Constructing an Efficient Bacillus subtilis Spore Display by Using Cohesin−Dockerin Interactions
Abstract
1. Introduction
2. Results
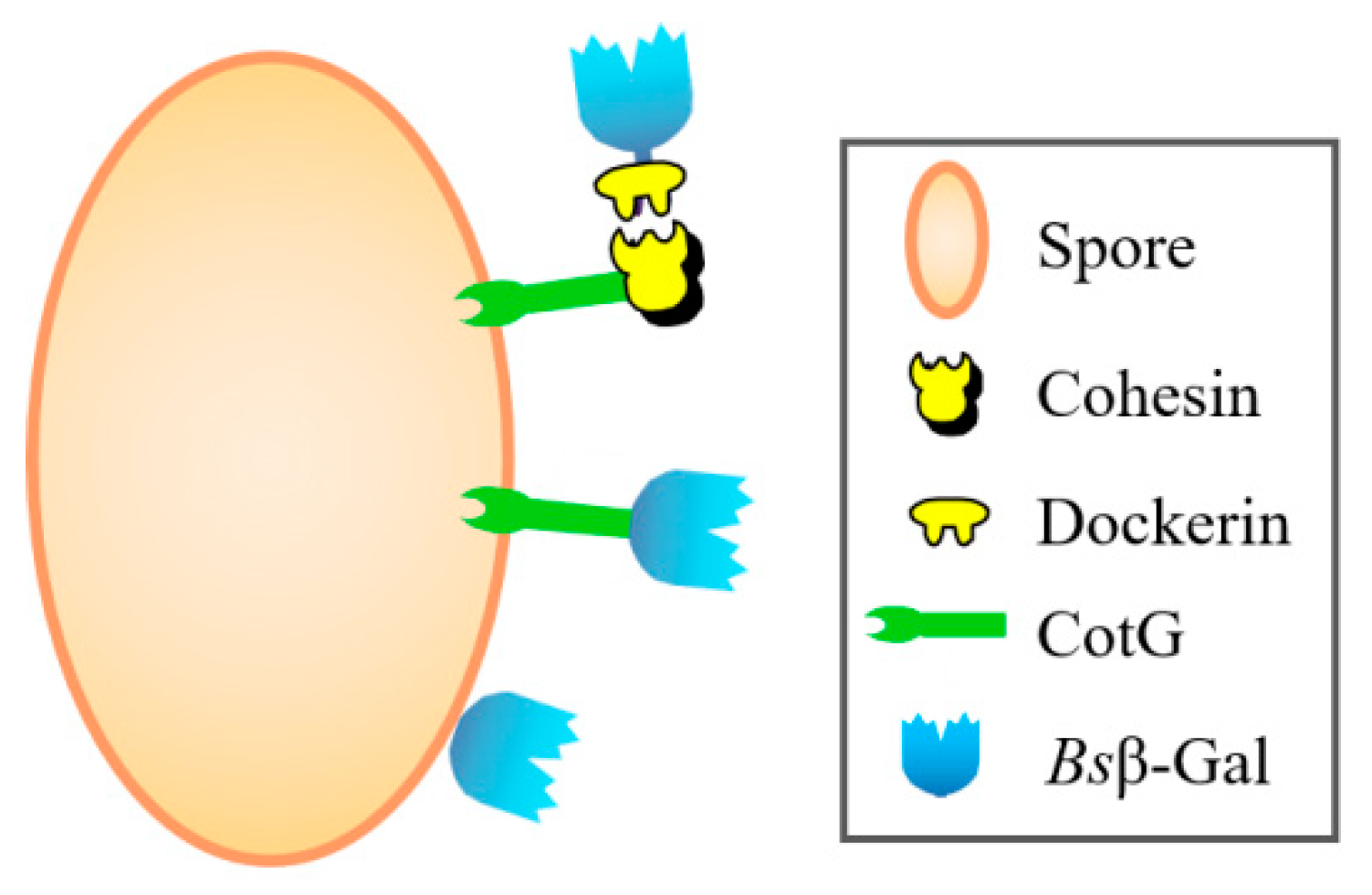
2.1. Generation of Coh–Doc Based Spore Surface Display Systems
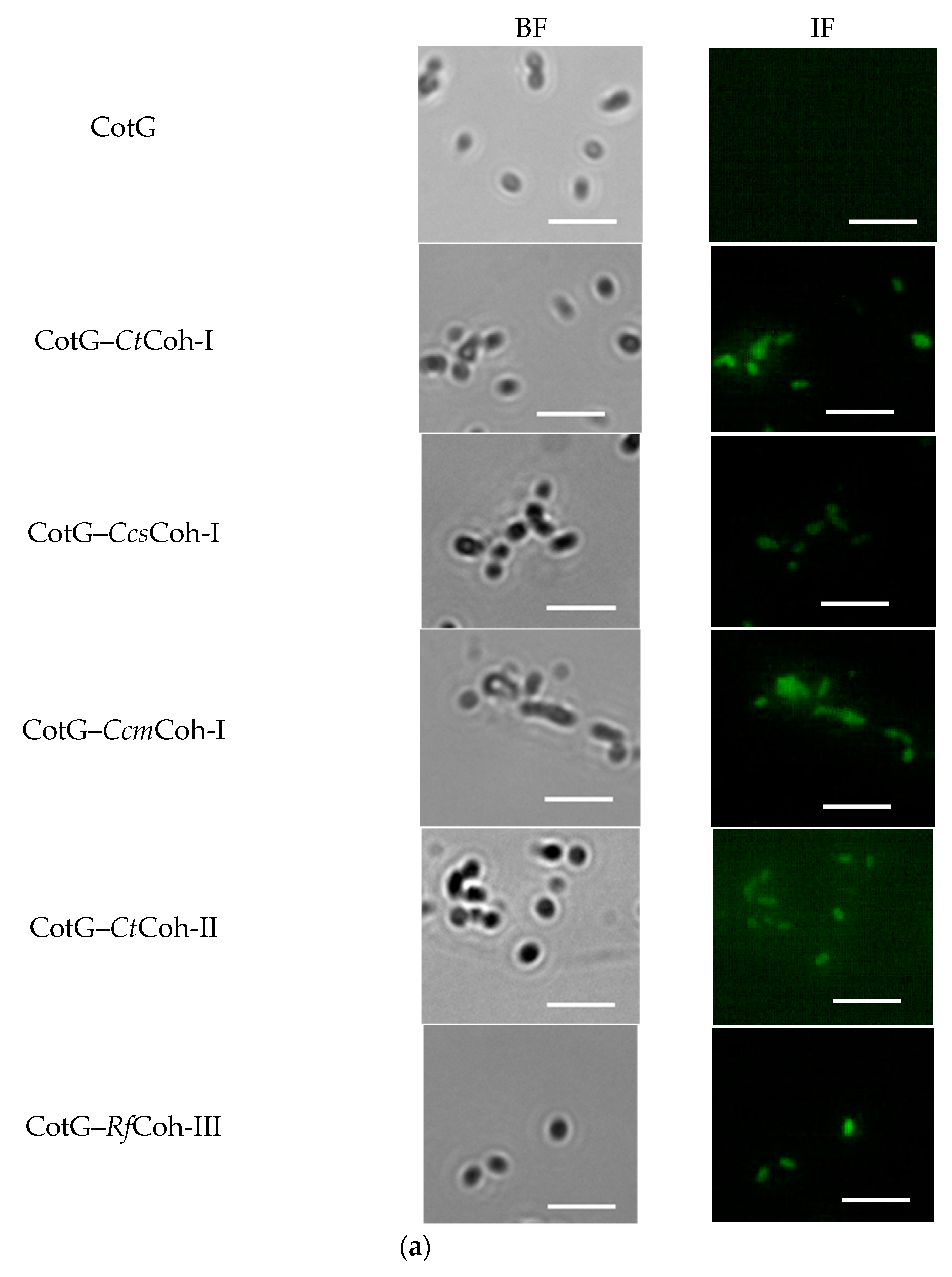
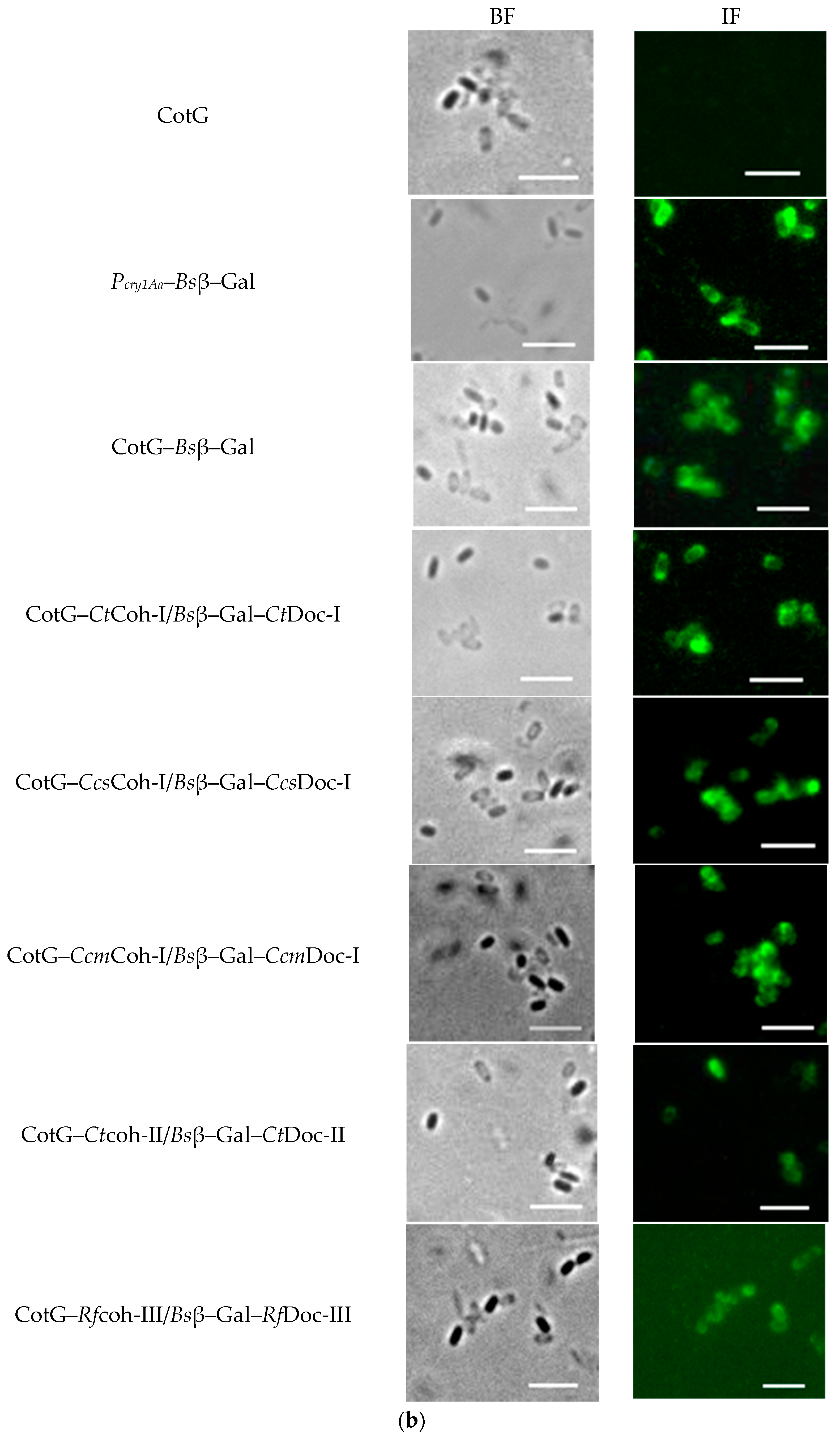
2.2. Demonstration of Coh–Doc Based Spore Surface Display Using Immunofluorescence Analysis
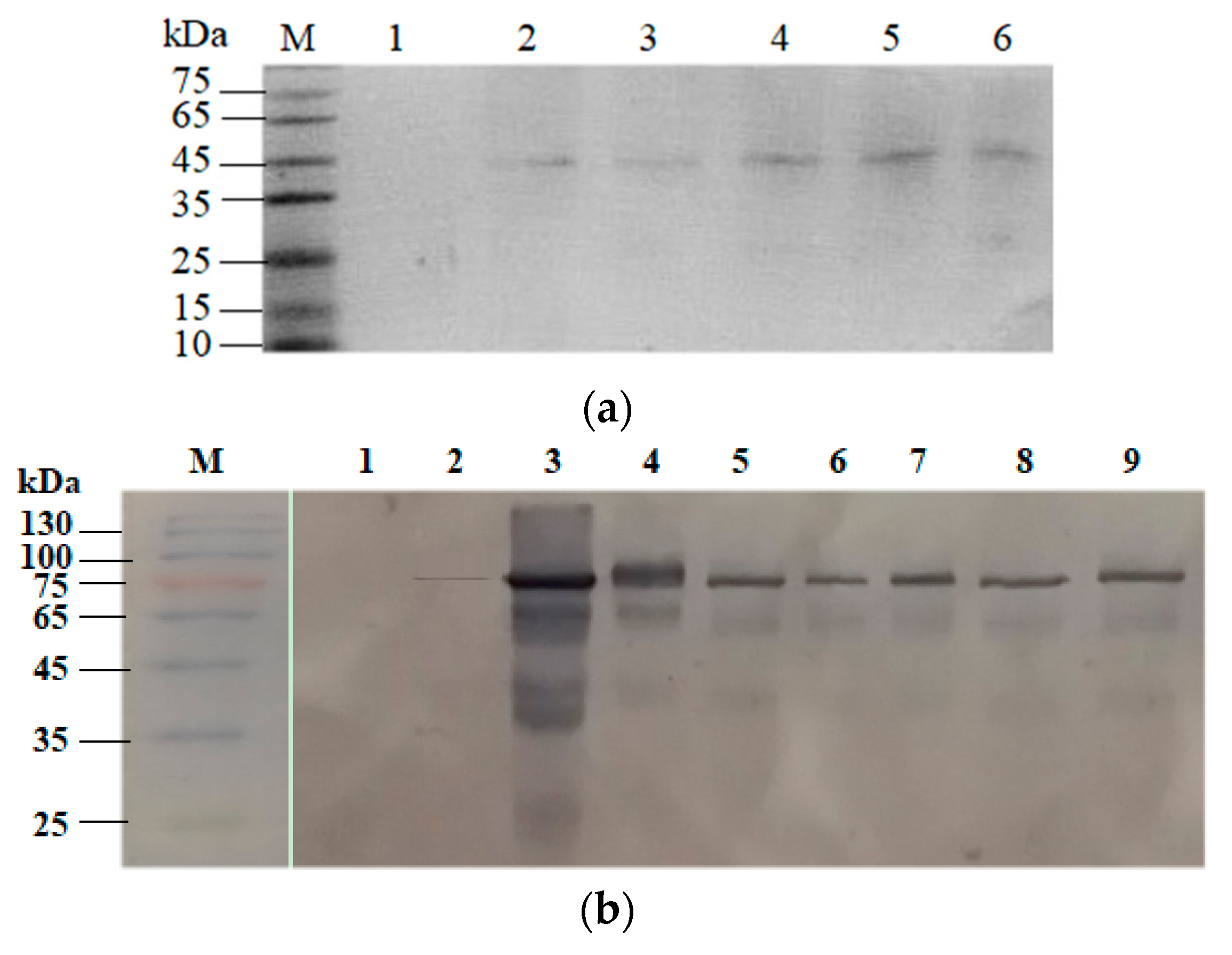
2.3. Confirmation of Coh–Doc Based Spore Surface Display
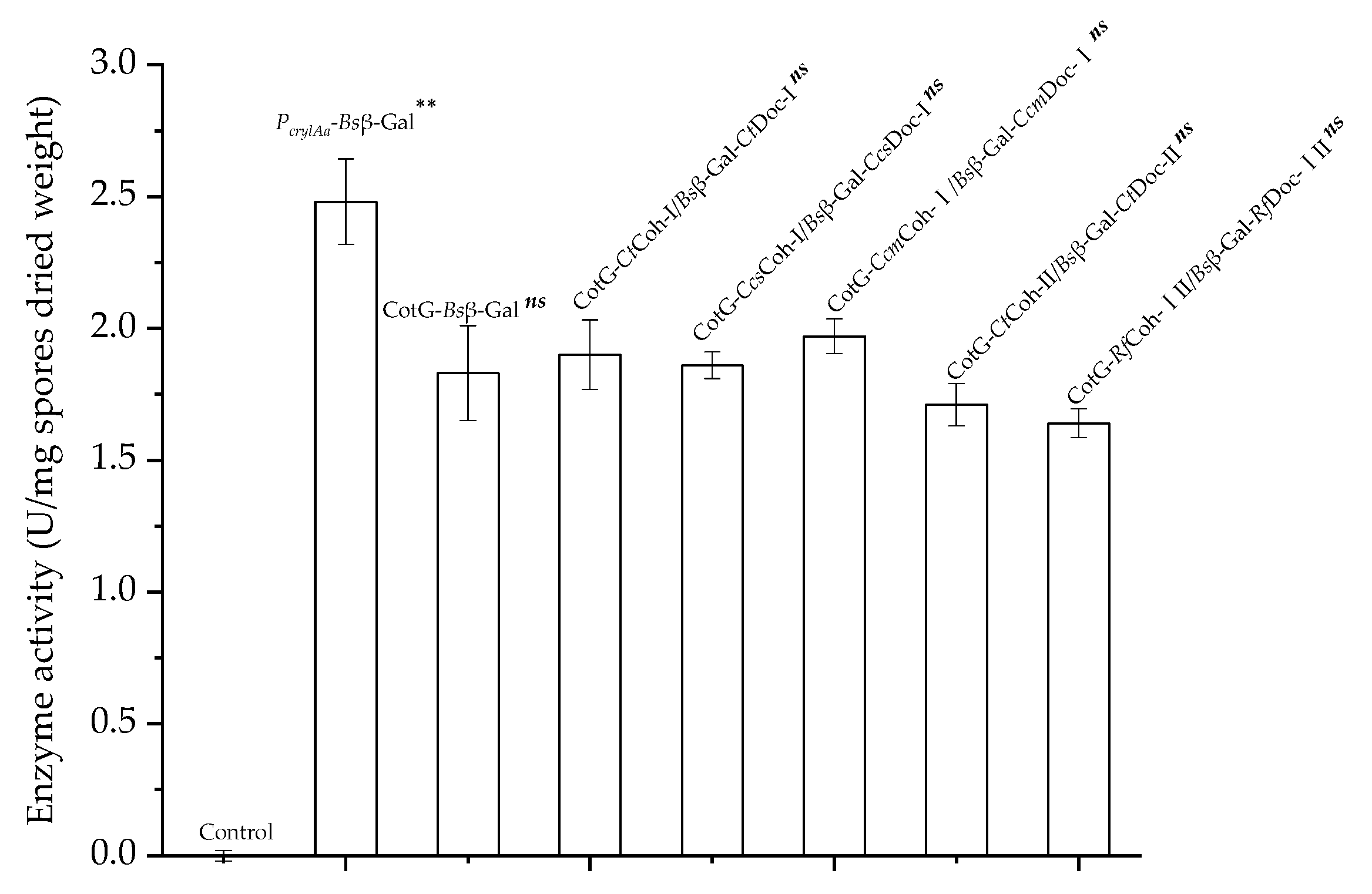
2.4. Effect of Coh–Doc Pair Type on Spore Display Efficiency
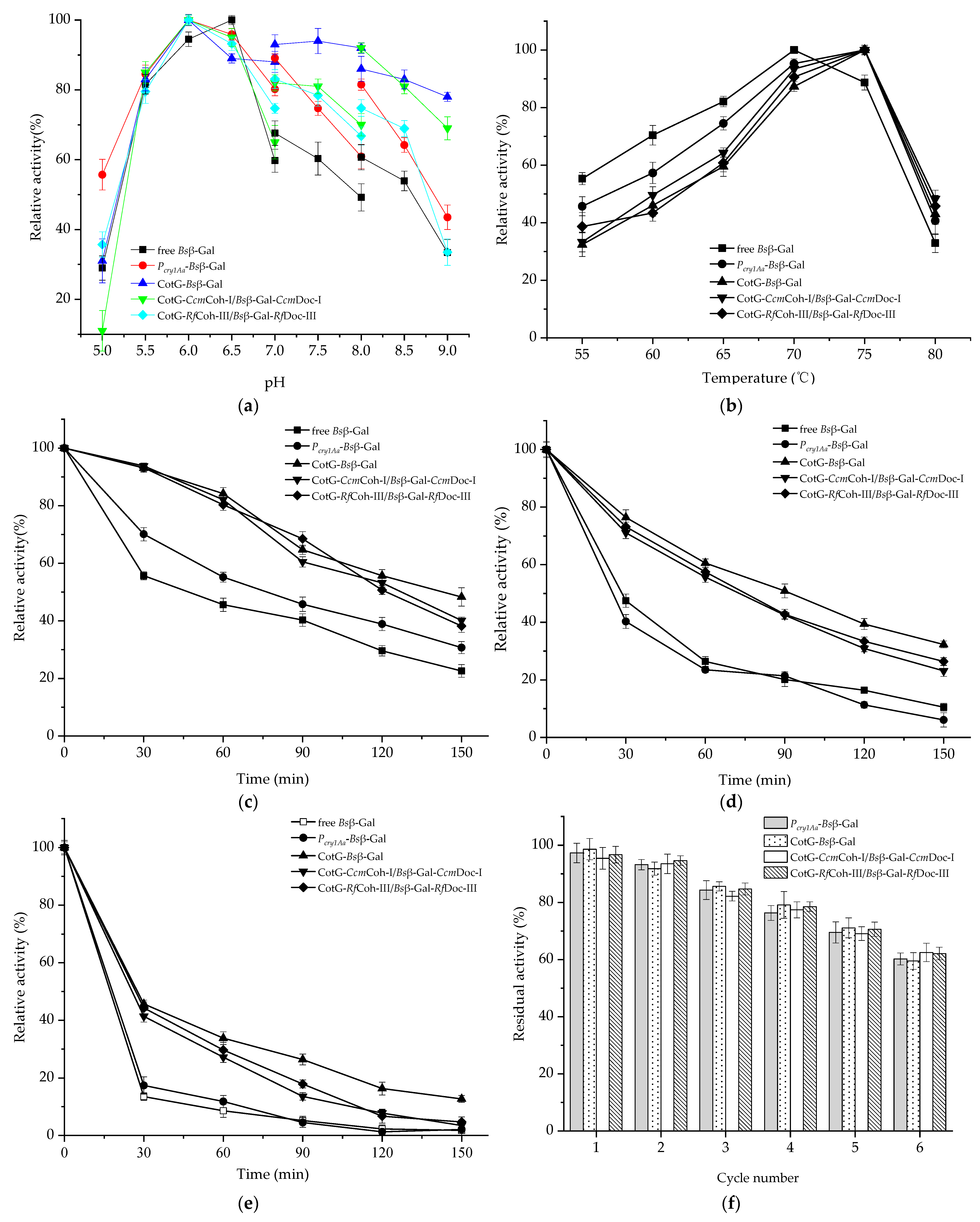
2.5. Enzymatic Properties of Spore Displayed Bsβ–Gal
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Construction of Spore Display Plasmid and Its Transformation
4.3. Expression and Purification of Bsβ–Gal–Doc
4.4. Bsβ–Gal Assembly on the Spore Surface
4.5. Western Blotting and Dot Blotting Analyses
4.6. Immunofluorescence Microscopy
4.7. Assay of Spore Displayed Bsβ–Gal Activity
4.8. Characterization of Spore Displayed Bsβ–Gal
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Isticato, R.; Cangiano, G.; Tran, H.T.; Ciabattini, A.; Medaglini, D.; Oggioni, M.R.; De Felice, M.; Del Pozzo, G.; Ricca, E. Surface display of recombinant proteins on Bacillus subtilis spores. J. Bacteriol. 2001, 183, 6294–6301. [Google Scholar] [CrossRef]
