set. Field Crop. Res. 2004, 86, 1–13. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. ABA Control of Plant Macroelement Membrane Transport Systems in Response to Water Deficit and High Salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Tocquin, P.; Corbesier, L.; Havelange, A.; Pieltain, A.; Kurtem, E.; Bernier, G.; Périlleux, C. A Novel High Efficiency, Low Maintenance, Hydroponic System for Synchronous Growth and Flowering of Arabidopsis Thaliana. BMC Plant Biol. 2003, 3, 2. [Google Scholar] [CrossRef]
- Deborde, C.; Fontaine, J.-X.; Jacob, D.; Botana, A.; Nicaise, V.; Richard-Forget, F.; Lecomte, S.; Decourtil, C.; Hamade, K.; Mesnard, F.; et al. Optimizing 1D 1H-NMR Profiling of Plant Samples for High Throughput Analysis: Extract Preparation, Standardization, Automation and Spectra Processing. Metabolomics 2019, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Gan, F.; Ruan, G.; Mo, J. Baseline Correction by Improved Iterative Polynomial Fitting with Automatic Threshold. Chemom. Intell. Lab. Syst. 2006, 82, 59–65. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Chen, S.; Liang, Y.-Z. Baseline Correction Using Adaptive Iteratively Reweighted Penalized Least Squares. Analyst 2010, 135, 1138–1146. [Google Scholar] [CrossRef] [PubMed]




| Roots | Stems | Leaves | ||||||
|---|---|---|---|---|---|---|---|---|
| Ratio Pi1AM/LuT | Ratio Pi1AM/LuT | Ratio Pi1AM/LuT | ||||||
| Metabolite | Stress | Control | Metabolite | Stress | Control | Metabolite | Stress | Control |
| Primary metabolites | Primary metabolites | Primary metabolites | ||||||
| Glutamic Acid | 0.5 *** | 0.65 ** | Glycerol | 1.3 * | 5.56 ** | Phenylalanine | 0.42 ** | 0.7 ** |
| Aspartic Acid | 0.53 *** | 0.55 *** | Ethanolamine | 1.38 ** | _ | GABA | 0.47 * | 0.63 * |
| GABA | 0.53 *** | _ | Galactose | 1.47 * | 1.19 ** | Threonine | 0.47 ** | _ |
| Tyrosine | 0.54 *** | _ | Fumaric Acid | 1.5 ** | _ | Alanine | 0.62 * | 0.64 *** |
| Glutamine | 0.56 *** | 0.63 *** | Uridine | 1.68 ** | _ | Serine | 0.64 * | 1.13 * |
| Alanine | 0.85 *** | _ | Fructose | 1.87 * | _ | Galactose | 0.66 ** | 0.75 *** |
| Threonine | 1.2 *** | 1.23 * | Tyrosine | 2.01 *** | 1.88 *** | Succinic Acid | 1.2 * | _ |
| Uridine | 1.34 *** | _ | Glucose | 2.31 ** | 1.42 ** | Glycerol | 1.36 * | 1.5 ** |
| Glycine | 1.38 *** | 1.41 *** | Phenylalanine | 3.05 *** | 1.6 ** | Fructose | 2.00 * | 1.72 *** |
| Succinic Acid | 1.42 *** | 1.64 ** | Coniferyl Alcohol | 7.72 *** | 8.65 *** | Choline | 2.05 ** | 1.75 *** |
| Formic Acid | 1.43 *** | _ | Alanine | _ | 0.7 ** | Glucose | 2.15 ** | 1.34 * |
| Sucrose | 1.45 *** | 2.07 *** | Succinic Acid | _ | 0.73 *** | Coniferyl Alcohol | 4.79 *** | 10.32 *** |
| Fructose | 1.69 *** | 1.73 *** | Tartaric Acid | _ | 0.74 ** | Uridine | _ | 0.55 * |
| Glucose | 1.79 *** | 1.85 *** | Asparagine | _ | 0.76 * | Adenosine | _ | 0.6 *** |
| Malic Acid | 2.03 *** | _ | Malic Acid | _ | 0.82 *** | Threonine | _ | 0.76 ** |
| Fumaric Acid | 2.11 *** | 1.34 * | Putrescine | _ | 0.84 ** | Ethanolamine | _ | 0.79 ** |
