Development of a Liquid Chromatography/Mass Spectrometry-Based Inhibition Assay for the Screening of Steroid 5-α Reductase in Human and Fish Cell Lines
Abstract
:1. Introduction
2. Results
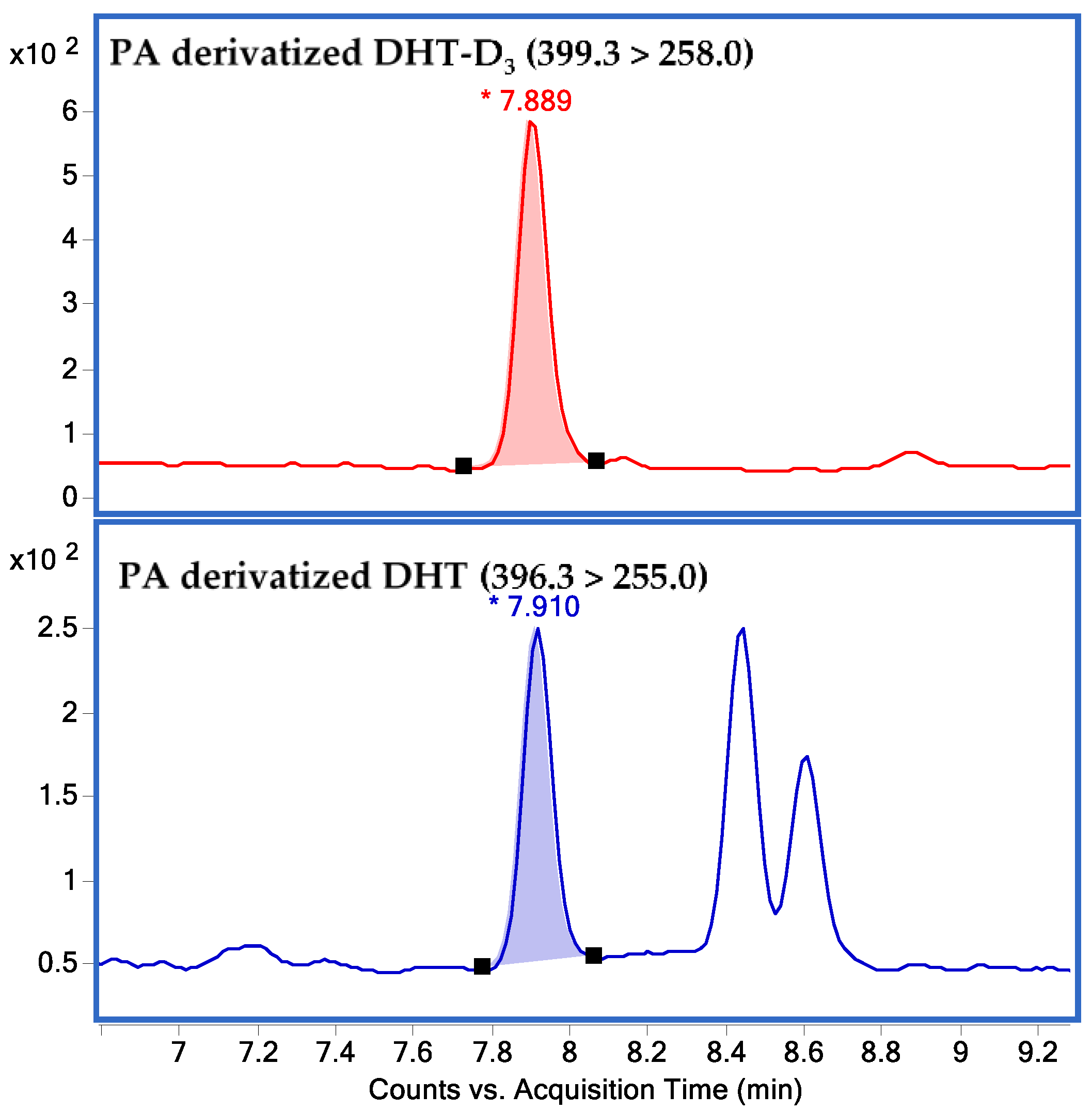
2.1. Method Validation
2.1.1. Linearity of the Calibration Curve and LLOQ
2.1.2. Accuracy and Precision
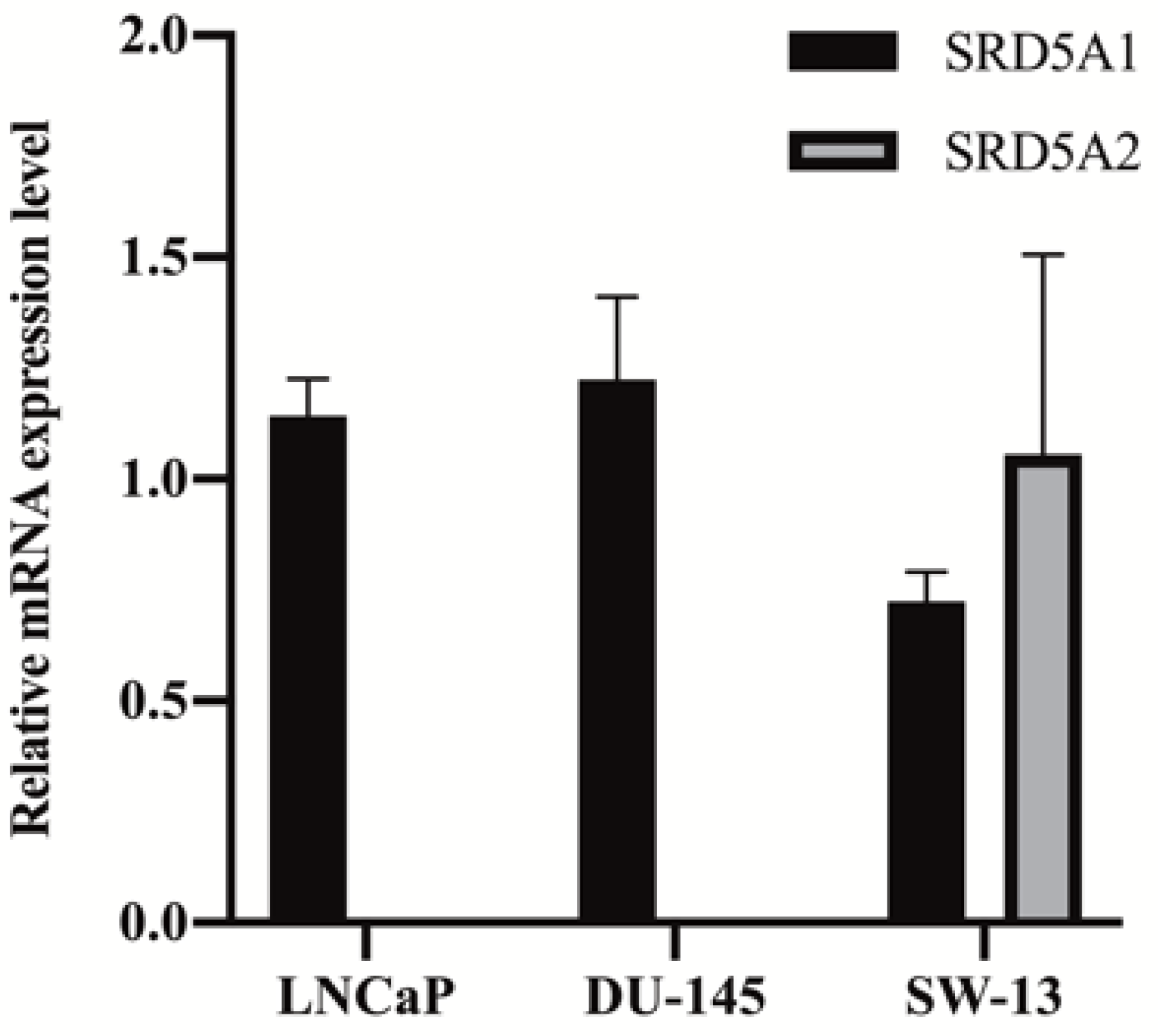
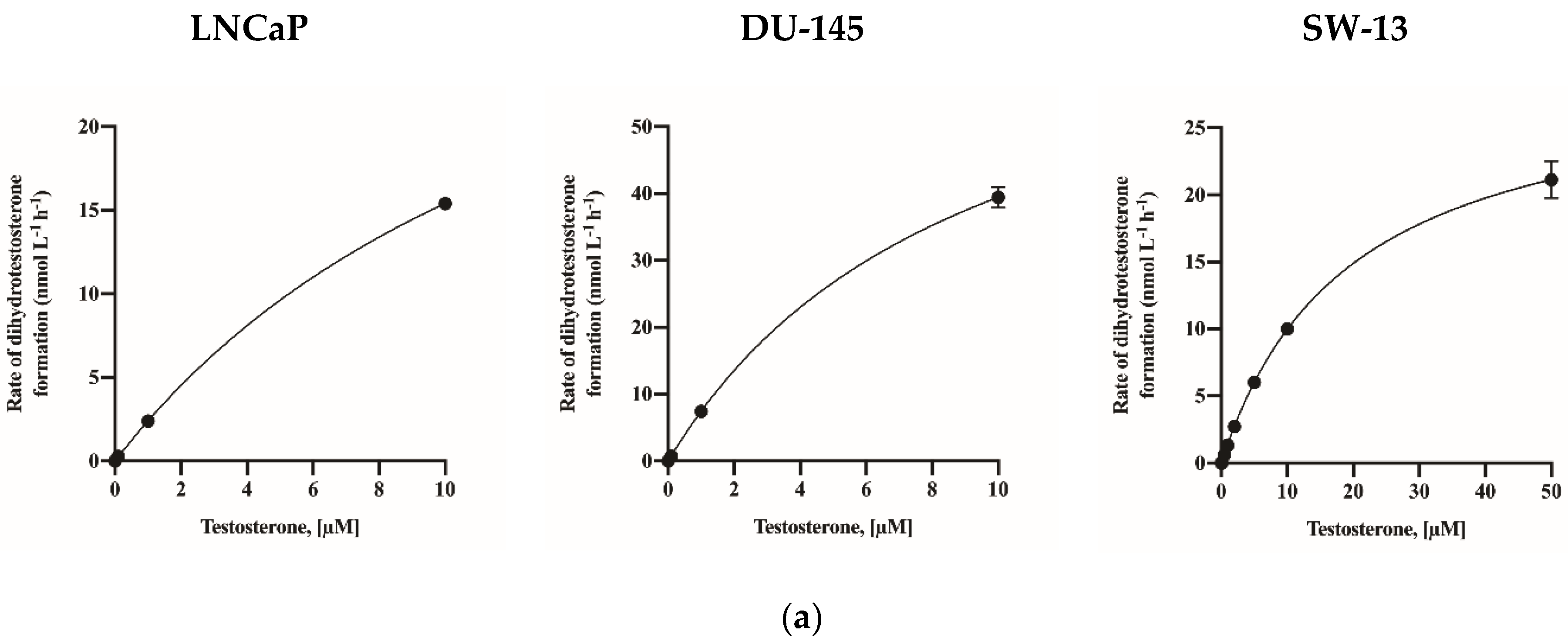
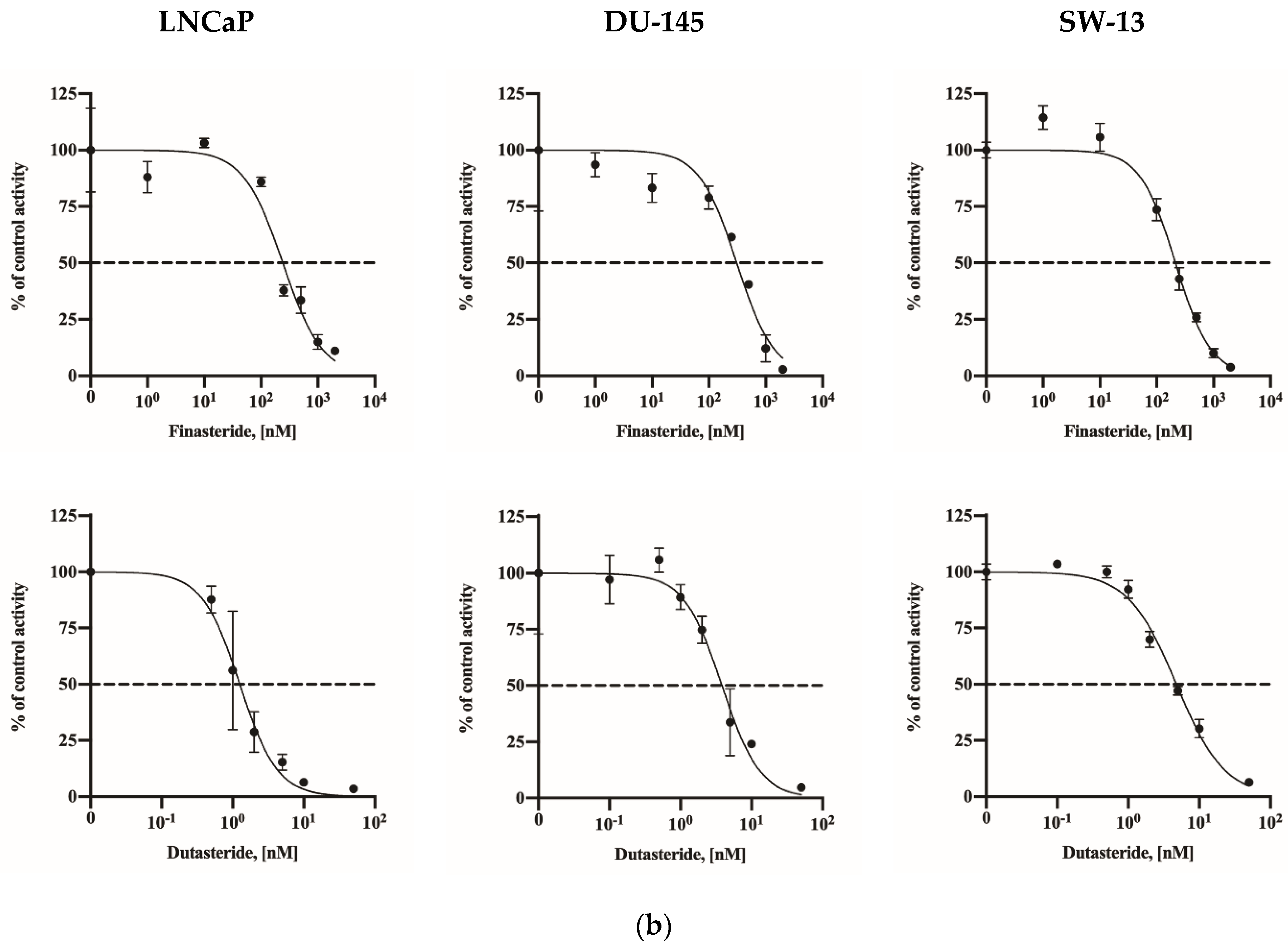
2.2. Assay Application in Human Cell Lines
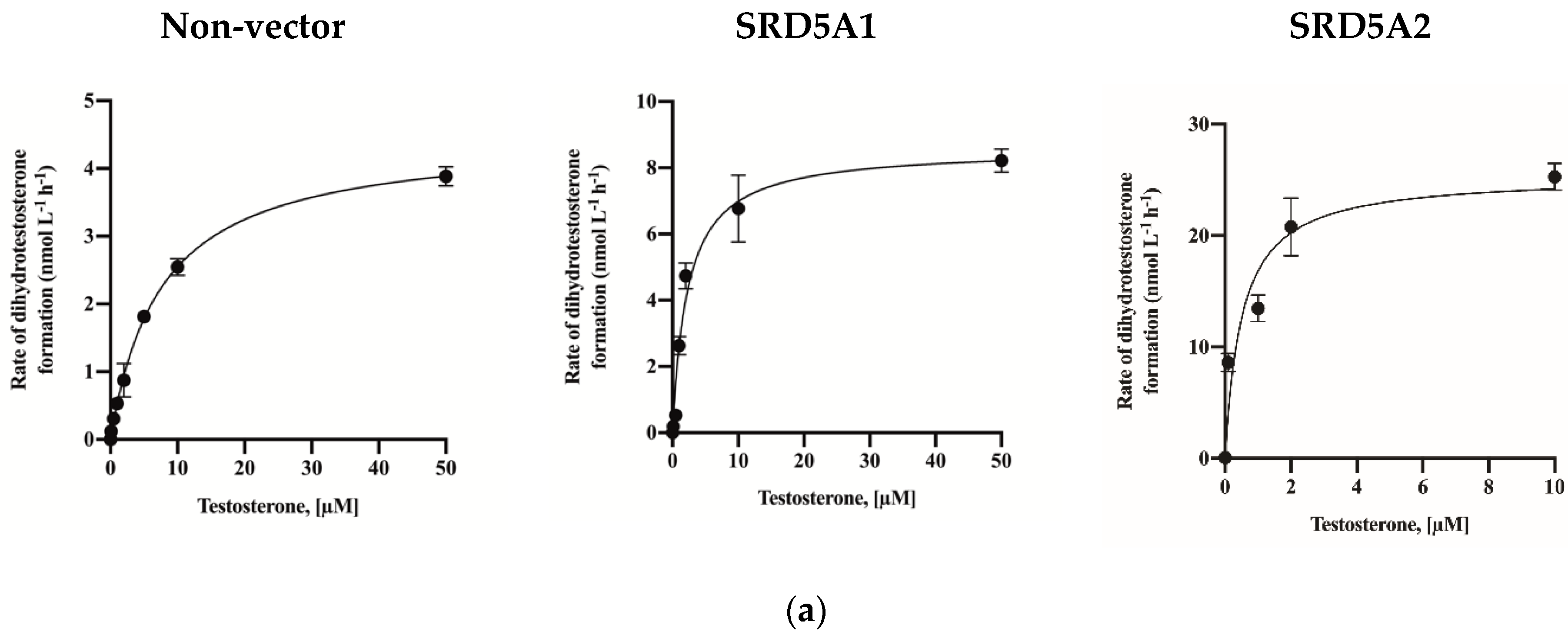
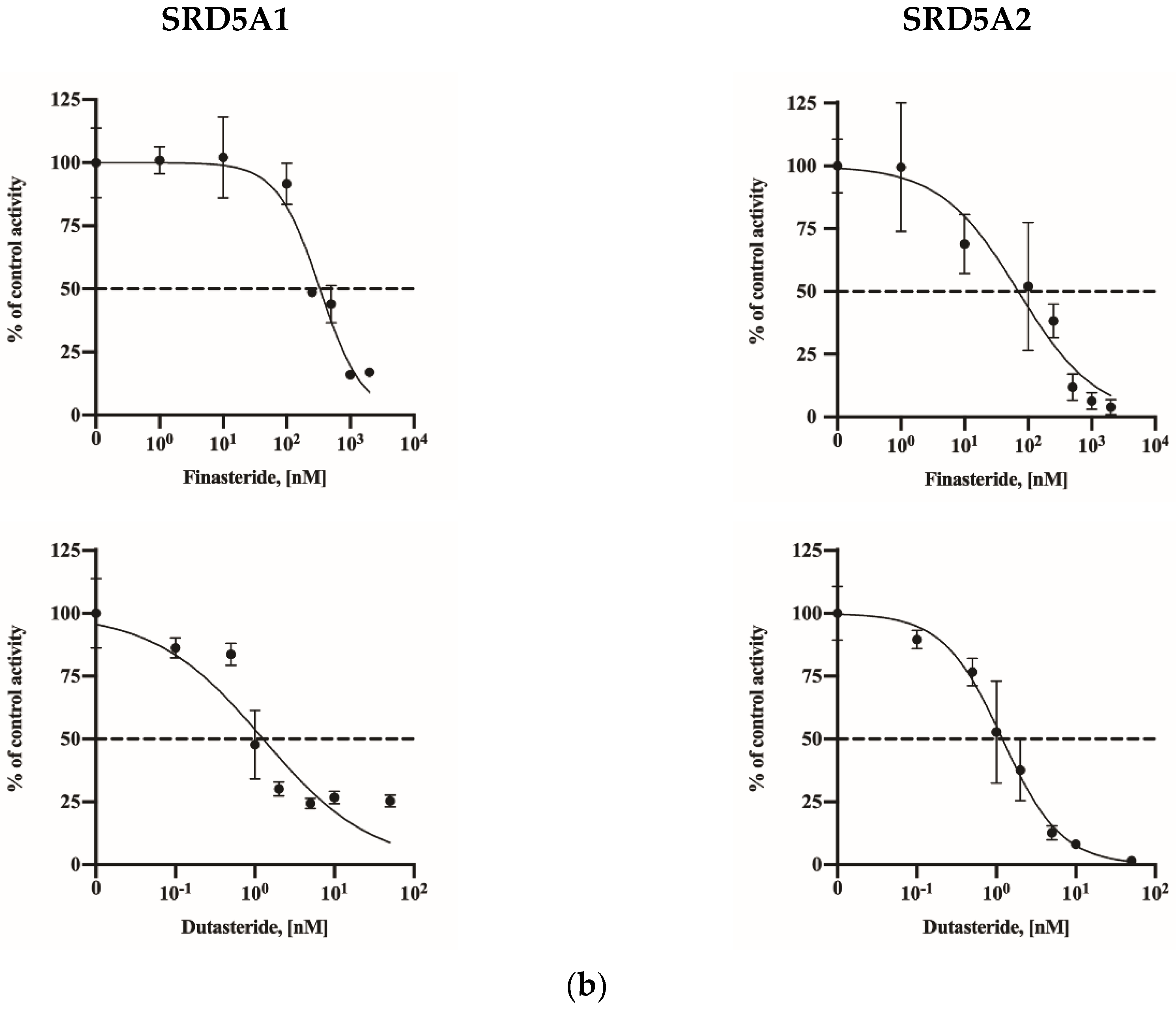