- Lin, P.; Yuan, H.; Du, J.; Liu, K.; Liu, H.; Wang, T. Progress in research and application development of surface display technology using Bacillus subtilis spores. Appl. Microbiol. Biotechnol. 2020, 104, 2319–2331. [Google Scholar] [CrossRef]
- Zhang, X.; Al-Dossary, A.; Hussain, M.; Setlow, P.; Li, J. Applications of Bacillus subtilis spores in biotechnology and advanced materials. Appl. Environ. Microbiol. 2020, 86, e01096-20. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Yang, R. Recent progress in Bacillus subtilis spore-surface display: Concept, progress, and future. Appl. Microbiol. Biotechnol. 2017, 101, 933–949. [Google Scholar] [CrossRef]
- Donadio, G.; Lanzilli, M.; Sirec, T.; Ricca, E.; Isticato, R. Localization of a red fluorescence protein adsorbed on wild type and mutant spores of Bacillus subtilis. Microb. Cell Fact. 2016, 15, 1–12. [Google Scholar] [CrossRef]
- Chen, L.; Holmes, M.; Schaefer, E.; Mulchandani, A.; Ge, X. Highly active spore biocatalyst by self-assembly of co-expressed anchoring scaffoldin and multimeric enzyme. Biotechnol. Bioeng. 2018, 115, 557–564. [Google Scholar] [CrossRef]
