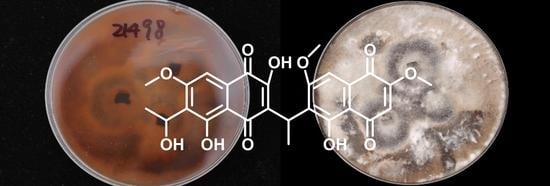
Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction and Isolation
3.4. Antimicrobial Activity of Fungal Cultures
3.5. Antimicrobial Testing of Extracts
3.6. Antimicrobial Assays of Pure Compounds
3.7. Cytotoxicity Assays
3.8. Haemolytic Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- WHO Antimicrobial Resistance: Global Report on Surveillance 2014. Available online: http://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 7 February 2020).
- Wright, G.D. Solving the Antibiotic Crisis. ACS Infect. Dis. 2015, 1, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. A Three-Step Plan for Antibiotics. Nature 2014, 509, 533. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The Fungi of New Zealand Volume 1: Introduction to Fungi of New Zealand. Available online: http://www.mwpress.co.nz/funginz/the-fungi-of-new-zealand-volume-1-introduction-to-fungi-of-new-zealand (accessed on 1 October 2020).
- Cadelis, M.M.; Geese, S.; Gris, L.; Weir, B.S.; Copp, B.R.; Wiles, S. A Revised Structure and Assigned Absolute Configuration of Theissenolactone A. Molecules 2020, 25, 4823. [Google Scholar] [CrossRef] [PubMed]
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug Resistant Bacterial Infections, Including Tuberculosis. Available online: http://www.who.int/medicines/areas/rational_use/prioritization-of-pathogens/en/ (accessed on 7 February 2020).
- Poch, G.K.; Gloer, J.B.; Shearer, C.A. New Bioactive Metabolites from a Freshwater Isolate of the Fungus Kirschsteiniothelia sp. J. Nat. Prod. 1992, 55, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Pittayakhajonwut, P.; Sohsomboon, P.; Dramae, A.; Suvannakad, R.; Lapanun, S.; Tantichareon, M. Antimycobacterial Substances from Phaeosphaeria sp BCC8292. Planta Med. 2008, 74, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, H.; Liu, Y.; Gu, Q.; Xu, J.; Huang, X.; She, Z. Ethylnaphthoquinone Derivatives as Inhibitors of Indoleamine-2, 3-Dioxygenase from the Mangrove Endophytic Fungus Neofusicoccum austral SYSU-SKS024. Fitoterapia 2018, 125, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Otomo, N.; Sato, H.; Sakamura, S. Novel Phytotoxins Produced by the Causal Fungus of the Shoot Blight of Larches. Agric. Biol. Chem. 1983, 47, 1115–1119. [Google Scholar] [CrossRef]
- Harrison, P.H.M.; Duspara, P.A.; Jenkins, S.I.; Kassam, S.A.; Liscombe, D.K.; Hughes, D.W. The Biosynthesis of Pramanicin in Stagonospora sp. ATCC 74235: A Modified Acyltetramic Acid. J. Chem. Soc. Perkin 1 2000, 24, 4390–4402. [Google Scholar] [CrossRef]
- Latypov, S.K.; Seco, J.M.; Quiñoá, E.; Riguera, R. MTPA vs MPA in the Determination of the Absolute Configuration of Chiral Alcohols by 1 H NMR. J. Org. Chem. 1996, 61, 8569–8577. [Google Scholar] [CrossRef]
- Aoyama, T.; Terasawa, S.; Sudo, K.; Shioiri, T. New Methods and Reagents in Organic Synthesis. 46. Trimethylsilyldiazomethane: A Convenient Reagent for the O-Methylation of Phenols and Enols. Chem. Pharm. Bull. 1984, 32, 3759–3760. [Google Scholar] [CrossRef]
- Presser, A.; Hüfner, A. Trimethylsilyldiazomethane-A Mild and Efficient Reagent for the Methylation of Carboxylic Acids and Alcohols in Natural Products. Mon. Chem./Chem. Mon. 2004, 135, 1015–1022. [Google Scholar] [CrossRef]
- Schwartz, R.E.; Helms, G.L.; Bolessa, E.A.; Wilson, K.E.; Giacobbe, R.A.; Tkacz, J.S.; Bills, G.F.; Liesch, J.M.; Zink, D.L.; Curotto, J.E.; et al. Pramanicin, a Novel Antimicrobial Agent from a Fungal Fermentation. Tetrahedron 1994, 50, 1675–1686. [Google Scholar] [CrossRef]
- Specimen Details. Available online: https://scd.landcareresearch.co.nz/Specimen/ICMP_21498 (accessed on 15 January 2020).
