Carrier-Free Immobilization of α-Galactosidase as Nano-Biocatalysts for Synthesizing Prebiotic α-Galacto-Oligosaccharides
Abstract
1. Introduction
2. Results and Discussion
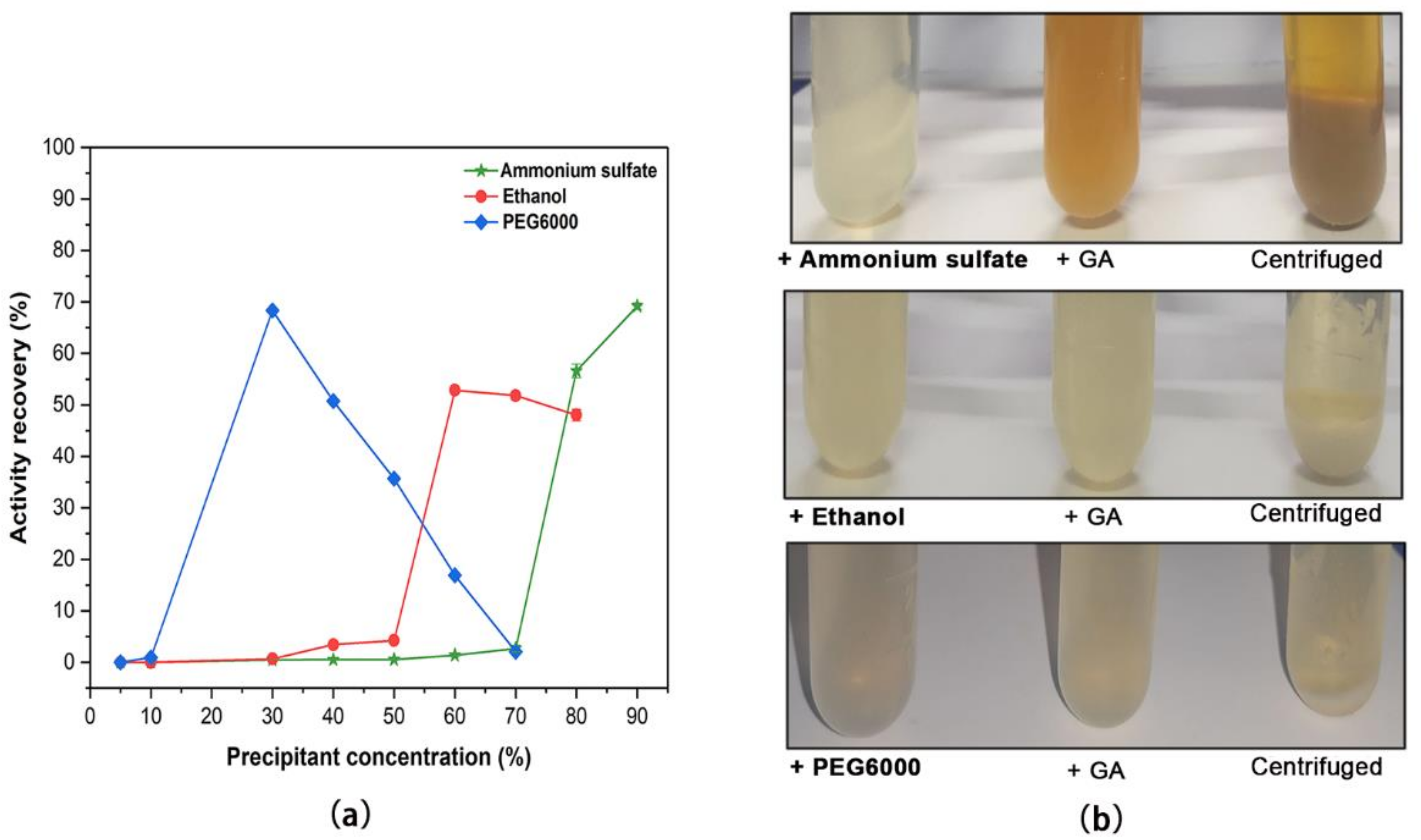
2.1. Screening of Precipitants for Inducing Aga-CLEAs Nano-Biocatalyst
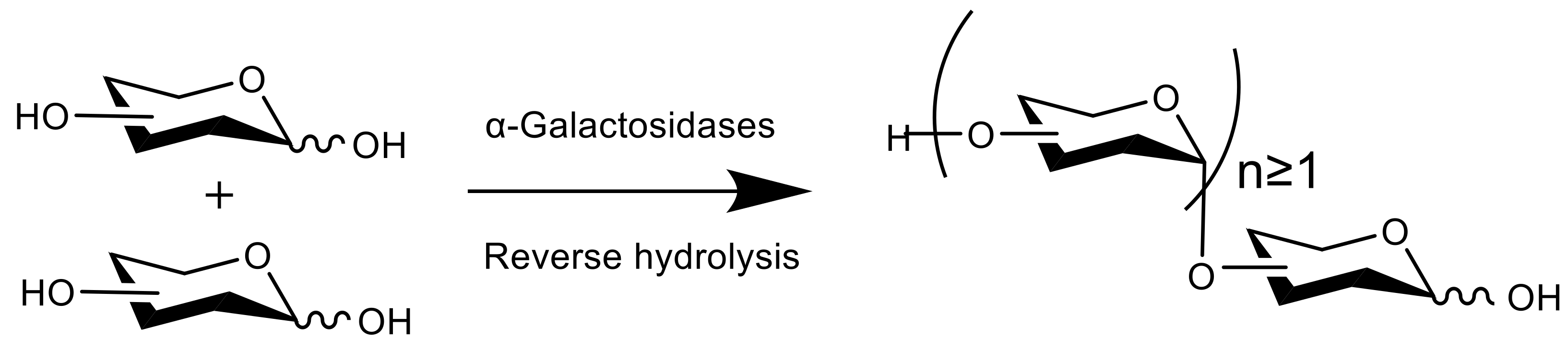
2.2. Optimization of the Cross-Linking Conditions for Aga-CLEAs by RSM
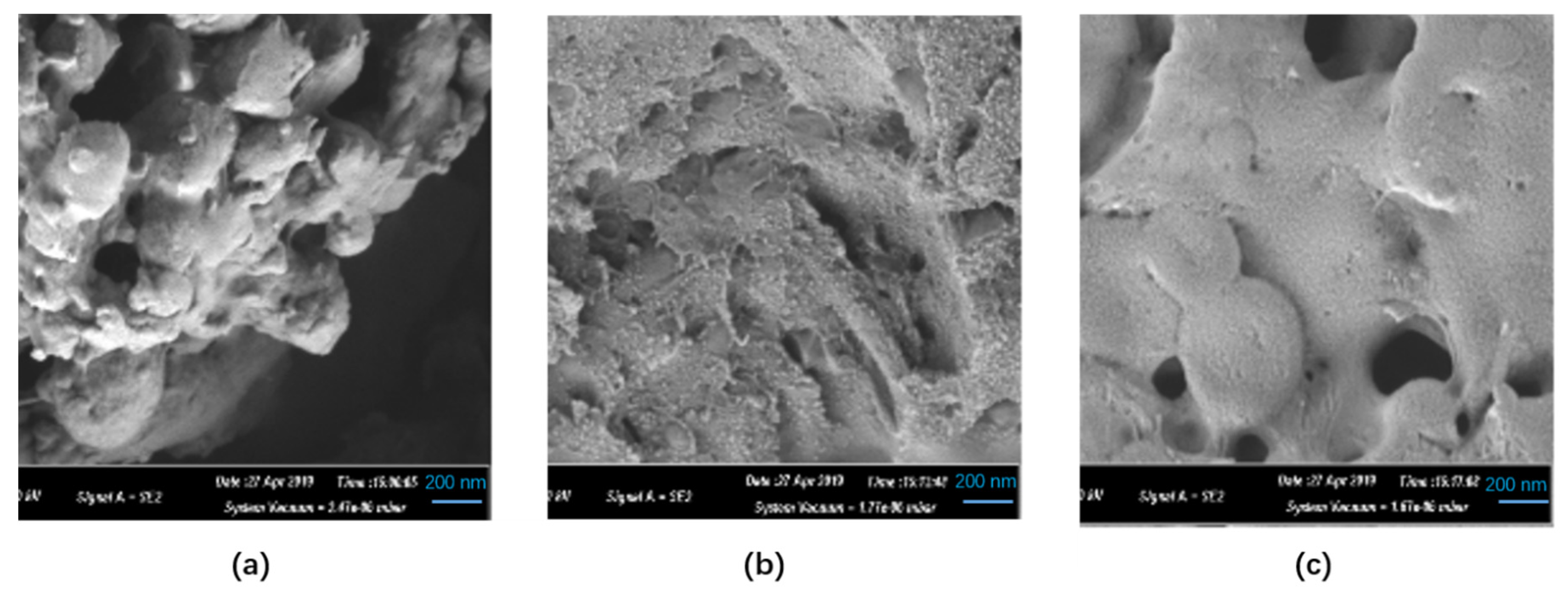
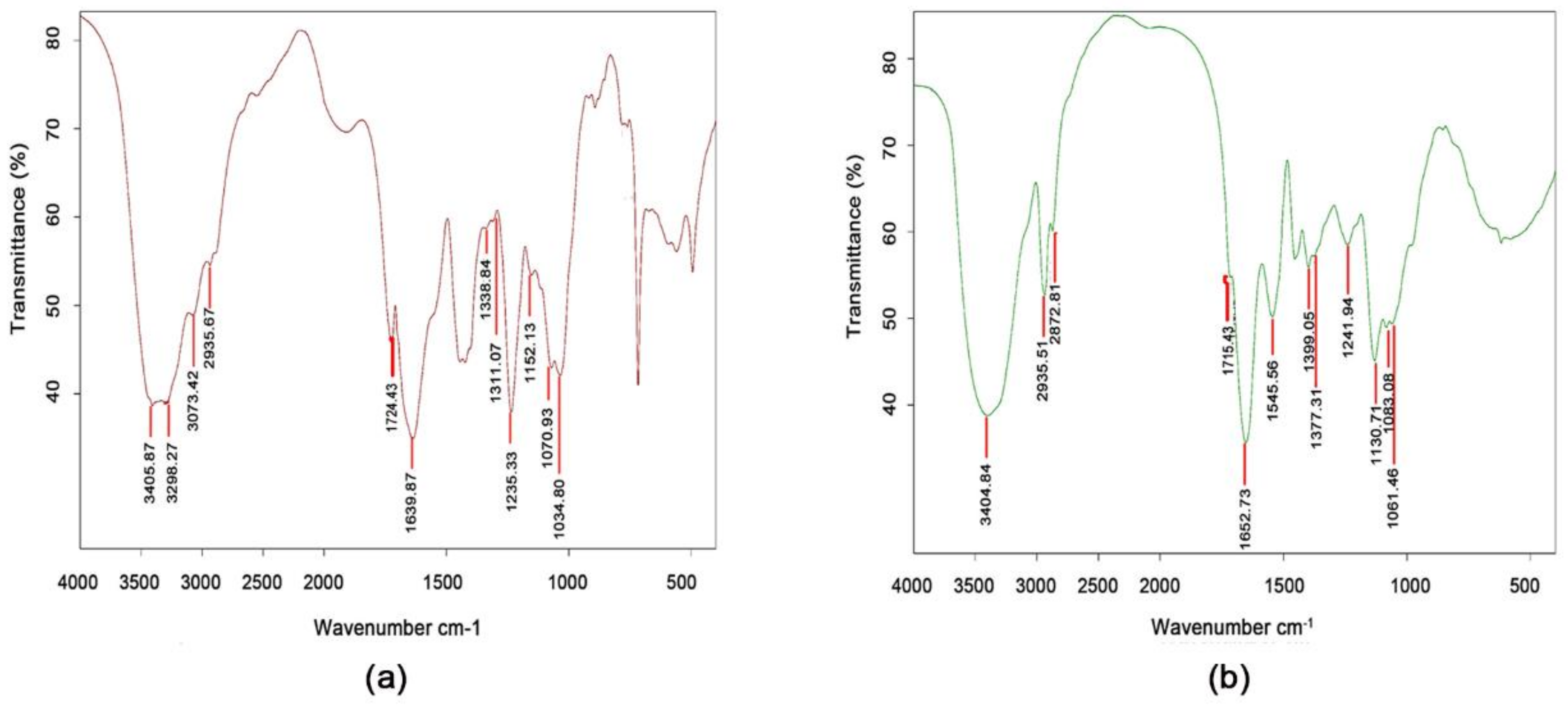
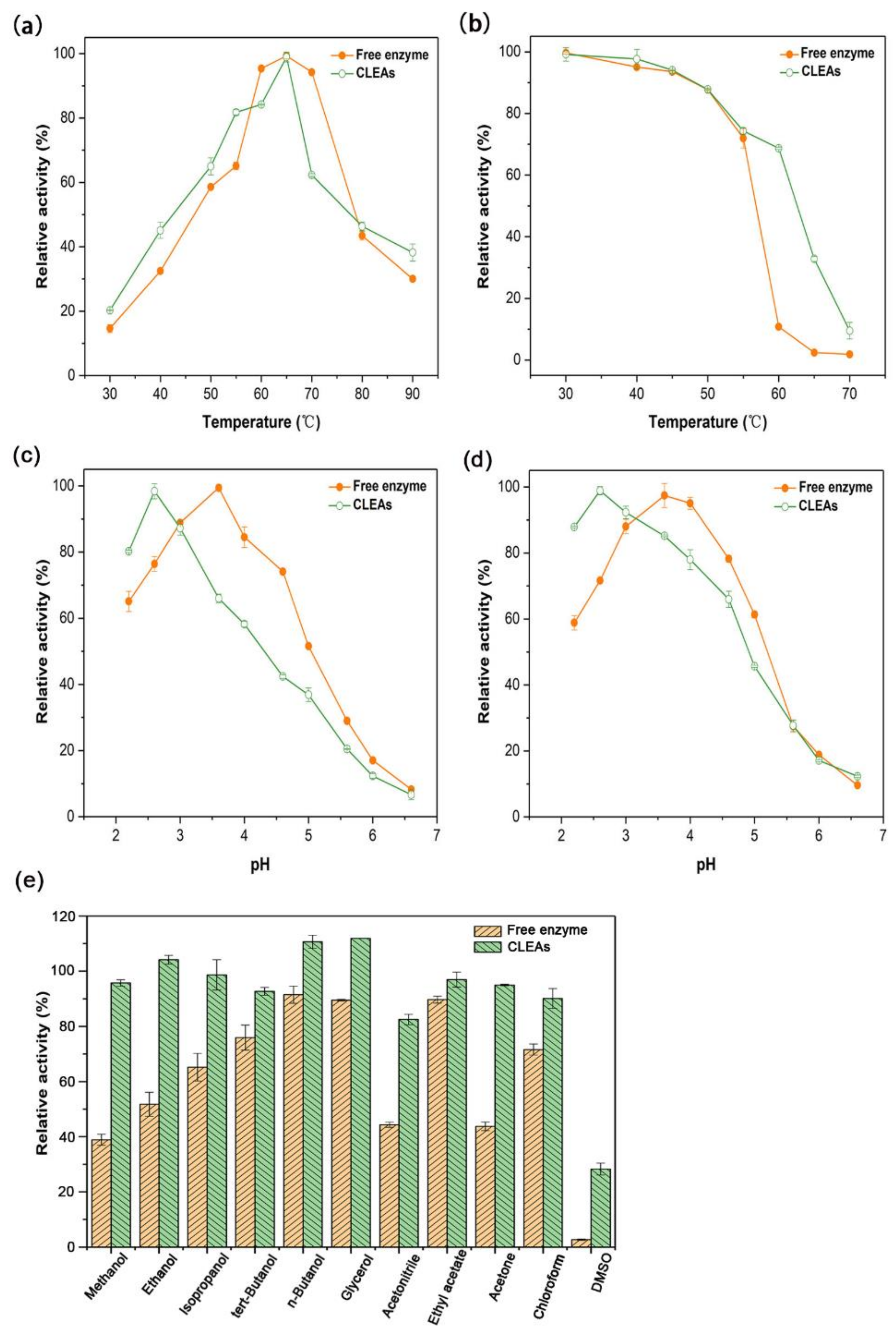
2.3. Biochemical Properties of Aga-CLEAs Nano-Biocatalyst
2.4. Storage Stability of Aga-CLEAs Nano-Biocatalyst
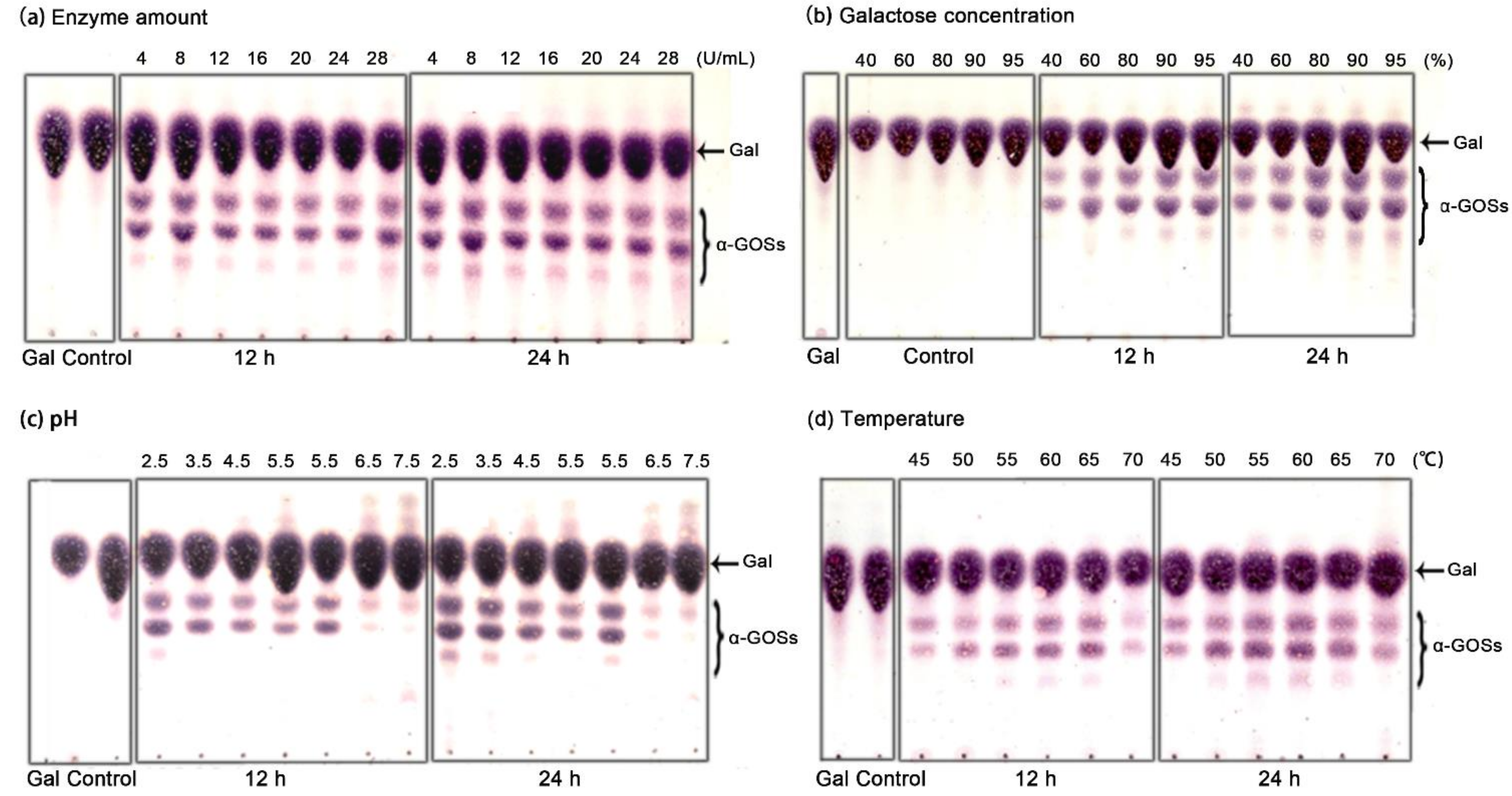
2.5. Batch Synthesis of α-GOSs by Recycling Aga-CLEAs Nano-Biocatalyst
3. Materials and Methods
3.1. Materials
3.2. Production of α-Galactosidase
3.3. Enzyme Activity Assay
3.4. Preparation of α-Galactosidase CLEAs
3.5. Structural Analysis of Aga-CLEAs
3.6. Optimization of Cross-Linking Conditions for Aga-CLEAs by Response Surface Methodology
3.7. Characterization of Aga-CLEAs Nano-Biocatalyst
3.8. Storage Stability of Aga-CLEAs Nano-Biocatalyst
3.9. Batch Synthesis of α-GOSs by Recycling Aga-CLEAs Nano-Biocatalyst