- Chen, L.; Mulchandani, A.; Ge, X. Spore-displayed enzyme cascade with tunable stoichiometry. Biotechnol. Prog. 2017, 33, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Tavassoli, S.; Hinc, K.; Iwanicki, A.; Obuchowski, M.; Ahmadian, G. Investigation of spore coat display of Bacillus subtilis β-galactosidase for developing of whole cell biocatalyst. Arch. Microbiol. 2013, 195, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.A.; Schumann, W. Use of IPTG-inducible promoters for anchoring recombinant proteins on the Bacillus subtilis spore surface. Protein Expr. Purif. 2014, 95, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Anandharaj, M.; Lin, Y.J.; Rani, R.P.; Nadendla, E.K.; Ho, M.C.; Huang, C.C.; Cheng, J.F.; Chang, J.J.; Li, W.H. Constructing a yeast to express the largest cellulosome complex on the cell surface. Proc. Natl. Acad. Sci. USA 2020, 117, 2385–2394. [Google Scholar] [CrossRef]
- Fan, S.; Liang, B.; Xiao, X.; Tang, X.; Lojou, E.; Cosnier, S.; Liu, A. Controllable display of sequential enzymes on yeast surface with enhanced biocatalytic activity toward efficient enzymatic biofuel cells. J. Am. Chem. Soc. 2020, 142, 3222–3230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, R.; Hua, X.; Zhang, W.; Zhao, W. An approach for lactulose production using the CotX-mediated spore-displayed β-galactosidase as a biocatalyst. J. Microbiol. Biotechnol. 2016, 26, 1267–1277. [Google Scholar] [CrossRef]