2.3. Assay Application in SRD5A2-Overexpressing HEK-293 Cells
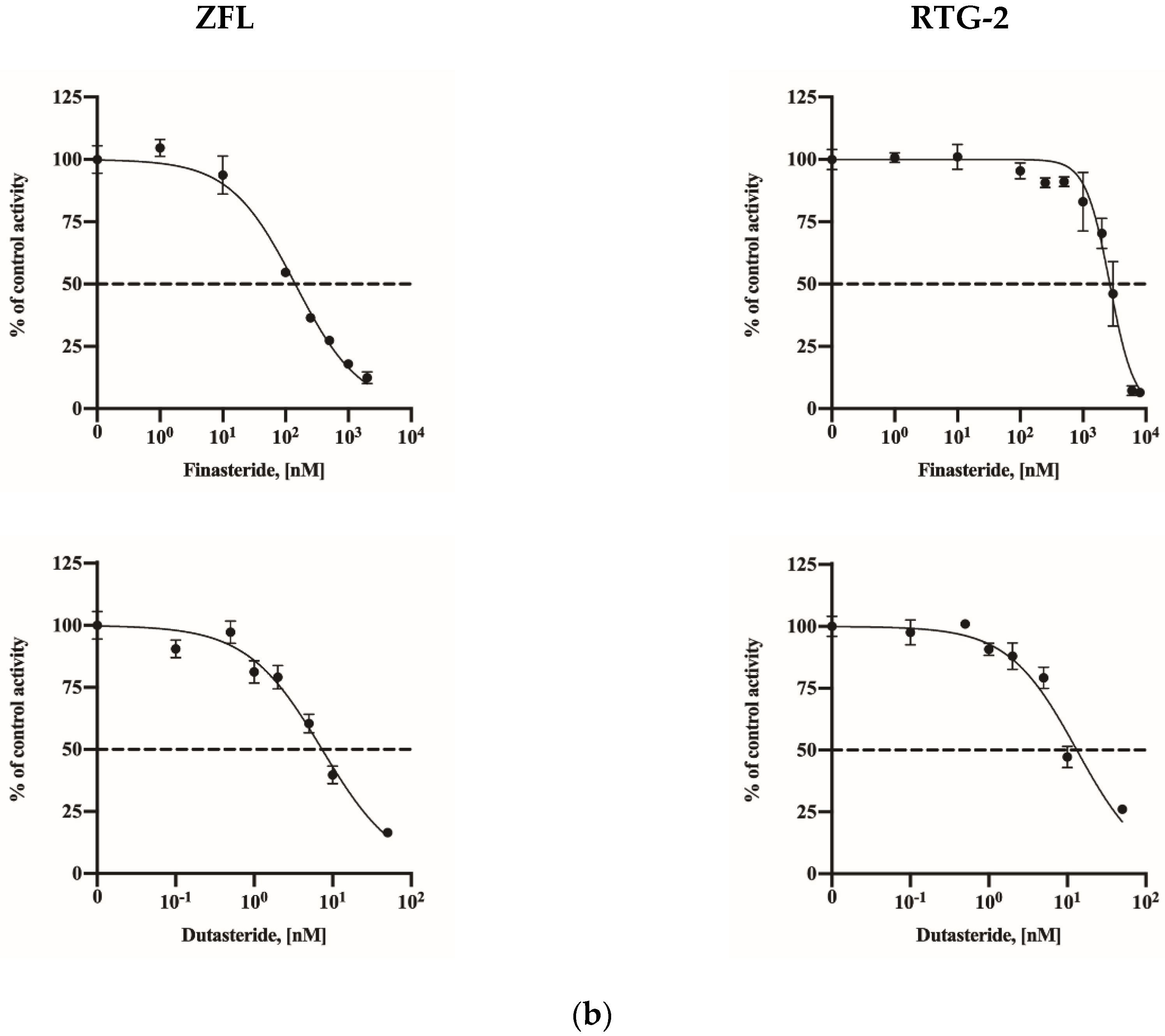
2.4. Assay Application in a Fish Cell Line
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Transient Overexpression
4.4. Cell Culture Assay Application
4.5. qRT PCR
4.6. Sample Preparation
4.7. Instrumental Conditions
4.8. Calibration Curve and LLOQ
4.9. Accuracy and Precision
4.10. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bruchovsky, N.; Wilson, J.D. The conversion of testosterone to 5α-androstan-17-beta-ol-3-one by rat