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Sample Availability
Appendix A
| Source | Sum of Square | Df | Mean Square | F Value | p-Value Prob ˃ F | |
|---|---|---|---|---|---|---|
| Model | 4316.99 | 9 | 479.67 | 20.33 | <0.0001 | Significant |
| X1 | 686.74 | 1 | 686.74 | 29.09 | 0.0003 | |
| X2 | 0.064 | 1 | 0.064 | 2.692 × 10−3 | 0.9596 | |
| X3 | 137.81 | 1 | 137.81 | 5.84 | 0.0363 | |
| X1 X2 | 4.56 | 1 | 4.56 | 0.19 | 0.6696 | |
| X1 X3 | 173.72 | 1 | 173.72 | 7.36 | 0.0218 | |
| X2 X3 | 293.55 | 1 | 293.55 | 12.44 | 0.0055 | |
| X12 | 627.86 | 1 | 627.86 | 26.60 | 0.0004 | |
| X22 | 2885.15 | 1 | 2885.15 | 122.23 | <0.0001 | |
| X32 | 70.20 | 1 | 70.20 | 2.97 | 0.1153 | |
| Residual | 236.05 | 10 | 23.60 | |||
| Lack of fit | 69.51 | 5 | 13.90 | 0.42 | 0.8202 | not significant |
| Pure error | 166.54 | 5 | 33.31 | |||
| Cor total | 4552.34 | 19 |
References
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.Y.; Youn, S.Y.; Park, M.S.; Baek, N.I.; Ji, G.E. Synthesis of stachyobifiose using Bifidobacterial α-galactosidase purified from recombinant Escherichia coli. J. Agric. Food Chem. 2018, 66, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lu, X.; Yang, X. Evaluation of clinical safety and beneficial effects of stachyose-enriched α-galacto-oligosaccharides on gut microbiota and bowel function in humans. Food Funct. 2017, 8, 262–269. [Google Scholar] [CrossRef]
- Pacifici, S.; Song, J.; Zhang, C.; Wang, Q.; Glahn, R.P.; Kolba, N.; Tako, E. Intra Amniotic Administration of Raffinose and Stachyose Affects the Intestinal Brush Border Functionality and Alters Gut Microflora Populations. Nutr. 2017, 9, 304. [Google Scholar] [CrossRef]
- Liu, Y.; Li, T.; Alim, A.; Ren, D.; Zhao, Y.; Yang, X. Regulatory effects of stachyose on colonic and hepatic inflammation, gut microbiota dysbiosis, and peripheral CD4+ T Cell distribution abnormality in high-fat diet-fed mice. J. Agric. Food Chem. 2019, 67, 11665–11674. [Google Scholar] [CrossRef]
- Liu, G.; Bei, J.; Liang, L.; Yu, G.; Li, L.; Li, Q. Stachyose improves inflammation through modulating gut microbiota of high-fat diet/streptozotocin-induced Type 2 diabetes in rats. Mol. Nutr. Food Res. 2018, 62, e1700954. [Google Scholar] [CrossRef]
- Huang, G.; Mao, J.; Ji, Z.; Ailati, A. Stachyose-induced apoptosis of Caco-2 cells via the caspase-dependent mitochondrial pathway. Food Funct. 2014, 6, 765–771. [Google Scholar] [CrossRef]