- Andrews, J.M. Determination of Minimum Inhibitory Concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Zuegg, J.; Elliott, A.G.; Cooper, M.A. Helping Chemists Discover New Antibiotics. ACS Infect. Dis. 2015, 1, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Cadelis, M.M.; Li, S.A.; Bourguet-Kondracki, M.-L.; Blanchet, M.; Douafer, H.; Brunel, J.M.; Copp, B.R. Spermine Derivatives of Indole-3-Carboxylic Acid, Indole-3-Acetic Acid and Indole-3-Acrylic Acid as Gram-Negative Antibiotic Adjuvants. ChemMedChem. 2020, 15. [Google Scholar] [CrossRef] [PubMed]
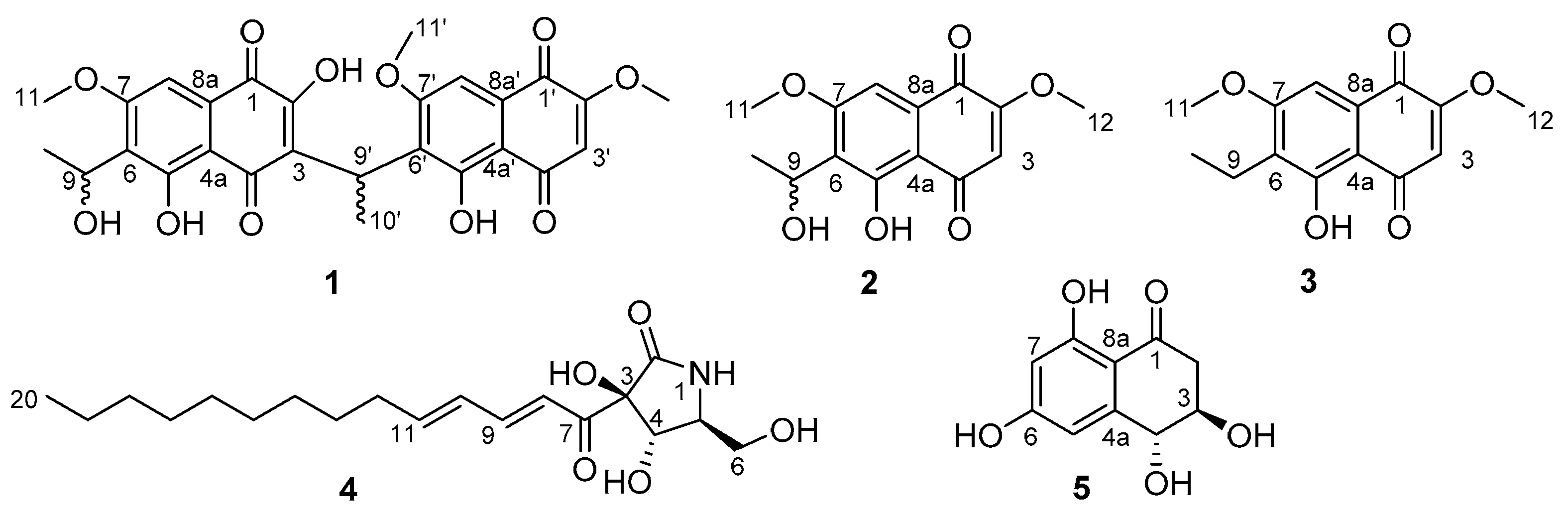
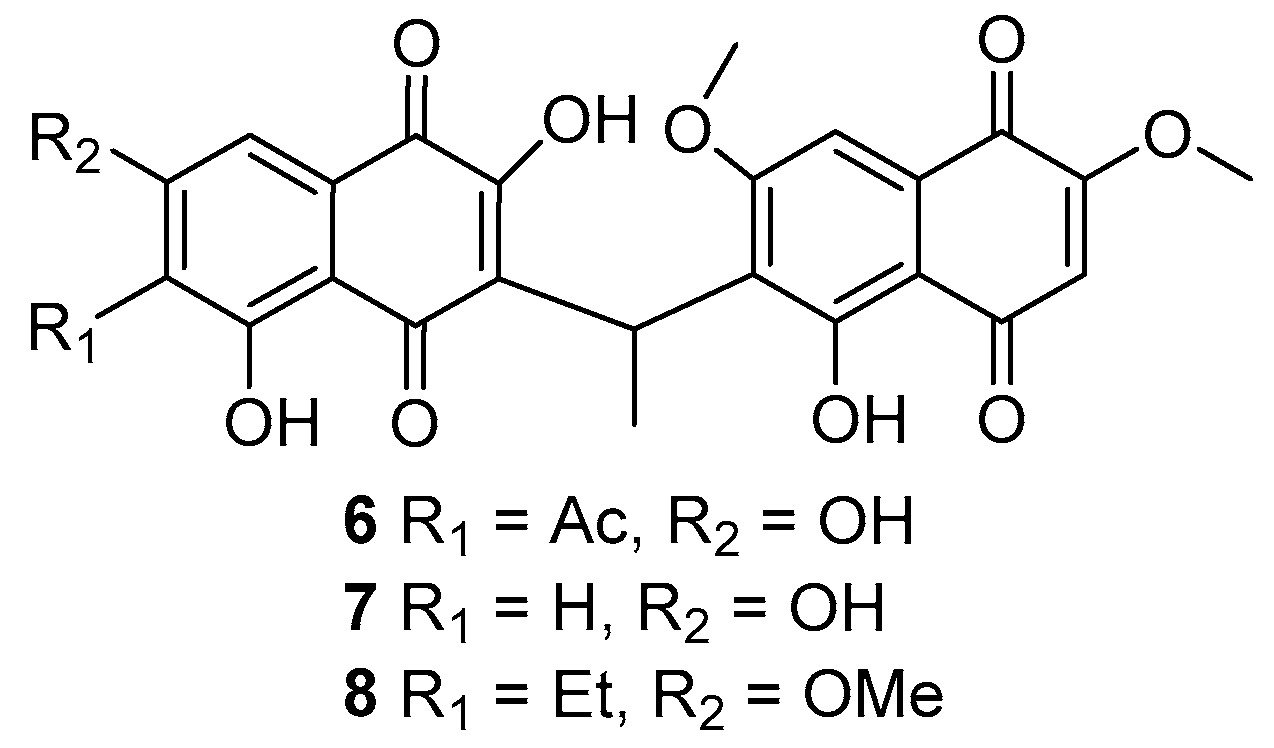

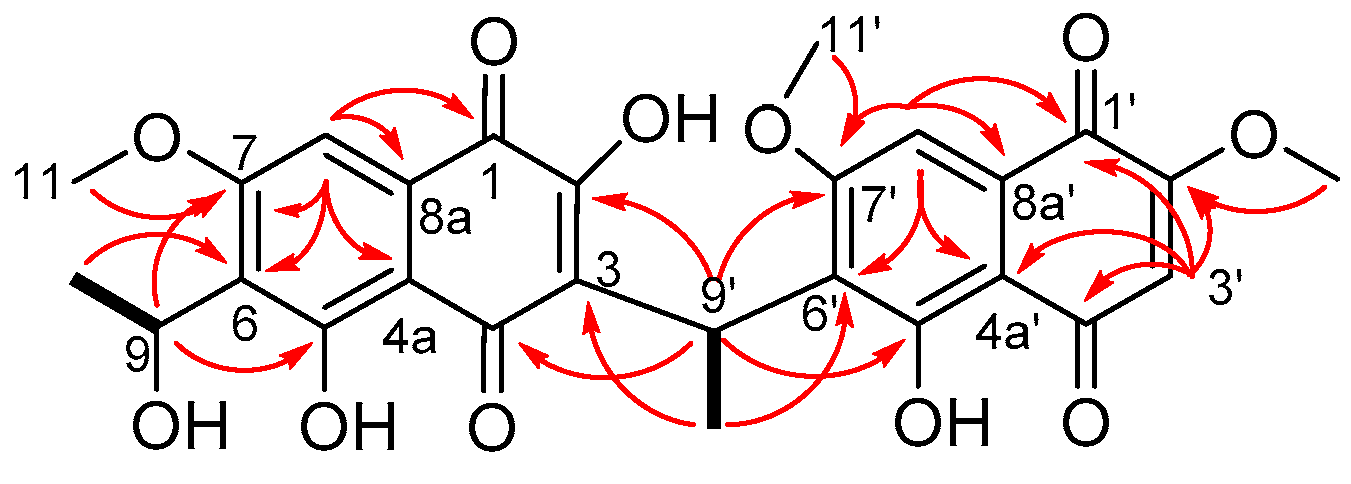
| δH (m, J in Hz) a | δC b | Selected HMBC Correlations | |
|---|---|---|---|
| 1 | 180.7 | ||
| 2 | - | 159.6 | |
| 3 | - | 123.5 | |
| 4 | - | 189.9 | |
| 4a | - | 108.6 | |
| 5 | - | 159.7 | |
| 6 | - | 126.6 | |
| 7 | - | 161.3 | |
| 8 | 7.11 (s) | 102.1 | 1, 4a, 6, 7, 8a, |
| 8a | - | 129.6 | |
| 9 | 5.20 (dq, 7.0, 6.7) | 60.6 | 5, 7, 10 |
| 10 | 1.41 (d, 6.7) | 21.7 | 6, 9 |
| 11 | 3.92 (s) | 56.2 | 7 |