- Kwon, S.J.; Jung, H.C.; Pan, J.G. Transgalactosylation in a water-solvent biphasic reaction system with β-galactosidase displayed on the surfaces of Bacillus subtilis spores. Appl. Environ. Microbiol. 2007, 73, 2251–2256. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, H.; Xia, Y.; Zhao, J.; Tian, F.; Zhang, H. Production, purification, and characterization of a potential thermostable galactosidase for milk lactose hydrolysis from Bacillus stearothermophilus. J. Dairy Sci. 2008, 91, 1751–1758. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Editorial for special issue: Enzyme immobilization and its applications. Molecules 2019, 24, 4619. [Google Scholar] [CrossRef] [PubMed]
- Lozančić, M.; Mrša, V.; Teparić, R. Surface display—an alternative to classic enzyme immobilization. Catalysts 2019, 9, 728. [Google Scholar] [CrossRef]
- Pan, J.G.; Choi, S.K.; Jung, H.C.; Kim, E.J. Display of native proteins on Bacillus subtilis spores. FEMS Microbiol. Lett. 2014, 358, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Israeli-Ruimy, V.; Bule, P.; Jindou, S.; Dassa, B.; Moraïs, S.; Borovok, I.; Barak, Y.; Slutzki, M.; Hamberg, Y.; Cardoso, V.; et al. Complexity of the Ruminococcus flavefaciens FD-1 cellulosome reflects an expansion of family-related protein-protein interactions. Sci. Rep. 2017, 7, 42355. [Google Scholar] [CrossRef]