prostate in vivo and in vitro. J. Biol. Chem. 1968, 243, 2012–2021. [Google Scholar] [CrossRef]
- Russell, D.W.; Wilson, J.D. Steroid 5α-reductase: Two genes/two enzymes. Annu. Rev. Biochem. 1994, 63, 25–61. [Google Scholar] [CrossRef]
- Langlois, V.S.; Zhang, D.; Cooke, G.M.; Trudeau, V.L. Evolution of steroid-5α-reductases and comparison of their function with 5beta-reductase. Gen. Comp. Endocrinol. 2010, 166, 489–497. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.D.; Wilson, J.D.; George, F.W.; Geller, J.; Pappas, F.; Stoner, E. Finasteride, an inhibitor of 5α-reductase, suppresses prostatic dihydrotestosterone in men with benign prostatic hyperplasia. J. Clin. Endocrinol. Metab. 1992, 74, 505–508. [Google Scholar] [CrossRef]
- Diani, A.R.; Mulholland, M.J.; Shull, K.L.; Kubicek, M.F.; Johnson, G.A.; Schostarez, H.J.; Brunden, M.N.; Buhl, A.E. Hair growth effects of oral administration of finasteride, a steroid 5α-reductase inhibitor, alone and in combination with topical minoxidil in the balding stumptail macaque. J. Clin. Endocrinol. Metab. 1992, 74, 345–350. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.D.; Roehrborn, C.G.; Bautista, O.M.; Andriole, G.L.; Dixon, C.M.; Kusek, J.W.; Lepor, H.; McVary, K.T.; Nyberg, L.M.; Clarke, H.S.; et al. The long-term effect of doxazosin, finasteride, and combination therapy on the clinical progression of benign prostatic hyperplasia. N. Engl. J. Med. 2003, 349, 2387–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, C.S.; Sung, B.; Baik, S.; Kim, Y.J.; Lee, Y. Inhibition of 5α-Reductase Leading to Impaired Fecundity in Female Fish. Available online: https://aopwiki.org/aops/289 (accessed on 11 December 2020).