- Kim, H.-S.; Cha, E.; Kim, Y.; Jeon, Y.H.; Olson, B.H.; Byun, Y.; Park, H.-D. Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa biofilm formation via binding to LecA and decreasing cellular cyclic diguanylate levels. Sci. Rep. 2016, 6, 25318. [Google Scholar] [CrossRef]
- Hayes, M.R.; Pietruszka, J. Synthesis of glycosides by glycosynthases. Mol. 2017, 22, 1434. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Singh, A.; Batra, N.; Singh, J. Microbial production and biotechnological applications of α-galactosidase. Int. J. Biol. Macromol. 2020, 150, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lu, L.; Xiao, M.; Wang, Q.; Lu, Y.; Liu, C.; Wang, P.; Kumagai, H.; Yamamoto, K. Cloning and characterization of a novel α-galactosidase fromBifidobacterium breve203 capable of synthesizing Gal-α-1,4 linkage. FEMS Microbiol. Lett. 2008, 285, 278–283. [Google Scholar] [CrossRef]
- Wang, C.; Wang, H.; Ma, R.; Shi, P.; Niu, C.; Luo, H.; Yang, P.; Yao, B. Biochemical characterization of a novel thermophilic α-galactosidase from Talaromyces leycettanus JCM12802 with significant transglycosylation activity. J. Biosci. Bioeng. 