| 1′ | - | 178.7 | |
| 2′ | - | 160.7 | |
| 3′ | 6.24 (s) | 109.3 | 1′, 2′, 4′, 4a’ |
| 4′ | - | 190.3 | |
| 4a′ | - | 108.3 | |
| 5′ | - | 160.2 | |
| 6′ | - | 126.8 | |
| 7′ | - | 162.5 | |
| 8′ | 7.12 (s) | 102.9 | 1′, 4a’, 6′, 7′, 8a’ |
| 8a′ | - | 130.0 | |
| 9′ | 4.80 (q, 7.0) | 27.5 | 2, 4, 5′, 7′ |
| 10′ | 1.60 (d, 7.0) | 17.6 | 3, 6′, 9′ |
| 11′ | 3.86 (s) | 56.3 | 7′ |
| 12′ | 3.86 (s) | 56.8 | 2′ |
| 2-OH | 10.89 (br s) | - | |
| 5-OH | 13.38 (br s) | - | |
| 9-OH | 4.61 (d, 7.0) | - | 9, 10 |
| 5′-OH | 12.95 (s) | - |
| Compound | MIC (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| S. aa | P. ab | E. cc | K. pd | A. be | C. af | C. ng | |
| 1 | 16 | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h |
| 2 | 16 | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h |
| 3 | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h |
| 5 | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h | >32 h |
| Compound | HEK-293 a CC50 (µg/mL) | HC10 (µg/mL) b |
|---|---|---|
| 1 | >32 c | >32 c |