- Wang, Y.; Leng, L.; Islam, M.K.; Liu, F.; Lin, C.S.K.; Leu, S.Y. Substrate-related factors affecting cellulosome-induced hydrolysis for lignocellulose valorization. Int. J. Mol. Sci. 2019, 20, 3354. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Miyake, H.; Kuroda, K.; Nakanishi, A.; Matsushima, C.; Doi, R.H.; Ueda, M. Comparison of the mesophilic cellulosome-producing Clostridium cellulovorans genome with other cellulosome-related clostridial genomes. Microb. Biotechnol. 2011, 4, 64–73. [Google Scholar] [CrossRef]
- Borne, R.; Bayer, E.A.; Pagès, S.; Perret, S.; Fierobe, H.P. Unraveling enzyme discrimination during cellulosome assembly independent of cohesin-dockerin affinity. FEBS J. 2013, 280, 5764–5779. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzym. Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Wang, F.; Song, T.; Jiang, H.; Pei, C.; Huang, Q.; Xi, H. Bacillus subtilis spore surface display of haloalkane dehalogenase DhaA. Curr. Microbiol. 2019, 76, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Wang, F.; Xiong, S.; Jiang, H. Surface display of organophosphorus-degrading enzymes on the recombinant spore of Bacillus subtilis. Biochem. Biophys. Res. Commun. 