Impact of Lignocellulose Pretreatment By-Products on S. cerevisiae Strain Ethanol Red Metabolism during Aerobic and An-aerobic Growth
Abstract
1. Introduction
2. Results
2.1. Assessment of the Capacity of S. cerevisiae Strain Ethanol Red to Metabolize Fermentation Inhibitors (By-Products of Pretreatment of Lignocellulosic Biomass)
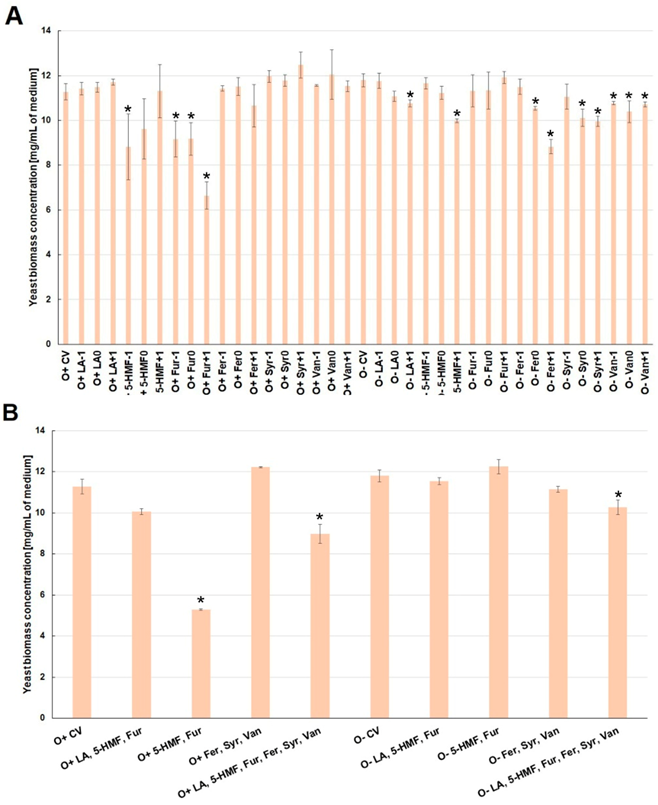
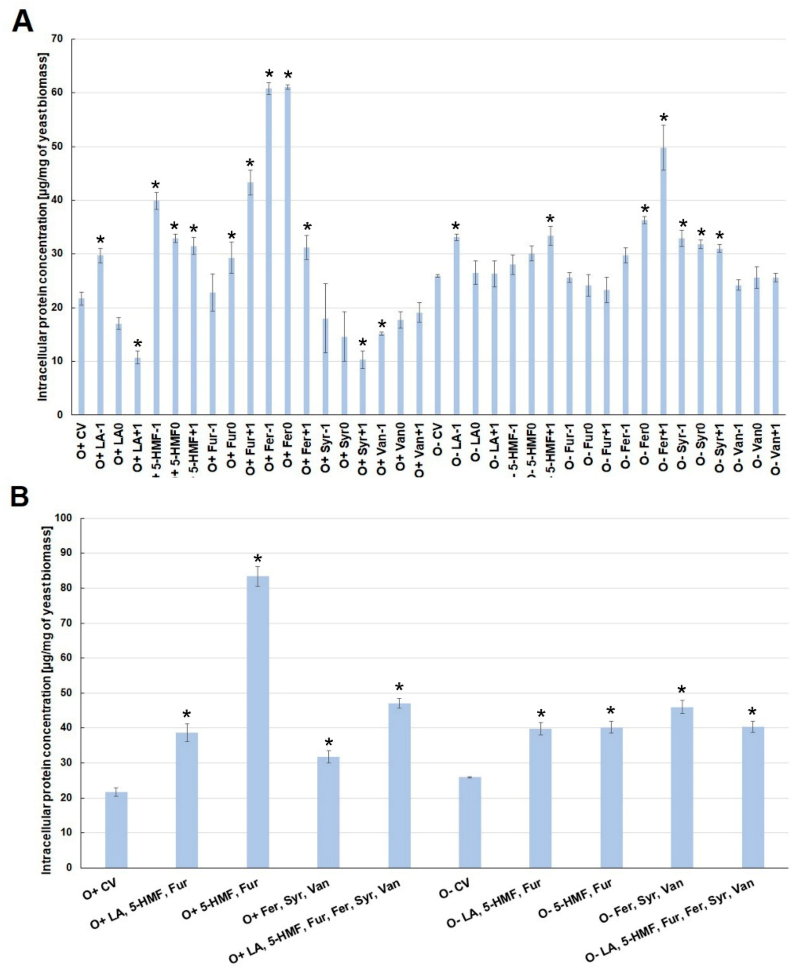
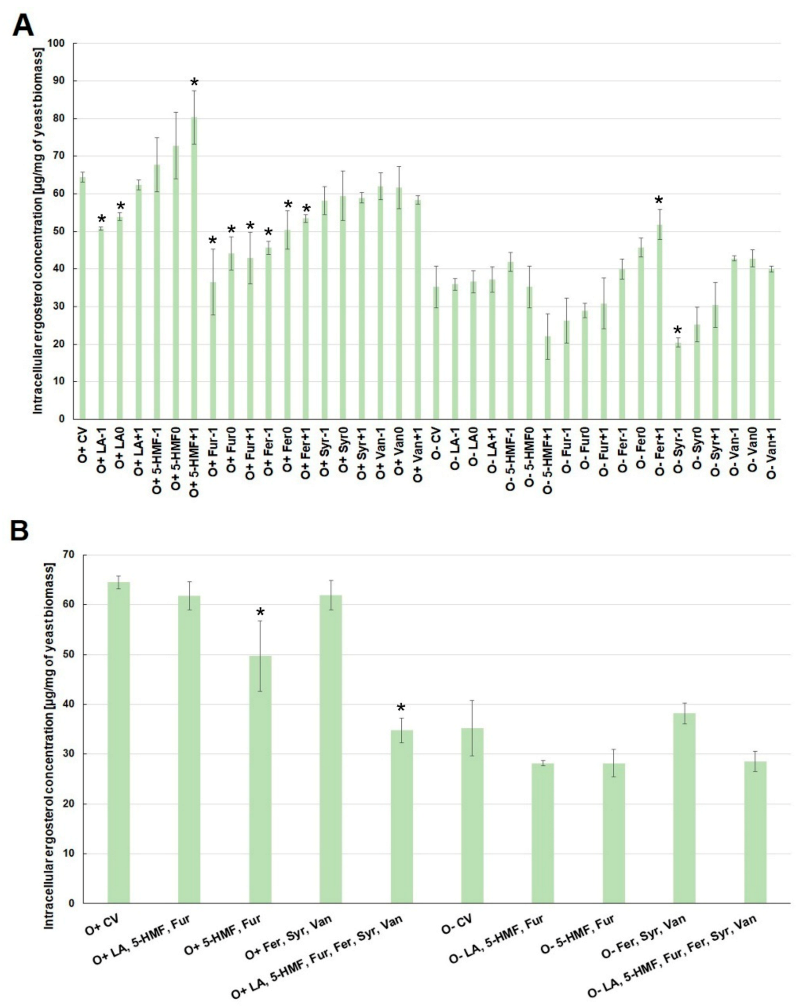
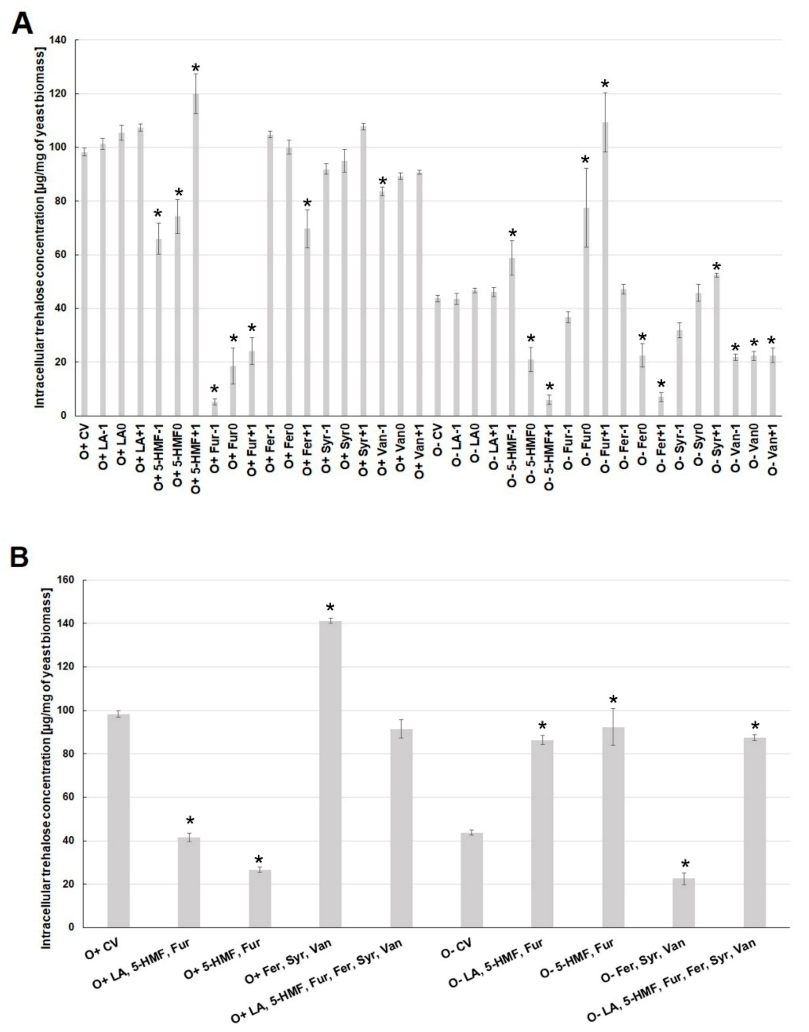
2.2. Effect of By-Products of Lignocellulose Pretreatment on the Growth of Yeast Biomass and the Concentration of Intracellular Metabolites under Various Culture Conditions
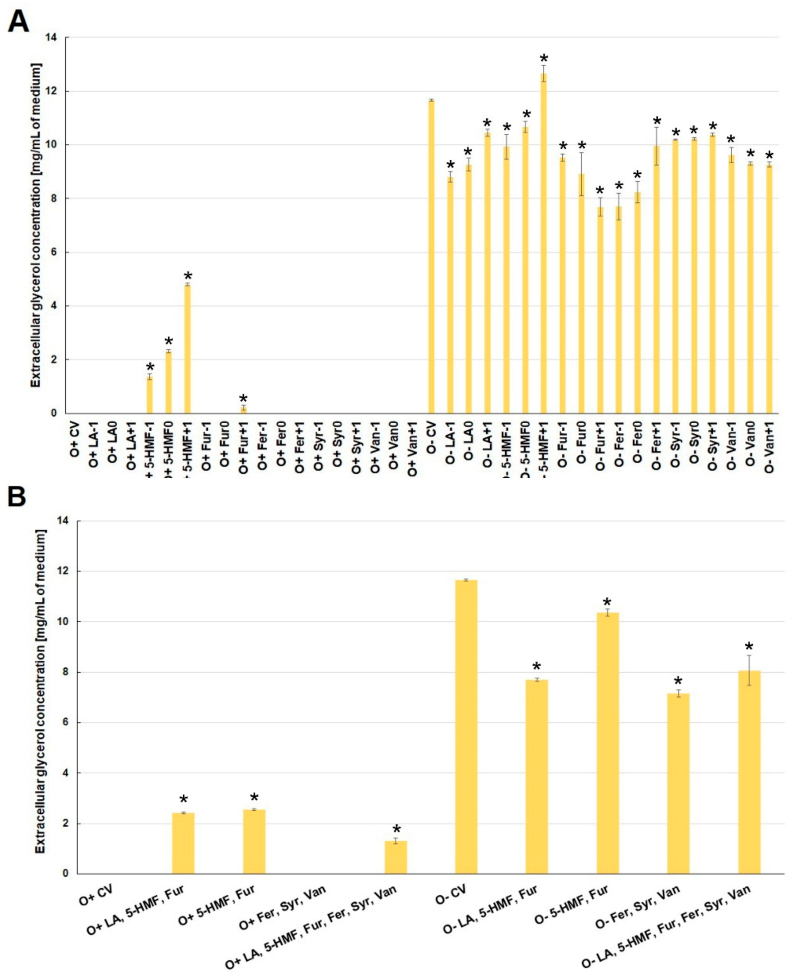
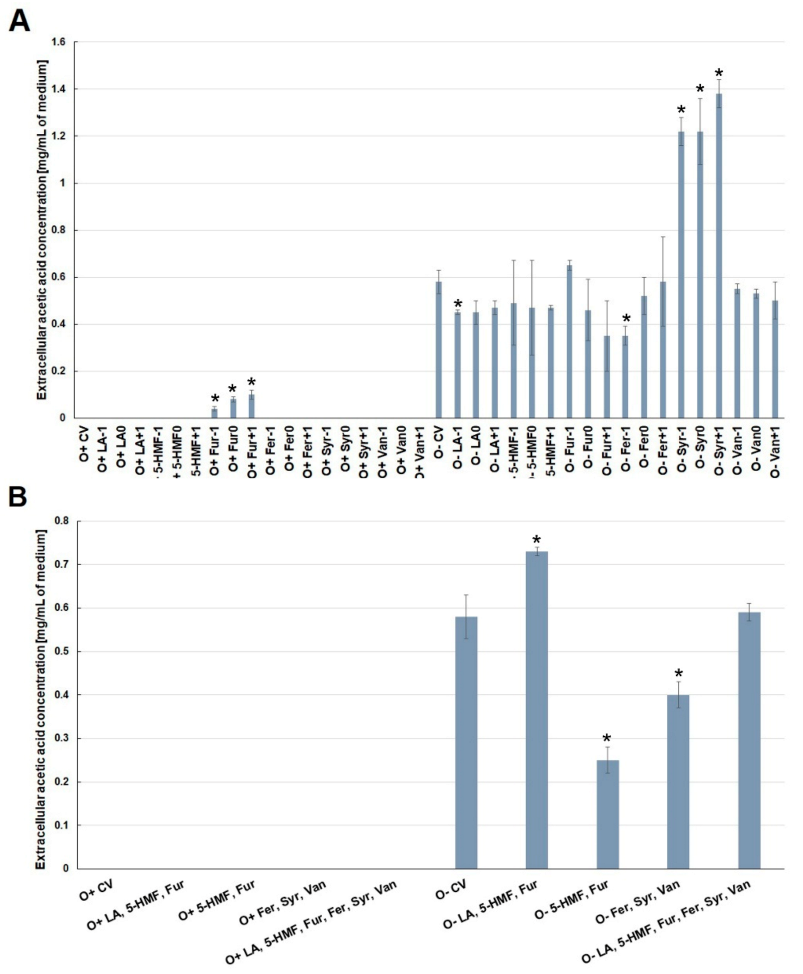
2.3. Effect of By-Products of Lignocellulose Pretreatment on the Production of Extracellular Metabolites (Acetic Acid and Glycerol) under Aerobic Conditions and during Fermentation
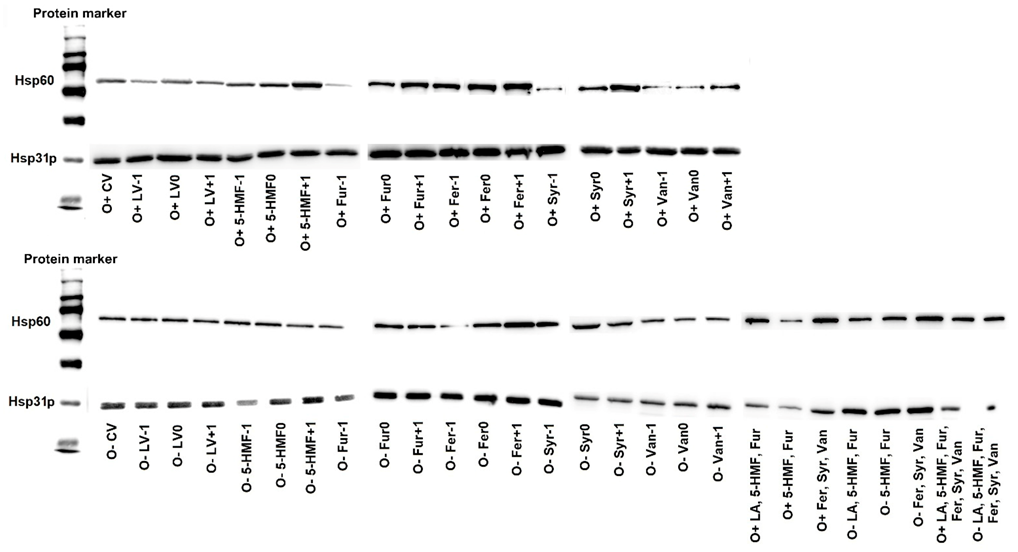
2.4. Production of Hsp31p and Hsp60 as a Metabolic Response of Yeast Cells to Stress Caused by the Presence of Lignocellulose Pretreatment By-Products
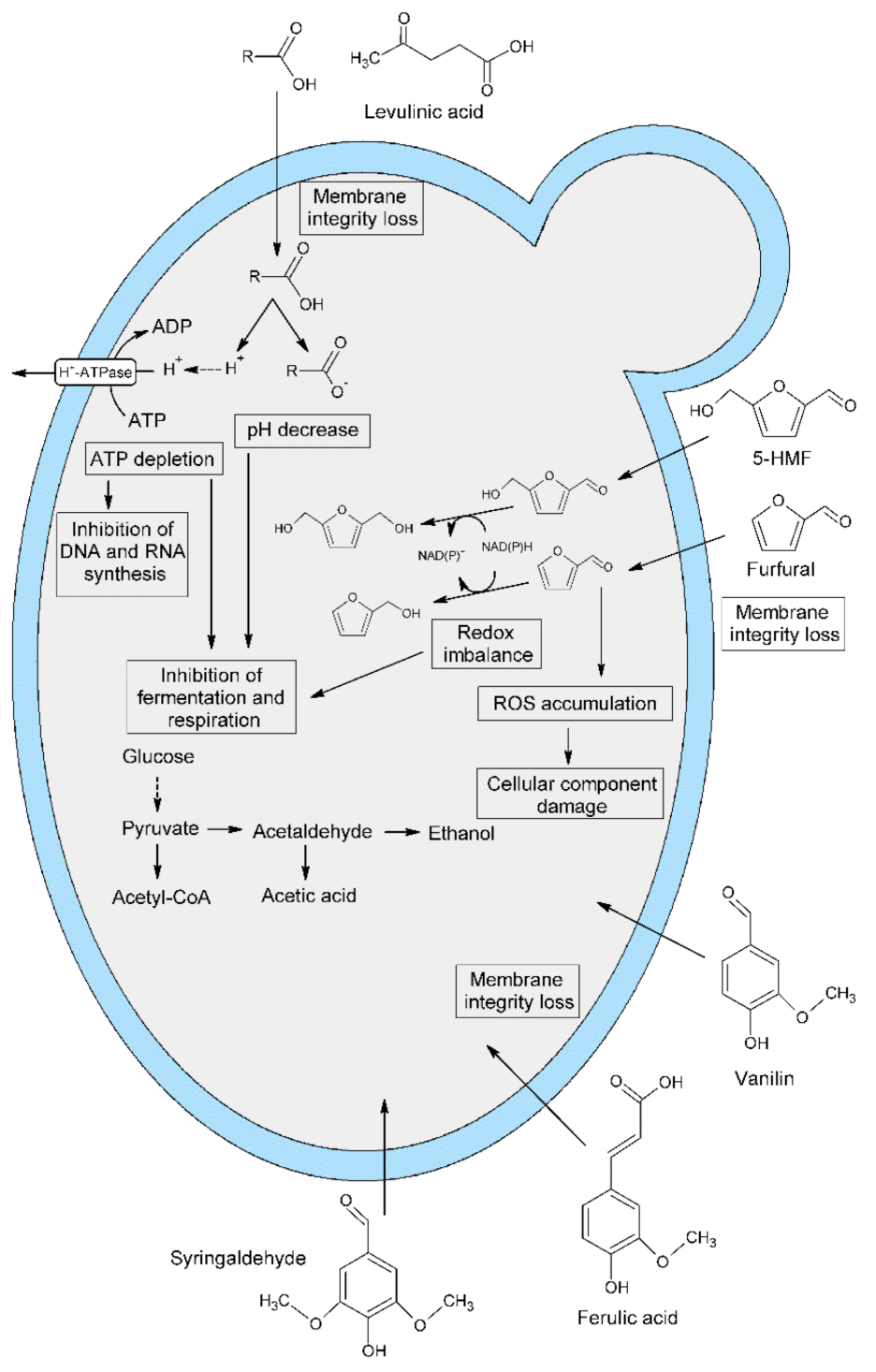
3. Discussion
4. Materials and Methods
4.1. Yeast
4.2. Materials
4.3. Research Plan
4.4. Culture Media
4.5. Analytical Methods
4.5.1. Determination of Yeast Biomass Concentration
4.5.2. Determination of Intracellular Concentration of Trehalase
4.5.3. Determination of the Intracellular Concentration of Ergosterol
4.5.4. Determination of Intracellular Protein Concentration
4.5.5. Determination of HSP Proteins by Western Blot
4.5.6. Determination of Extracellular Concentrations of Acetic Acid and Glycerol
4.5.7. Measuring the Concentration of Lignocellulose Pretreatment By-Products in Culture Media
4.6. Statistical Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-HMF | 5-Hydroxymethylfurfural |
| VFAs | Volatile fatty acids |
| gTME | Global transcription engineering technique |
| HSP | Heat shock protein |
| HPLC | High performance liquid chromatography |
| ROS | Reactive oxygen species |
| CFU | Colony-forming unit |
| ESTD | External standard |
| SDS | Sodium dodecyl sulfate |
| HRP | Horseradish peroxidase |
| PVDF | Polyvinylidene fluoride |
| TBST | Tris-buffered saline Tween |
References
- Sarkar, N.; Ghosh, S.K.; Bannerjee, S.; Aikat, K. Bioethanol production from agricultural wastes: An overview. Renew. Energy 2012, 37, 19–27. [Google Scholar] [CrossRef]
- Sharma, B.; Larroche, C.; Dussap, C.-G. Comprehensive assessment of 2G bioethanol production. Bioresour. Technol. 2020, 313, 123630. [Google Scholar] [CrossRef]