2016, 121, 7–12. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, L.; Fan, S.; Jin, L.; Gu, G.; Xu, L.; Xiao, M. One-step synthesis of α-Gal epitope and globotriose derivatives by an engineered α-galactosidase. RSC Adv. 2015, 5, 22361–22364. [Google Scholar] [CrossRef]
- Gong, W.; Xu, L.; Gu, G.; Lu, L.; Xiao, M. Efficient and regioselective synthesis of globotriose by a novel α-galactosidase from Bacteroides fragilis. Appl. Microbiol. Biotechnol. 2016, 100, 6693–6702. [Google Scholar] [CrossRef]
- Yamashita, A.; Hashimoto, H.; Fujita, K.; Okada, M.; Mori, S.; Kitahata, S. Reverse reaction of Aspergillus niger APC-9319 α-galactosidase in a supersaturated substrate solution: Production of α-linked galactooligosaccharide (α-GOS). Biosci. Biotechnol. Biochem. 2005, 69, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Lyu, W.; Xiang, X.; Tang, Y.; Hu, B.; Ou, S.; Zeng, X. Immunomodulatory effects of enzymatic-synthesized α-galactooligosaccharides and evaluation of the structure–activity relationship. J. Agric. Food Chem. 2018, 66, 9070–9079. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Katayama, C.; Goto, M.; Okinaga, T.; Kitahata, S. Enzymatic synthesis of α-linked galactooligosaccharides using the reverse reaction of a cell-bound α-galactosidase from Candida guilliermondii H-404. Biosci. Biotechnol. Biochem. 1995, 59, 179–183. [Google Scholar] [CrossRef]
- Goulas, A.K.; Tzortzis, G.; Gibson, G.R. Development of a process for the production and purification of α- and β-galactooligosaccharides from Bifidobacterium bifidum NCIMB 41171. Int. Dairy, J. 2007, 17, 648–656. [Google Scholar] [CrossRef]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef]
- Wang, M.; Qi, W.; Su, R.; He, Z. Advances in carrier-bound and carrier-free immobilized nanobiocatalysts. Chem. Eng. Sci. 2015, 135, 21–32. [Google Scholar] [CrossRef]
- Rehm, F.B.H.; Chen, S.; Rehm, B.H.A. Enzyme Engineering for In Situ Immobilization. Mol. 2016, 21, 1370. [Google Scholar] [CrossRef]
- Guimarães, J.R.; Miranda, L.P.; Fernandez-Lafuente, R.; Tardioli, P.W. Immobilization of Eversa® Transform via CLEA Technology Converts It in a Suitable Biocatalyst for Biolubricant Production Using Waste Cooking Oil. Mol. 2021, 26, 193. [Google Scholar] [CrossRef]
