Bioinformatics and Functional Analyses Implicate Potential Roles for EOGT and L-fringe in Pancreatic Cancers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Contribution of EOGT and LFNG to Notch Signaling in PDAC
2.2. Expression of EOGT in PDAC Cell Lines
2.3. CRISPR/CAS9-Mediated Lentiviral Knockout of EOGT in a PDAC Cell Line
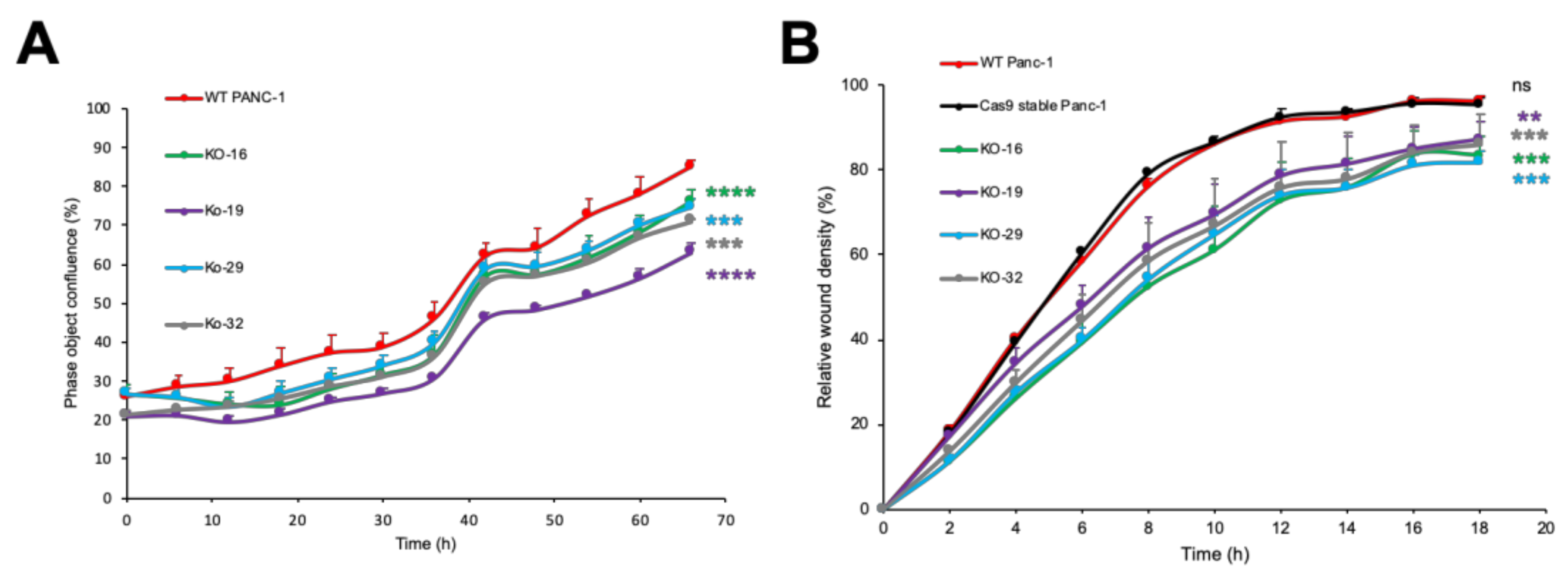
2.4. EOGT Knockout in Panc-1 Cells Impairs Cell Proliferation and Migration
2.5. CRISPR/Cas9-Mediated Lentiviral Knockout of LFNG in a PDAC Cell Line
2.6. LFNG Knockout in Panc-1 Cells Impairs Cell Proliferation and Migration
2.7. Low Expression of Both EOGT and LFNG Predicts Better Overall Survival in PDAC Patients
3. Materials and Methods
3.1. Antibodies and Reagents
3.2. Cell Culture
3.3. Vectors
3.4. Cell Growth Inhibition by DAPT
3.5. Lentivirus Vector Construction
3.6. CRISPR/Cas9-Mediated Lentiviral Knockout of EOGT and LFNG Genes
3.7. Evaluation of CRISPR/Cas9-mediated Genome Editing by T7 Endonuclease Assay and Sequencing Analysis
3.8. Immunostaining
3.9. Western Blotting
3.10. Cell Proliferation Assay
3.11. Cell Migration Assay
3.12. Statistical Analysis and Database Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; E Neale, R.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Bliss, L.A.; Witkowski, E.R.; Yang, C.J.; Tseng, J.F. Outcomes in operative management of pancreatic cancer. J. Surg. Oncol. 2014, 110, 592–598. [Google Scholar] [CrossRef]
- Ntziachristos, P.; Lim, J.S.; Sage, J.; Aifantis, I. From Fly Wings to Targeted Cancer Therapies: A Centennial for Notch Signaling. Cancer Cell 2014, 25, 318–334. [Google Scholar] [CrossRef] [Green Version]
- Tashima, Y.; Okajima, T. Congenital diseases caused by defective O-glycosylation of Notch receptors. Nagoya J. Med Sci. 2018, 80, 299–307. [Google Scholar]
- Espinoza, I.; Miele, L. Notch inhibitors for cancer treatment. Pharmacol. Ther. 2013, 139, 95–110. [Google Scholar] [CrossRef] [Green Version]
- Miele, L.; Espinoza, I.; Pochampally, R.; Watabe, K.; Xing, F. Notch signaling: Targeting cancer stem cells and epithelial-to-mesenchymal transition. OncoTargets Ther. 2013, 6, 1249–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila, J.L.; Kissil, J.L. Notch signaling in pancreatic cancer: Oncogene or tumor suppressor? Trends Mol. Med. 2013, 19, 320–327. [Google Scholar] [CrossRef] [Green Version]
- Nakhai, H.; Siveke, J.T.; Klein, B.; Mendoza-Torres, L.; Mazur, P.K.; Algül, H.; Radtke, F.; Strobl, L.; Zimber-Strobl, U.; Schmidt, G. Conditional ablation of Notch signaling in pancreatic development. Development 2008, 135, 2757–2765. [Google Scholar] [CrossRef] [Green Version]
- Hanlon, L.; Avila, J.L.; Demarest, R.M.; Troutman, S.; Allen, M.; Ratti, F.; Rustgi, A.K.; Stanger, B.Z.; Radtke, F.; Adsay, V.; et al. Notch1 Functions as a Tumor Suppressor in a Model of K-ras–Induced Pancreatic Ductal Adenocarcinoma. Cancer Res. 2010, 70, 4280–4286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J.; et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef] [Green Version]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [Green Version]