- Anu; Kumar, A.; Rapoport, A.; Kunze, G.; Kumar, S.; Singh, D.; Sing, B. Multifarious pretreatment strategies for the lignocellulosic substrates for the generation of renewable and sustainable biofuels: A review. Renew. Energ. 2020, 160, 1228–1252. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Dien, B.S.; Berhow, M.A.; Kurtzman, C.P.; Gorsich, S.W. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 2004, 31, 345–352. [Google Scholar] [CrossRef]
- Ra, C.H.; Jeong, G.-T.; Shin, M.K.; Kim, S.-K. Biotransformation of 5-hydroxymethylfurfural (HMF) by Scheffersomyces stipitis during ethanol fermentation of hydrolysate of the seaweed Gelidium amansii. Bioresour. Technol. 2013, 140, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, B.; Ezeji, T.C. Biotransformation of furfural and 5-hydroxymethyl furfural (HMF) by Clostridium acetobutylicum ATCC 824 during butanol fermentation. New Biotechnol. 2012, 29, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.Z.; Wang, X.; Yang, Y.; Yuan, Y.-J. Comparative metabolic profiling of parental and inhibitors-tolerant yeasts during lig-nocellulosic ethanol fermentation. Metabolomics 2012, 8, 232–243. [Google Scholar] [CrossRef]
- Palmqvist, E.; Hahn-Hägerdal, B. Fermentation of lignocellulosic hydrolysates. II: Inhibitors and mechanisms of inhibition. Bioresour. Technol. 2000, 74, 25–33. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Alriksson, B.; Nilvebrant, N.-O. Bioconversion of lignocellulose: Inhibitors and detoxification. Biotechnol. Biofuels 2013, 6, 16. [Google Scholar] [CrossRef]
- Bearson, S.; Bearson, B.; Foster, J.W. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 1997, 147, 173–180. [Google Scholar] [CrossRef]
- Fiedurek, J.; Trytek, M. The efect of acid and osmotic stress on metabolite production by microorganisms. Post. Mikrobiol. 2016, 55, 195–204. (In Polish) [Google Scholar]
- Mollapour, M.; Piper, P.W. Hog1 Mitogen-Activated Protein Kinase Phosphorylation Targets the Yeast Fps1 Aquaglyceroporin for Endocytosis, Thereby Rendering Cells Resistant to Acetic Acid. Mol. Cell. Biol. 2007, 27, 6446–6456. [Google Scholar] [CrossRef]
- Giannattasio, S.; Guaragnella, N.; Ždralević, M.; Marra, E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front. Microbiol. 2013, 4, 33. [Google Scholar] [CrossRef]
- Ullah, A.; Orij, R.; Brul, S.; Smits, G.J. Quantitative analysis of the modes of growth inhibition by weak organic acids in yeast. Appl. Environ. Microbiol. 2012, 78, 8377–8387. [Google Scholar] [CrossRef] [PubMed]