| 2 | >32 c | >32 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadelis, M.M.; Geese, S.; Uy, B.B.; Mulholland, D.R.; van de Pas, S.J.; Grey, A.; Weir, B.S.; Copp, B.R.; Wiles, S. Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe. Molecules 2021, 26, 1094. https://doi.org/10.3390/molecules26041094
Cadelis MM, Geese S, Uy BB, Mulholland DR, van de Pas SJ, Grey A, Weir BS, Copp BR, Wiles S. Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe. Molecules. 2021; 26(4):1094. https://doi.org/10.3390/molecules26041094
Chicago/Turabian StyleCadelis, Melissa M., Soeren Geese, Benedict B. Uy, Daniel R. Mulholland, Shara J. van de Pas, Alex Grey, Bevan S. Weir, Brent R. Copp, and Siouxsie Wiles. 2021. "Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe" Molecules 26, no. 4: 1094. https://doi.org/10.3390/molecules26041094
APA StyleCadelis, M. M., Geese, S., Uy, B. B., Mulholland, D. R., van de Pas, S. J., Grey, A., Weir, B. S., Copp, B. R., & Wiles, S. (2021). Antimicrobial Metabolites against Methicillin-Resistant Staphylococcus aureus from the Endophytic Fungus Neofusicoccum australe. Molecules, 26(4), 1094. https://doi.org/10.3390/molecules26041094