2019, 510, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Wiencek, K.M.; Klapes, N.A.; Foegeding, P.M. Hydrophobicity of Bacillus and Clostridium spores. Appl. Environ. Microbiol. 1990, 56, 2600–2605. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.A.; Kim, E.J.; Pan, J.G. Adsorption immobilization of Escherichia coli phytase on probiotic Bacillus polyfermenticus spores. Enzym. Microb. Technol. 2011, 49, 66–71. [Google Scholar] [CrossRef]
- Wang, H.; Yang, R.; Hua, X.; Zhao, W.; Zhang, W. Functional display of active β-galactosidase on Bacillus subtilis spores using crust proteins as carriers. Food Sci. Biotechnol. 2015, 24, 1755–1759. [Google Scholar] [CrossRef]
- Cutting, S.M.; Vander-Horn, P.B. Genetic analysis. In Molecular biological methods for Bacillus, 1st ed.; Harwood, C.R., Cutting, S.M., Eds.; Wiley: Chichester, UK, 1990; Volume 1, pp. 27–74. [Google Scholar]
| Bsβ–Gal Source | Amount of Sample Used | Density in OD/mm2 (± SD) | β–Gal Concentration (ng) | Number of Fusion Protein Molecules per Spore |
|---|---|---|---|---|
| Free Bsβ–Gal | 10.0 ng | 50.3 (± 0.12) | NA | |
| 5.0 ng | 25.8 (± 0.04) | NA | ||
| 2.5 ng | 12.9 (± 0.06) | NA | ||
| CotG–CcmCoh-I/Bsβ–Gal–CcmDoc-I | 2.0 × 106 | 42.2 (± 0.07) | 8.4 | 3.0 × 104 |
| 1.0 × 106 | 22.1 (± 0.02) | 4.4 | ||
| 5.0 × 105 | 10.9 (± 0.11) | 2.2 | ||
| CotG–RfCoh-III/Bsβ–Gal–RfDoc-III | 2.0 × 106 | 39.8 (± 0.13) | 7.9 | 2.8 × 104 |