- Ndukwe, J.K.; Aliyu, G.O.; Onwosi, C.O.; Chukwu, K.O.; Ezugworie, F.N. Mechanisms of weak acid-induced stress tolerance in yeasts: Prospects for improved bioethanol production from lignocellulosic biomass. Process. Biochem. 2020, 90, 118–130. [Google Scholar] [CrossRef]
- Liu, Z.L. Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl. Microbiol. Biotechnol. 2006, 73, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Liu, Z.L. Engineered NADH-dependent GRE2 from Saccharomyces cerevisiae by directed enzyme evolution enhances HMF reduction using additional cofactor NADPH. Enzym. Microb. Technol. 2012, 50, 115–120. [Google Scholar] [CrossRef]
- Palmqvist, E.; Meinander, Q.; Grage, H.; Hahn-Hägerdal, B. Main and interaction effects of acetic acid, furfural and p-hy-droxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol. Bioeng. 1999, 63, 46–55. [Google Scholar] [CrossRef]
- Nguyen, T.T.M.; Iwaki, A.; Ohya, Y.; Izawa, S. Vanillin causes the activation of Yap1 and mitochondrial fragmentation in Saccha-romyces cerevisiae. J. Biosci. Bioeng. 2014, 117, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Alriksson, B.; Horváth, I.S.; Jönsson, L.J. Overexpression of Saccharomyces cerevisiae transcription factor and multidrug resistance genes conveys enhanced resistance to lignocellulose-derived fermentation inhibitors. Process. Biochem. 2010, 45, 264–271. [Google Scholar] [CrossRef]
- Gorsich, S.W.; Dien, B.S.; Nichols, N.N.; Slininger, P.J.; Liu, Z.L.; Skory, C.D. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2006, 71, 339–349. [Google Scholar] [CrossRef]
- Petersson, A.; Almeida, J.R.M.; Modig, T.; Karhumaa, K.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.-F.; Lidén, G. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 2006, 23, 455–464. [Google Scholar] [CrossRef]
- Kumari, R.; Pramanik, K. Improvement of multiple stress tolerance in yeast strain by sequential mutagenesis for enhanced bioethanol production. J. Biosci. Bioeng. 2012, 114, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.-S.; Cate, J.H.D. Metabolic engineering of yeast for lignocellulosic biofuel production. Curr. Opin. Chem. Biol. 2017, 41, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Wang, X.; Weber, S.A. Tolerant industrial yeast Saccharomyces cerevisiae possess a more robust cell wall integrity signaling pathway against 2-furaldehyde and 5-(hydroxymethyl)-2-furaldehyde. J. Biotechnol. 2018, 2018, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Mashego, M.R.; Jansen, M.L.A.; Vinke, J.L.; van Gulik, W.M.; Heijnen, J.J. Changes in the metabolome of Saccharomyces cerevisiae associated with evolution in aerobic glucose-limited chemostats. FEMS Yeast Res. 2005, 5, 419–430. [Google Scholar] [CrossRef]