- Plentz, R.; Park, J.; Rhim, A.D.; Abravanel, D.; Hezel, A.F.; Sharma, S.V.; Gurumurthy, S.; Deshpande, V.; Kenific, C.; Settleman, J.; et al. Inhibition of γ-Secretase Activity Inhibits Tumor Progression in a Mouse Model of Pancreatic Ductal Adenocarcinoma. Gastroenterology 2009, 136, 1741–1749.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, Y.; Maitra, A.; Ghosh, B.; Zechner, U.; Argani, P.; A Iacobuzio-Donahue, C.; Sriuranpong, V.; Iso, T.; Meszoely, I.M.; Wolfe, M.S.; et al. Notch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 2003, 3, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Wen, J.; Ning, Y.; Li, Y. Higher notch expression implies poor survival in pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Pancreatology 2018, 18, 954–961. [Google Scholar] [CrossRef]
- Thomas, M.M.; Zhang, Y.; Mathew, E.; Kane, K.T.; Maillard, I.; Di Magliano, M.P. Epithelial Notch signaling is a limiting step for pancreatic carcinogenesis. BMC Cancer 2014, 14, 862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Wang, Y.; Lan, H.; Zhang, Y. Expression of Notch receptors and their ligands in pancreatic ductal adenocarcinoma. Exp. Ther. Med. 2018, 16, 53–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Xia, J.; Miele, L.; Sarkar, F.H.; Wang, Z. Notch Signaling Pathway in Pancreatic Cancer Progression. Pancreat. Disord. Ther. 2013, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, A.; Ito, M.; Sakaidani, Y.; Kondo, T.; Murakami, K.; Furukawa, K.; Nadano, D.; Matsuda, T.; Okajima, T. O-LinkedN-Acetylglucosamine Is Present on the Extracellular Domain of Notch Receptors. J. Biol. Chem. 2008, 283, 35486–35495. [Google Scholar] [CrossRef] [Green Version]
- Tashima, Y.; Stanley, P. Antibodies That Detect O-Linked β-d-N-Acetylglucosamine on the Extracellular Domain of Cell Surface Glycoproteins. J. Biol. Chem. 2014, 289, 11132–11142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, I.; Silberstein, E.; Perez, Y.; Landau, D.; Elbedour, K.; Langer, Y.; Kadir, R.; Volodarsky, M.; Sivan, S.; Narkis, G.; et al. Autosomal recessive Adams–Oliver syndrome caused by homozygous mutation in EOGT, encoding an EGF domain-specific O-GlcNAc transferase. Eur. J. Hum. Genet. 2013, 22, 374–378. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, M.; Okajima, T. Structure and function of extracellular O-GlcNAc. Curr. Opin. Struct. Biol. 2019, 56, 72–77. [Google Scholar] [CrossRef]
- Hassed, S.; Li, S.; Mulvihill, J.; Aston, C.; Palmer, S. Adams-Oliver syndrome review of the literature: Refining the diagnostic phenotype. Am. J. Med Genet. Part A 2017, 173, 790–800. [Google Scholar] [CrossRef]
- Sawaguchi, S.; Varshney, S.; Ogawa, M.; Sakaidani, Y.; Yagi, H.; Takeshita, K.; Murohara, T.; Kato, K.; Sundaram, S.; Stanley, P.; et al. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. eLife 2017, 6, 7280. [Google Scholar] [CrossRef] [PubMed]
- Dunwoodie, S.L. Mutation of the fucose-specific β1,3 N-acetylglucosaminyltransferase LFNG results in abnormal formation of the spine. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2009, 1792, 100–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakuda, S.; Lopilato, R.K.; Ito, A.; Haltiwanger, R.S. Canonical Notch ligands and Fringes have distinct effects on NOTCH1 and NOTCH2. J. Biol. Chem. 2020, 295, 14710–14722. [Google Scholar] [CrossRef]