| 1.0 × 106 | 19.6 (± 0.12) | 3.9 | ||
| 5.0 × 105 | 11.4 (± 0.08) | 2.3 | ||
| CotG–Bsβ–Gal | 1.0 × 106 | 36.6 (± 0.06) | 7.3 | 4.3 × 104 |
| 5.0 × 105 | 19.1 (± 0.11) | 3.8 | ||
| 2.5 × 105 | 9.8 (± 0.04) | 2.0 | ||
| Pcry1Aa–Bsβ–Gal | 5.0 × 105 | 42.3 (± 0.06) | 8.4 | 1.3 × 105 |
| 2.5 × 105 | 21.1 (± 0.09) | 4.2 | ||
| 1.3 × 105 | 11.9 (± 0.05) | 2.4 |
| Solvent | logP Value of Solvent | Relative Activity (%) | ||||
|---|---|---|---|---|---|---|
| Free Bsβ-Gal | Pcry1Aa–Bsβ–Gal | CotG–Bsβ–Gal | CotG–CcmCoh-I/Bsβ–Gal–CcmDoc-I | CotG–RfCoh-III/Bsβ–Gal–RfDoc-III | ||
| Control | 100 ± 0.8 | 100 ± 1.3 | 100 ± 1.8 | 100 ± 0.9 | 100 ± 1.6 | |
| n-Hexane | 3.66 | 93.4 ± 2.1 | 93.7 ± 2.7 | 102.4 ± 3.4 | 98.3 ± 3.5 | 97.2 ± 2.4 |
| 1-Chloroheptane | 3.37 | 90.6 ± 2.5 | 91.8 ± 2.9 | 107.8 ± 1.7 | 99.3 ± 2.2 | 98.5 ± 3.1 |
| Toluene | 2.39 | 33.1 ± 1.1 | 37.5 ± 1.6 | 73.3 ± 2.5 | 67.4 ± 1.4 | 71.2 ± 1.8 |
| Cyclohexanone | 1.40 | 37.2 ± 1.3 | 39.3 ± 1.9 | 67.8 ± 2.2 | 62.9 ± 2.7 | 66.1 ± 2.0 |
| Ethyl ether | 1.05 | 21.4 ± 0.7 | 27.9 ± 1.4 | 56.1 ± 2.6 | 47.2 ± 1.3 | 46.5 ± 1.5 |
| Acetonitrile | 0.47 | 12.3 ± 0.4 | 11.4 ± 0.2 | 30.6 ± 0.5 | 28.2 ± 0.6 | 29.5 ± 1.0 |
| Ethanol | 0.06 | 0 | 1.7 ± 0.1 | 13.2 ± 0.3 | 6.5 ± 0.4 | 8.3 ± 0.2 |
| 1,4-Dioxane | −0.23 | 0 | 0 | 3.6 ± 0.1 | 0.4 ± 0.1 | 1.1 ± 0.1 |