- Heer, D.; Sauer, U. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb. Biotechnol. 2008, 1, 497–506. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Gorsich, S.W. Enhanced Biotransformation of Furfural and Hydroxymethylfurfural by Newly Developed Ethanologenic Yeast Strains. Appl. Biochem. Biotechnol. 2005, 121, 0451–0460. [Google Scholar] [CrossRef]
- Laluce, C.; Schenberg, A.C.G.; Gallardo, J.C.M.; Coradello, L.F.C.; Sponchiado, S.R.P. Advances and Developments in Strategies to Improve Strains of Saccharomyces cerevisiae and Processes to Obtain the Lignocellulosic Ethanol−A Review. Appl. Biochem. Biotechnol. 2012, 166, 1908–1926. [Google Scholar] [CrossRef]
- Mikulski, D.; Kłosowski, G. Hydrotropic pretreatment on distillery stillage for efficient cellulosic ethanol production. Bioresour. Technol. 2020, 300, 122661. [Google Scholar] [CrossRef]
- Mikulski, D.; Kłosowski, G. Microwave-assisted dilute acid pretreatment in bioethanol production from wheat and rye stillages. Biomass- Bioenergy 2020, 136, 105528. [Google Scholar] [CrossRef]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Alcohols and Other Volatile Compounds. In Handbook of Enology; Wiley: West Sussex, UK, 2005; Volume 2. [Google Scholar]
- Siderius, M.; Van Wuytswinkel, O.; Reijenga, K.A.; Kelders, M.; Mager, W.H. The control of intracellular glycerol in Saccharomyces cerevisiae influences osmotic stress response and resistance to increased temperature. Mol. Microbiol. 2002, 36, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Biochemistry of Alcoholic Fermentation and Metabolic Pathways of Wine Yeasts. In Handbook of Enology; Wiley: West Sussex, UK, 2005; Volume 1. [Google Scholar]
- Skoneczna, A.; Micialkiewicz, A.; Skoneczny, M. Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free. Radic. Biol. Med. 2007, 42, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, B.A.; Kolesar, J.E.; Perlman, P.S.; Butow, R.A. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell Biol. 2003, 163, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocellulose hydrolysates. Appl. Microbiol. Biotechnol. 2011, 90, 809–825. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, H.-O.; Cho, Y.-J.; Kim, J.; Chun, J.; Choi, J.; Lee, Y.; Jung, W.H. A Vanillin Derivative Causes Mitochondrial Dysfunction and Triggers Oxidative Stress in Cryptococcus neoformans. PLoS ONE 2014, 9, e89122. [Google Scholar] [CrossRef]