- Zhang, S.; Chung, W.-C.; Xu, K. Lunatic Fringe is a potent tumor suppressor in Kras-initiated pancreatic cancer. Oncogene 2016, 35, 2485–2495. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Senoo, Y.; Ikeda, K.; Takeuchi, H.; Okajima, T. Structural Divergence in O-GlcNAc Glycans Displayed on Epidermal Growth Factor-like Repeats of Mammalian Notch1. Molecules 2018, 23, 1745. [Google Scholar] [CrossRef] [Green Version]
- Varshney, S.; Stanley, P. Multiple roles for O-glycans in Notch signalling. FEBS Lett. 2018, 592, 3819–3834. [Google Scholar] [CrossRef] [Green Version]
- Kakuda, S.; Haltiwanger, R.S. Deciphering the Fringe-Mediated Notch Code: Identification of Activating and Inhibiting Sites Allowing Discrimination between Ligands. Dev. Cell 2017, 40, 193–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haltom, A.R.; Jafar-Nejad, H. The multiple roles of epidermal growth factor repeatO-glycans in animal development. Glycobiology 2015, 25, 1027–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juiz, N.; Elkaoutari, A.; Bigonnet, M.; Gayet, O.; Roques, J.; Nicolle, R.; Iovanna, J.L.; Dusetti, N. Basal-like and classical cells coexist in pancreatic cancer revealed by single-cell analysis on biopsy-derived pancreatic cancer organoids from the classical subtype. FASEB J. 2020, 34, 12214–12228. [Google Scholar] [CrossRef] [PubMed]
- Nandagopal, N.; Santat, L.A.; Lebon, L.; Sprinzak, D.; Bronner, M.E.; Elowitz, M.B. Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell 2018, 172, 869–880.e19. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Fernandez, A.; Rodriguez-Baena, D.; Gomez-Vela, F.; Divina, F.; Garcia-Torres, M. A multi-GPU biclustering algorithm for binary datasets. J. Parallel Distrib. Comput. 2021, 147, 209–219. [Google Scholar] [CrossRef]
- Orzechowski, P.; Sipper, M.; Huang, X.; Moore, J.H. EBIC: An evolutionary-based parallel biclustering algorithm for pattern discovery. Bioinformatics 2018, 34, 3719–3726. [Google Scholar] [CrossRef]
- Xie, J.; Ma, A.; Zhang, Y.; Liu, B.; Cao, S.; Wang, C.; Xu, J.; Zhang, C.; Ma, Q. QUBIC2: A novel and robust biclustering algorithm for analyses and interpretation of large-scale RNA-Seq data. Bioinformatics 2020, 36, 1143–1149. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Cui, Y. A GPU-accelerated algorithm for biclustering analysis and detection of condition-dependent coexpression network modules. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Cheng, Z.; Li, Y.; Zhou, Z.-G.; Yang, L.; Zhang, M.-M. Suppressive effects of gamma-secretase inhibitor DAPT on the proliferation of pancreatic cancer cells. Sichuan da xue xue bao. Yi xue ban J. Sichuan Univ. Med Sci. Ed. 2013, 44, 699–702. [Google Scholar]
- Harbuzariu, A.; Rampoldi, A.; Daley-Brown, D.S.; Candelaria, P.; Harmon, T.L.; Lipsey, C.C.; Beech, D.J.; Quarshie, A.; Ilies, G.O.; Gonzalez, R.R. Leptin-Notch signaling axis is involved in pancreatic cancer progression. Oncotarget 2016, 8, 7740–7752. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Su, H.; Ling, C.; Guoliang, Q.; Cheng, L.; Qin, R.; Qing, G.; Liu, H. The NOTCH Ligand JAGGED2 Promotes Pancreatic Cancer Metastasis Independent of NOTCH Signaling Activation. Mol. Cancer Ther. 2015, 14, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Wang, H.; Bao, Y.; Xie, K. Notch signaling molecule is involved in the invasion of MiaPaCa2 cells induced by CoCl2 via regulating epithelial‑mesenchymal transition. Mol. Med. Rep. 2018, 17, 4965–4972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieber, M.; Mazzetta, J.; Nelson-Rees, W.; Kaplan, M.; Todaro, G. Establishment of a continuous tumor-cell line (PANC-1) from a human carcinoma of the exocrine pancreas. Int. J. Cancer 1975, 15, 741–747. [Google Scholar] [CrossRef]
- Watanabe, M. Metabolic Profiling Comparison of Human Pancreatic Ductal Epithelial Cells and Three Pancreatic Cancer Cell Lines using NMR Based Metabonomics. J. Mol. Biomarkers Diagn. 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Single, A.; Beetham, H.; Telford, B.J.; Guilford, P.; Chen, A. A Comparison of Real-Time and Endpoint Cell Viability Assays for Improved Synthetic Lethal Drug Validation. J. Biomol. Screen. 2015, 20, 1286–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2014, 343, 80–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narimatsu, Y.; Joshi, H.J.; Yang, Z.; Gomes, C.; Chen, Y.-H.; Lorenzetti, F.C.; Furukawa, S.; Schjoldager, K.T.; Hansen, L.; Clausen, H.; et al. A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology 2018, 28, 295–305. [Google Scholar] [CrossRef]
- Giuliano, C.J.; Lin, A.; Girish, V.; Sheltzer, J.M. Generating Single Cell–Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 2019, 128, e100. [Google Scholar] [CrossRef] [Green Version]
- Abrahimi, P.; Chang, W.G.; Kluger, M.S.; Qyang, Y.; Tellides, G.; Saltzman, W.M.; Pober, J.S. Efficient Gene Disruption in Cultured Primary Human Endothelial Cells by CRISPR/Cas9. Circ. Res. 2015, 117, 121–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alam, S.M.D.; Tsukamoto, Y.; Ogawa, M.; Senoo, Y.; Ikeda, K.; Tashima, Y.; Takeuchi, H.; Okajima, T. N-Glycans on EGF domain-specific O-GlcNAc transferase (EOGT) facilitate EOGT maturation and peripheral endoplasmic reticulum localization. J. Biol. Chem. 2020, 295, 8560–8574. [Google Scholar] [CrossRef] [PubMed]
- Libério, M.S.; Sadowski, M.C.; Soekmadji, C.; Davis, R.A.; Nelson, C.C. Differential Effects of Tissue Culture Coating Substrates on Prostate Cancer Cell Adherence, Morphology and Behavior. PLOS ONE 2014, 9, e112122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, 556–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barua, R.; Mizuno, K.; Tashima, Y.; Ogawa, M.; Takeuchi, H.; Taguchi, A.; Okajima, T. Bioinformatics and Functional Analyses Implicate Potential Roles for EOGT and L-fringe in Pancreatic Cancers. Molecules 2021, 26, 882. https://doi.org/10.3390/molecules26040882
Barua R, Mizuno K, Tashima Y, Ogawa M, Takeuchi H, Taguchi A, Okajima T. Bioinformatics and Functional Analyses Implicate Potential Roles for EOGT and L-fringe in Pancreatic Cancers. Molecules. 2021; 26(4):882. https://doi.org/10.3390/molecules26040882
Chicago/Turabian StyleBarua, Rashu, Kazuyuki Mizuno, Yuko Tashima, Mitsutaka Ogawa, Hideyuki Takeuchi, Ayumu Taguchi, and Tetsuya Okajima. 2021. "Bioinformatics and Functional Analyses Implicate Potential Roles for EOGT and L-fringe in Pancreatic Cancers" Molecules 26, no. 4: 882. https://doi.org/10.3390/molecules26040882