- García-García, M.; Sánchez-Hernández, M.; García-Hernández, M.P.; García-Ayala, A.; Chaves-Pozo, E. Role of 5α-dihydrotestosterone in testicular development of gilthead seabream following finasteride administration. J. Steroid Biochem. Mol. Biol. 2017, 174, 48–55. [Google Scholar] [CrossRef]
- Margiotta-Casaluci, L.; Courant, F.; Antignac, J.P.; Le Bizec, B.; Sumpter, J.P. Identification and quantification of 5α-dihydrotestosterone in the teleost fathead minnow (Pimephales promelas) by gas chromatography-tandem mass spectrometry. Gen. Comp. Endocrinol. 2013, 191, 202–209. [Google Scholar] [CrossRef]
- Andersson, S.; Bishop, R.W.; Russell, D.W. Expression cloning and regulation of steroid 5α-reductase, an enzyme essential for male sexual differentiation. J. Biol. Chem. 1989, 264, 16249–16255. [Google Scholar] [CrossRef]
- Raynaud, J.P.; Cousse, H.; Martin, P.M. Inhibition of type 1 and type 2 5α-reductase activity by free fatty acids, active ingredients of permixon. J. Steroid Biochem. Mol. Biol. 2002, 82, 233–239. [Google Scholar] [CrossRef]
- Iwai, A.; Yoshimura, T.; Wada, K.; Watabe, S.; Sakamoto, Y.; Ito, E.; Miura, T. Spectrophotometric method for the assay of steroid 5α-reductase activity of rat liver and prostate microsomes. Anal. Sci. 2013, 29, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, H.; Sato, N.; Yamazaki, M.; Naruto, S.; Kubo, M. Testosterone 5α-reductase inhibitory active constituents from Anemarrhenae Rhizoma. Biol. Pharm. Bull. 2001, 24, 586–587. [Google Scholar] [CrossRef] [Green Version]
- Mitamura, K.; Narukawa, H.; Mizuguchi, T.; Shimada, K. Degradation of estrogen conjugates using titanium dioxide as a photocatalyst. Anal. Sci. 2004, 20, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Abe, M.; Ito, Y.; Oyunzul, L.; Oki-Fujino, T.; Yamada, S. Pharmacologically relevant receptor binding characteristics and 5α-reductase inhibitory activity of free fatty acids contained in saw palmetto extract. Biol. Pharm. Bull. 2009, 32, 646–650. [Google Scholar] [CrossRef] [Green Version]
- Gorityala, S.; Yang, S.; Montano, M.M.; Xu, Y. Simultaneous determination of dihydrotestosterone and its metabolites in mouse sera by LC-MS/MS with chemical derivatization. J. Chromatogr. B 2018, 1090, 22–35. [Google Scholar] [CrossRef]
- Yamashita, K.; Takahashi, M.; Tsukamoto, S.; Numazawa, M.; Okuyama, M.; Honma, S. Use of novel picolinoyl derivatization for simultaneous quantification of six corticosteroids by liquid chromatography-electrospray ionization tandem mass spectrometry. J. Chromatogr. A 2007, 1173, 120–128. [Google Scholar] [CrossRef]
- Srivilai, J.; Rabgay, K.; Khorana, N.; Waranuch, N.; Nuengchamnong, N.; Ingkaninan, K. A new label-free screen for steroid 5α-reductase inhibitors using LC-MS. Steroids 2016, 116, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Lu, Y.; Cong, Y.; Liu, Y.; Li, Y.; Wang, H.; Zhang, Q.; Huang, W.; Liu, J.; Dong, Y.; et al. Simultaneous quantitation of four androgens and 17-hydroxyprogesterone in polycystic ovarian syndrome patients by LC-MS/MS. J. Clin. Lab. Anal. 2020, 34, 23539. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.J.Y.; Pan, J.; Sun, L.; Zhang, J.; Wu, C.; Kang, L. Bioactives in Chinese proprietary medicine modulates 5α-reductase activity and gene expression associated with androgenetic alopecia. Front. Pharmacol. 2017, 8, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Zhang, M. Rapid quantitation of testosterone hydroxyl metabolites by ultra-performance liquid chromatography and mass spectrometry. J. Chromatogr. B 2007, 855, 290–294. [Google Scholar] [CrossRef]
- Jain, R.; Monthakantirat, O.; Tengamnuay, P.; De-Eknamkul, W. Identification of a new plant extract for androgenic alopecia treatment using a non-radioactive human hair dermal papilla cell-based assay. BMC Complementary Altern. Med. 2015, 16, 18. [Google Scholar] [CrossRef] [Green Version]
- Nouri, M.-Z.; Kroll, K.J.; Webb, M.; Denslow, N.D. Quantification of steroid hormones in low volume plasma and tissue homogenates of fish using LC-MS/MS. Gen. Comp. Endocrinol. 2020, 296, 113543. [Google Scholar] [CrossRef] [PubMed]
- Licea-Perez, H.; Wang, S.; Szapacs, M.E.; Yang, E. Development of a highly sensitive and selective UPLC/MS/MS method for the simultaneous determination of testosterone and 5α-dihydrotestosterone in human serum to support testosterone re-placement therapy for hypogonadism. Steroids 2008, 73, 601–610. [Google Scholar] [CrossRef]