- Endo, A.; Nakamura, T.; Shima, J. Involvement of ergosterol in tolerance to vanillin, a potential inhibitor of bioethanol fermentation, in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2009, 299, 95–99. [Google Scholar] [CrossRef]
- Wang, X.; Liang, Z.; Hou, J.; Bao, X.; Shen, Y. Identification and functional evaluation of the reductases and dehydrogenases from Saccharomyces cerevisiae involved in vanillin resistance. BMC Biotechnol. 2016, 16, 31. [Google Scholar] [CrossRef]
- Thompson, O.; Hawkins, G.M.; Gorsich, S.W.; Doran-Peterson, J. Phenotypic characterization and comparative transcriptomics of evolved Saccharomyces cerevisiae strains with improved tolerance to lignocellulosic derived inhibitors. Biotechnol. Biofuels 2016, 9, 200. [Google Scholar] [CrossRef]
- Ma, M.; Liu, Z.L. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genom. 2010, 11, 660. [Google Scholar] [CrossRef]
- Higgins, V.J.; Beckhouse, A.G.; Oliver, A.D.; Rogers, P.J.; Dawes, I.W. Yeast Genome-Wide Expression Analysis Identifies a Strong Ergosterol and Oxidative Stress Response during the Initial Stages of an Industrial Lager Fermentation. Appl. Environ. Microbiol. 2003, 69, 4777–4787. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, Z.; Olsson, L. Physiological response of Saccharomyces cerevisiae to weak acids present in lignocellulosic hydrolysate. FEMS Yeast Res. 2014, 14, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Cabiscol, E.; Bellí, G.; Tamarit, J.; Echave, P.; Herrero, E.; Ros, J. Mitochondrial Hsp60, Resistance to Oxidative Stress, and the Labile Iron Pool Are Closely Connected in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 44531–44538. [Google Scholar] [CrossRef]
- Treweek, T.M.; Linder, R.A.; Mariani, M.; Carver, J.A. The small heat-shock chaperone protein, α-crystallin, does not recognise stable molten globule states of cytosolic proteins. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 2000, 1481, 175–188. [Google Scholar] [CrossRef]
- Swan, T.M.; Watson, K. Stress tolerance in a yeast sterol auxotroph: Role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 1998, 169, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Odumeru, J.A.; D’Amore, T.; Russell, I.; Stewart, G.G. Effects of heat shock and ethanol stress on the viability of a Saccharomyces uvarum (carlsbergensis) brewing yeast strain during fermentation of high gravity wort. J. Ind. Microbiol. Biotechnol. 1992, 10, 111–116. [Google Scholar] [CrossRef]