| Coniferyl Alcohol | 5.51 *** | 7.91 *** | Threonine | _ | 0.87 * | Asparagine | _ | 0.85 * |
| Maltose | _ | 0.87 * | Aspartic Acid | _ | 1.27 * | Tartaric Acid | _ | 0.91 * |
| Serine | _ | 1.48 *** | Glutamic Acid | _ | 1.39 * | Raffinose | _ | 1.28 ** |
| Putrescine | _ | 1.6 *** | Secondary metabolites | Sucrose | _ | 1.32 ** | ||
| Secondary metabolites | LMG | 0.46 *** | 0.73 *** | Tyrosine | _ | 3.73 *** | ||
| SMG | 0.06 *** | 0.15 *** | LDG | 0.64 *** | 0.17 *** | Secondary metabolites | ||
| LMG | 0.28 *** | 0.31 ** | Triticuside-A | 0.8 * | 0.8 ** | LMG | 0.27 *** | 0.50 *** |
| Chicoric Acid | 0.71 *** | _ | Vicenin-2 | 1.36 *** | _ | Vitexin | 0.65 *** | 0.69 *** |
| Trigonelline | 1.21 *** | _ | CAFG | 1.45 ** | 1.42 ** | Vicenin-2 | 0.7 ** | 0.77** |
| Lotaustralin | 1.81 ** | 1.49 ** | Vitexin | 1.54 *** | _ | Schaftoside | 0.9 *** | _ |
| Linamarin | 1.75 ** | 1.41 ** | Chlorogenic Acid | 1.63 ** | 1.32 ** | Lucenin-2 | 1.27* | 1.21 ** |
| Coniferin | 1.83 *** | 2.32 *** | FAG | 1.81 *** | 1.21 * | Chicoric Ac | 1.33 *** | 1.23 *** |
| PDG | 12.66 *** | 12.27 *** | Neolunistatin | 2.04 ** | 1.84 ** | CAFG | 1.42 * | 1.11 ** |
| PMG | 18.63 *** | 18.13 *** | Orientin | 2.67 *** | 1.92 *** | linamarin | 1.45 *** | _ |
| Lucenin-2 | 3.07 *** | 2.57 *** | Carlinoide | 1.5 *** | 1.45 *** | |||
| Carlinoside | 3.51 ** | 2.8 *** | FAG | 1.66 *** | 1.24 * | |||
| Coniferin | 3.63 *** | 2.4 *** | Chlorogenic Acid | 2.19 *** | 1.51 ** | |||
| PDG | 3.83 *** | 4.73 *** | Coniferin | 6.45 | 7.68 | |||
| PMG | 4.25 *** | 5.31 *** | PMG | 14.1 | 13.78 *** | |||
| Linamarin | _ | 0.76 *** | PDG | 14.5 *** | 15.03 *** | |||
| Lotaustralin | _ | 0.76* | Linustatin | _ | 1.27 ** | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamade, K.; Fliniaux, O.; Fontaine, J.-X.; Molinié, R.; Otogo Nnang, E.; Bassard, S.; Guénin, S.; Gutierrez, L.; Lainé, E.; Hano, C.; et al. NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax. Molecules 2021, 26, 767. https://doi.org/10.3390/molecules26030767
Hamade K, Fliniaux O, Fontaine J-X, Molinié R, Otogo Nnang E, Bassard S, Guénin S, Gutierrez L, Lainé E, Hano C, et al. NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax. Molecules. 2021; 26(3):767. https://doi.org/10.3390/molecules26030767
Chicago/Turabian StyleHamade, Kamar, Ophélie Fliniaux, Jean-Xavier Fontaine, Roland Molinié, Elvis Otogo Nnang, Solène Bassard, Stéphanie Guénin, Laurent Gutierrez, Eric Lainé, Christophe Hano, and et al. 2021. "NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax" Molecules 26, no. 3: 767. https://doi.org/10.3390/molecules26030767
APA StyleHamade, K., Fliniaux, O., Fontaine, J.-X., Molinié, R., Otogo Nnang, E., Bassard, S., Guénin, S., Gutierrez, L., Lainé, E., Hano, C., Pilard, S., Hijazi, A., El Kak, A., & Mesnard, F. (2021). NMR and LC-MS-Based Metabolomics to Study Osmotic Stress in Lignan-Deficient Flax. Molecules, 26(3), 767. https://doi.org/10.3390/molecules26030767