- Yamashita, K.; Miyashiro, Y.; Maekubo, H.; Okuyama, M.; Honma, S.; Takahashi, M.; Numazawa, M. Development of highly sensitive quantification method for testosterone and dihydrotestosterone in human serum and prostate tissue by liquid chro-matography-electrospray ionization tandem mass spectrometry. Steroids 2009, 74, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Koseki, J.; Matsumoto, T.; Matsubara, Y.; Tsuchiya, K.; Mizuhara, Y.; Sekiguchi, K.; Nishimura, H.; Watanabe, J.; Kaneko, A.; Hattori, T.; et al. Inhibition of Rat 5α-Reductase activity and testosterone-induced sebum synthesis in hamster sebocytes by an extract of Quercus acutissima Cortex. Evid. Based Complementary Altern. Med. 2015, 2015, 853846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorgan, J.F.; Fears, T.R.; McMahon, R.P.; Friedman, L.A.; Patterson, B.H.; Greenhut, S.F. Measurement of steroid sex hormones in serum: A comparison of radioimmunoassay and mass spectrometry. Steroids 2002, 67, 151–158. [Google Scholar] [CrossRef]
- Bauman, T.M.; Sehgal, P.D.; Johnson, K.A.; Pier, T.; Bruskewitz, R.C.; Ricke, W.A.; Huang, W. Finasteride treatment alters tissue specific androgen receptor expression in prostate tissues. Prostate 2014, 74, 923–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.G.; Liu, X.M.; Kreis, W.; Budman, D.R. Down-regulation of prostate-specific antigen expression by finasteride through inhibition of complex formation between androgen receptor and steroid receptor-binding consensus in the promoter of the PSA gene in LNCaP cells. Cancer Res. 1997, 57, 714–719. [Google Scholar]
- Audet-Walsh, É.; Yee, T.; Tam, I.S.; Giguère, V. Inverse regulation of DHT synthesis enzymes 5α-reductase Types 1 and 2 by the androgen receptor in prostate cancer. Endocrinology 2017, 158, 1015–1021. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Wan, Y.; Hu, J. Determination and source apportionment of five classes of steroid hormones in urban rivers. Environ. Sci. Technol. 2009, 43, 7691–7698. [Google Scholar] [CrossRef]
- Schmid, S.; Willi, R.A.; Fent, K. Effects of environmental steroid mixtures are regulated by individual steroid receptor signaling. Aquat. Toxicol. 2020, 226, 105562. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.-Y.; Chuang, B.-F.; Wang, Q.; Sartor, O.; Balk, S.P.; Brown, M.; Kantoff, P.W.; Lee, G.-S.M. Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate 2008, 68, 839–848. [Google Scholar] [CrossRef] [Green Version]
- Span, P.N.; Smals, A.G.; Sweep, C.G.; Benraad, T.J. Rat steroid 5α-reductase kinetic characteristics: Extreme pH-dependency of the type II isozyme in prostate and epididymis homogenates. J. Steroid. Biochem. Mol. Biol. 1995, 54, 185–192. [Google Scholar] [CrossRef]
- Normington, K.; Russell, D.W. Tissue distribution and kinetic characteristics of rat steroid 5α-reductase isozymes. Evidence for distinct physiological functions. J. Biol. Chem. 1992, 267, 19548–19554. [Google Scholar] [CrossRef]
- Martyniuk, C.J.; Bissegger, S.; Langlois, V.S. Current perspectives on the androgen 5α-dihydrotestosterone (DHT) and 5 α-reductases in teleost fishes and amphibians. Gen. Comp. Endocrinol. 2013, 194, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Pasmanik, M.; Schlinger, B.A.; Callard, G.V. In Vivo steroid regulation of aromatase and 5α-reductase in goldfish brain and pituitary. Gen. Comp. Endocrinol. 1988, 71, 175–182. [Google Scholar] [CrossRef]
- Latz, M.; Reinboth, R. Androgen metabolism in the skin of the rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 1993, 11, 281–286. [Google Scholar] [CrossRef]
- Bramson, H.N.; Hermann, D.; Batchelor, K.W.; Lee, F.W.; James, M.K.; Frye, S.V. Unique preclinical characteristics of GG745, a potent dual inhibitor of 5AR. J. Pharmacol. Exp. Ther. 1997, 282, 1496–1502. [Google Scholar]
- Tian, G.; Mook, R.; Moss, M.L.; Frye, S.V. Mechanism of time-dependent inhibition of 5α-reductases by. DELTA.1-4-Azasteroids: Toward perfection of rates of time-dependent inhibition by using ligand-binding energies. Biochemistry 1995, 34, 13453–13459. [Google Scholar] [CrossRef]