- Lee, S.J.; Ramesh, R.; de Boor, V.; Gebler, J.M.; Silva, R.C.; Sattlegger, E. Cost-effective and rapid lysis of Saccharomyces cerevisiae cells for quantitative western blot analysis of proteins, including phosphorylated eIF2α. Yeast 2017, 34, 371–382. [Google Scholar] [CrossRef]
- Liu, H.-J.; Liu, D.; Zhong, J.-J. Interesting physiological response of the osmophilic yeast Candida krusei to heat shock. Enzym. Microb. Technol. 2005, 36, 409–416. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid sensitive method for the quntitation of quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Cho, D.H.; Lee, Y.J.; Um, Y.; Sang, B.-I.; Kim, Y.H. Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl. Microbiol. Biotechnol. 2009, 83, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
| Research Variant | Concentration [mg/L] of By-Products Resulting from the Pretreatment of Lignocellulosic Biomass | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Levulinic Acid | 5-HMF | Furfural | Ferulic Acid | Syringaldehyde | Vanillin | |||||||||||||
| 0 h | 72 h | %reduc. | 0 h | 72 h | %reduc. | 0 h | 72 h | %reduc. | 0 h | 72 h | %reduc. | 0 h | 72 h | %reduc. | 0 h | 72 h | %reduc. | |
| O+ CV | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ LA−1 | 408.1 ± 7.9 | 233.0 ± 9.3 | 42.9 ± 3.1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ LA0 | 812.6 ± 23.8 | 565.1 ± 10.1 | 30.4 ± 2.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ LA+1 | 1606.4 ± 29.2 | 1247.7 ± 25.9 | 22.3 ± 2.8 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ 5-HMF−1 | nf | nf | - | 1344.4 ± 157.0 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ 5-HMF0 | nf | nf | - | 2573.9 ± 82.1 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ 5-HMF+1 | nf | nf | - | 5704.8 ± 249.3 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ Fur−1 | nf | nf | - | nf | nf | - | 126.4 ± 9.7 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ Fur0 | nf | nf | - | nf | nf | - | 545.6 ± 22.0 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ Fur+1 | nf | nf | - | nf | nf | - | 1433.5 ± 40.7 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ Fer−1 | nf | nf | - | nf | nf | - | nf | nf | - | 83.4 ± 0.9 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - |
| O+ Fer0 | nf | nf | - | nf | nf | - | nf | nf | - | 176.0± 2.4 | 18.0 ± 1.5 | 89.8 ± 0.8 | nf | nf | - | nf | nf | - |
| O+ Fer+1 | nf | nf | - | nf | nf | - | nf | nf | - | 376.3± 2.9 | 240.8 ± 26.7 | 36.0 ± 6.9 | nf | nf | - | nf | nf | - |