| Recombinant Spores | Vmax/(μmol·min−1·mg·spores−1 (dry weight)) | Km/(mM) |
|---|---|---|
| Bsβ–Gal | 7.24 ± 0.12 a | 2.97 ± 0.05 a |
| Pcry1Aa–Bsβ–Gal | 1.01 ± 0.16 b | 5.71 ± 0.09 b |
| CotG–Bsβ–Gal | 0.77 ± 0.13 b | 4.43 ± 0.16 c |
| CotG–CcmCoh-I/Bsβ–Gal–CcmDoc-I | 1.04 ± 0.08 b | 3.83 ± 0.10 d |
| CotG–RfCoh-III/Bsβ–Gal–RfDoc-III | 0.78 ± 0.11 b | 3.71 ± 0.17 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Jiang, X.; Qian, Y.; Yin, L. Constructing an Efficient Bacillus subtilis Spore Display by Using Cohesin−Dockerin Interactions. Molecules 2021, 26, 1186. https://doi.org/10.3390/molecules26041186
Wang H, Jiang X, Qian Y, Yin L. Constructing an Efficient Bacillus subtilis Spore Display by Using Cohesin−Dockerin Interactions. Molecules. 2021; 26(4):1186. https://doi.org/10.3390/molecules26041186
Chicago/Turabian StyleWang, He, Xiaomin Jiang, Yongchang Qian, and Lianghong Yin. 2021. "Constructing an Efficient Bacillus subtilis Spore Display by Using Cohesin−Dockerin Interactions" Molecules 26, no. 4: 1186. https://doi.org/10.3390/molecules26041186
APA StyleWang, H., Jiang, X., Qian, Y., & Yin, L. (2021). Constructing an Efficient Bacillus subtilis Spore Display by Using Cohesin−Dockerin Interactions. Molecules, 26(4), 1186. https://doi.org/10.3390/molecules26041186