- Azzolina, B.; Ellsworth, K.; Andersson, S.; Geissler, W.; Bull, H.G.; Harris, G.S. Inhibition of rat alpha-reductases by finasteride: Evidence for isozyme differences in the mechanism of inhibition. J. Steroid. Biochem. Mol. Biol. 1997, 61, 55–64. [Google Scholar] [CrossRef]
- Levy, M.A.; Brandt, M.; Sheedy, K.M.; Holt, D.A.; Heaslip, J.I.; Trill, J.J.; Ryan, P.J.; Morris, R.A.; Garrison, L.M.; Bergsma, D.J. Cloning, expression and functional characterization of type 1 and type 2 steroid 5α-reductases from cynomolgus monkey: Comparisons with human and rat isoenzymes. J. Steroid Biochem. Mol. Biol. 1995, 52, 307–319. [Google Scholar] [CrossRef]
- Häußler, A.; Allegrini, P.; Biollaz, M.; Batzl, C.; Scheidegger, E.; Bhatnagar, A. CGP 53153: A new potent inhibitor of 5α-reductase. J. Steroid Biochem. Mol. Biol. 1996, 57, 187–195. [Google Scholar] [CrossRef]
- Hirosumi, J.; Nakayama, O.; Fagan, T.; Sawada, K.; Chida, N.; Inami, M.; Takahashi, S.; Kojo, H.; Notsu, Y.; Okuhara, M. FK143, a novel nonsteroidal inhibitor of steroid 5α-reductase: (1) In vitro effects on human and animal prostatic enzymes. J. Steroid Biochem. Mol. Biol. 1995, 52, 357–363. [Google Scholar] [CrossRef]
- Di Salle, E.; Giudici, D.; Radice, A.; Zaccheo, T.; Ornati, G.; Nesi, M.; Panzeri, A.; Delos, S.; Martin, P. PNU 157706, a novel dual type I and II5α-reductase inhibitor. J. Steroid Biochem. Mol. Biol. 1998, 64, 179–186. [Google Scholar] [CrossRef]
- Xu, Y.; Dalrymple, S.L.; Becker, R.E.; Denmeade, S.R.; Isaacs, J.T. Pharmacologic basis for the enhanced efficacy of dutasteride against prostatic cancers. Clin. Cancer Res. 2006, 12, 4072–4079. [Google Scholar] [CrossRef] [Green Version]
- Mitamura, K.; Ogasawara, C.; Shiozawa, A.; Terayama, E.; Shimada, K. Determination method for steroid 5α-reductase activity using liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Sci. 2005, 21, 1241–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Low QC | Medium QC | High QC | |
|---|---|---|---|
| CV%—inter day a | 1.3 | 0.7 | 1.6 |
| CV%—intra day b | 0.9 | 2.5 | 1.3 |
| Accuracy%—inter day | 102.3 | 104.0 | 95.0 |
| Accuracy%—intra day | 101.0 | 98.9 | 95.5 |
| Vmax (nmol L−1 h−1) | KM (nM) | ||
|---|---|---|---|
| Human cell lines | LNCaP | 38.67 (34.36–45.16) * | 15.10 (12.35–19.25) |
| DU-145 | 75.55 (66.61–90.45) | 9.15 (7.01–12.81) | |
| SW-13 | 29.35 (27.94–30.90) | 19.42 (17.31–21.88) | |
| Overexpression lines | Non vector | 4.473 (4.29–4.66) | 7.56 (6.74–8.46) |
| SRD5A1 | 8.584 (7.86–9.35) | 2.29 (1.70–3.09) | |
| SRD5A2 | 22.52 (20.01–25.42) | 0.36 (0.18–0.65) | |
| Fish cell lines | ZFL | 116.60 (106.9–126.9) | 0.46 (0.30–0.68) |
| RTG-2 | 13.09 (11.88–14.37) | 1.12 (0.78–1.61) |
| IC50 Value | |||
|---|---|---|---|
| Finasteride (nM; 95% CI *) | Dutasteride (nM; 95% CI) | ||
| Human cell lines | LNCaP | 241.0 (185.8–303.9) | 1.26 (1.02–1.57) |
| DU-145 | 308.5 (217.0–415.5) | 3.83 (3.10–4.78) | |
| SW-13 | 213.5 (180.2–250.7) | 4.75 (4.26–5.32) | |
| Overexpression lines | SRD5A1 | 332.8 (260.9–424.8) | 1.27 (0.76–2.06) |
| SRD5A2 | 69.83 (33.65–133.3) | 1.19 (0.96–1.47) | |
| Fish cell lines | ZFL | 142.4 (121.5–165.7) | 7.33 (6.12–8.77) |
| RTG-2 | 2667 (2394–2952) | 13.19 (10.73–16.54) | |
| Zebrafish | Rainbow Trout | ||
| srd5a1 | srd5a1 | ||
| Human srd5a1 | 51.7 | 50.2 | |
| Zebrafish | Rainbow Trout | ||
| srd5a2a | srd5a2b | srd5a2a | |
| Human srd5a2 | 52.3 | 42.4 | 50.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Cho, H.; Eggers, R.; Kim, S.K.; Ryu, C.S.; Kim, Y.J. Development of a Liquid Chromatography/Mass Spectrometry-Based Inhibition Assay for the Screening of Steroid 5-α Reductase in Human and Fish Cell Lines. Molecules 2021, 26, 893. https://doi.org/10.3390/molecules26040893
Kim D, Cho H, Eggers R, Kim SK, Ryu CS, Kim YJ. Development of a Liquid Chromatography/Mass Spectrometry-Based Inhibition Assay for the Screening of Steroid 5-α Reductase in Human and Fish Cell Lines. Molecules. 2021; 26(4):893. https://doi.org/10.3390/molecules26040893
Chicago/Turabian StyleKim, Dahye, Hyunki Cho, Ruth Eggers, Sang Kyum Kim, Chang Seon Ryu, and Young Jun Kim. 2021. "Development of a Liquid Chromatography/Mass Spectrometry-Based Inhibition Assay for the Screening of Steroid 5-α Reductase in Human and Fish Cell Lines" Molecules 26, no. 4: 893. https://doi.org/10.3390/molecules26040893