| O+ Syr−1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 19.5 ± 0.2 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O+ Syr0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 41.2 ± 0.9 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O+ Syr+1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 83.8 ± 0.4 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O+ Van−1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 19.4 ± 0.7 | 0.0 ± 0.0 | 100.0± 0.0 |
| O+ Van0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 39.1 ± 0.4 | 0.0 ± 0.0 | 100.0± 0.0 |
| O+ Van+1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 81.5 ± 0.5 | 0.0 ± 0.0 | 100.0± 0.0 |
| O− CV | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− LA−1 | 588.7 ± 47.4 | 494.5 ± 40.2 | 16.0 ± 3.4 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− LA0 | 863.8 ± 24.1 | 756.7 ± 31.6 | 12.4 ± 1.5 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− LA+1 | 1709.1 ± 27.7 | 1595.5 ± 28.8 | 6.6 ± 1.8 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− 5-HMF−1 | nf | nf | - | 1410.6 ± 39.6 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− 5-HMF0 | nf | nf | - | 2676.5 ± 57.6 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− 5-HMF+1 | nf | nf | - | 5386.3 ± 254.4 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - |
| O− Fur−1 | nf | nf | - | nf | nf | - | 128.6 ± 17.3 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O− Fur0 | nf | nf | - | nf | nf | - | 524.8 ± 75.5 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O− Fur+1 | nf | nf | - | nf | nf | - | 1350.9 ± 62.4 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O− Fer−1 | nf | nf | - | nf | nf | - | nf | nf | - | 108.0± 1.1 | 48.3 ± 7.9 | 55.3 ± 7.3 | nf | nf | - | nf | nf | - |
| O− Fer0 | nf | nf | - | nf | nf | - | nf | nf | - | 209.3± 1.7 | 114.8 ± 15.8 | 45.2 ± 7.1 | nf | nf | - | nf | nf | - |
| O− Fer+1 | nf | nf | - | nf | nf | - | nf | nf | - | 395.9± 5.4 | 307.7 ± 13.0 | 22.2 ± 3.7 | nf | nf | - | nf | nf | - |
| O− Syr−1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 19.1 ± 1.2 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O− Syr0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 40.6 ± 1.7 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O− Syr+1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 85.6 ± 2.2 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - |
| O− Van−1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 18.1 ± 0.6 | 0.0 ± 0.0 | 100.0± 0.0 |
| O− Van0 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 39.1 ± 1.0 | 0.0 ± 0.0 | 100.0± 0.0 |
| O− Van+1 | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | nf | nf | - | 83.3 ± 1.3 | 0.0 ± 0.0 | 100.0± 0.0 |
| O+ LA, 5-HMF, Fur | 822.3 ± 9.8 | 752.2 ± 27.2 | 8.5 ± 4.4 | 2727.5 ± 17.2 | 849.2 ± 16.6 | 68.9 ± 0.8 | 482.6 ± 30.0 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ 5-HMF, Fur | nf | nf | - | 2785.5 ± 38.5 | 143.4 ± 1.5 | 94.9 ± 0.1 | 479.4 ± 10.1 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O+ Fer, Syr, Van | nf | nf | - | nf | nf | - | nf | nf | - | 201.6± 2.6 | 114.9 ± 12.2 | 43.0 ± 6.4 | 39.5 ± 1.3 | 0.0 ± 0.0 | 100.0± 0.0 | 39.5 ± 1.3 | 0.0 ± 0.0 | 100.0± 0.0 |
| O− LA, 5-HMF, Fur | 807.2 ± 3.3 | 733.9 ± 43.9 | 9.1 ± 5.4 | 2719.7 ± 56.6 | 0.0 ± 0.0 | 100.0± 0.0 | 509.2 ± 19.0 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O− 5-HMF, Fur | nf | nf | - | 2692.8 ± 53.8 | 0.0 ± 0.0 | 100.0± 0.0 | 503.8 ± 20.1 | 0.0 ± 0.0 | 100.0± 0.0 | nf | nf | - | nf | nf | - | nf | nf | - |
| O− Fer, Syr, Van | nf | nf | - | nf | nf | - | nf | nf | - | 201.6± 1.9 | 113.2 ± 14.0 | 43.9 ± 6.9 | 38.0 ± 0.6 | 0.0 ± 0.0 | 100.0± 0.0 | 39.5 ± 0.7 | 0.0 ± 0.0 | 100.0± 0.0 |
| O+ LA, 5-HMF, Fur, Fer, Syr, Van | 798.7 ± 14.2 | 741.3 ± 23.3 | 7.2 ± 3.8 | 2650.8 ± 90.0 | 489.5 ± 21.5 | 81.5 ± 1.3 | 459.4 ± 22.2 | 0.0 ± 0.0 | 100.0± 0.0 | 209.0± 4.3 | 173.2 ± 9.2 | 17.1 ± 4.5 | 41.7 ± 0.8 | 0.0 ± 0.0 | 100.0± 0.0 | 40.4 ± 1.5 | 0.0 ± 0.0 | 100.0± 0.0 |
| O− LA, 5-HMF, Fur, Fer, Syr, Van | 769.1 ± 13.2 | 680.1 ± 14.1 | 11.5 ± 2.9 | 2705.1 ± 4.5 | 0.0 ± 0.0 | 100.0± 0.0 | 431.1 ± 19.6 | 0.0 ± 0.0 | 100.0± 0.0 | 208.3± 3.8 | 156.9 ± 15.1 | 24.6 ± 7.9 | 41.7 ± 0.7 | 0.0 ± 0.0 | 100.0± 0.0 | 39.7 ± 1.4 | 0.0 ± 0.0 | 100.0± 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kłosowski, G.; Mikulski, D. Impact of Lignocellulose Pretreatment By-Products on S. cerevisiae Strain Ethanol Red Metabolism during Aerobic and An-aerobic Growth. Molecules 2021, 26, 806. https://doi.org/10.3390/molecules26040806
Kłosowski G, Mikulski D. Impact of Lignocellulose Pretreatment By-Products on S. cerevisiae Strain Ethanol Red Metabolism during Aerobic and An-aerobic Growth. Molecules. 2021; 26(4):806. https://doi.org/10.3390/molecules26040806
Chicago/Turabian StyleKłosowski, Grzegorz, and Dawid Mikulski. 2021. "Impact of Lignocellulose Pretreatment By-Products on S. cerevisiae Strain Ethanol Red Metabolism during Aerobic and An-aerobic Growth" Molecules 26, no. 4: 806. https://doi.org/10.3390/molecules26040806
APA StyleKłosowski, G., & Mikulski, D. (2021). Impact of Lignocellulose Pretreatment By-Products on S. cerevisiae Strain Ethanol Red Metabolism during Aerobic and An-aerobic Growth. Molecules, 26(4), 806. https://doi.org/10.3390/molecules26040806