- Bayraktar, H.; Önal, S. Cross-linked α-galactosidase aggregates: Optimization, characterization and application in the hydrolysis of raffinose-type oligosaccharides in soymilk. J. Sci. Food Agric. 2019, 99, 4748–4760. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for op-timization in analytical chemistry. Talanta. 2008, 76, 965–977. [Google Scholar] [CrossRef]
- Schoevaart, R.; Wolbers, M.W.; Golubovic, M.; Ottens, M.; Kieboom, A.P.G.; Van Rantwijk, F.; Van Der Wielen, L.; Sheldon, R.A. Preparation, optimization, and structures of cross-linked enzyme aggregates (CLEAs). Biotechnol. Bioeng. 2004, 87, 754–762. [Google Scholar] [CrossRef]
- Talekar, S.; Joshi, G.; Chougle, R.; Nainegali, B.; Desai, S.; Joshi, A.; NKadmbale, S.; Kamat, P.; Haripurkar, R.; Jadhav, S.; et al. Preparation of stable cross-linked enzyme ag-gre-gates (CLEAs) of NADH-dependent nitrate reductase and its use for silver nanoparticle synthesis from silver nitrate. Catal. Commun. 2014, 53, 62–66. [Google Scholar] [CrossRef]
- Yang, Q.; Dou, F.; Liang, B.; Shen, Q. Studies of cross-linking reaction on chitosan fiber with glyoxal. Carbohydr. Polym. 2005, 59, 205–210. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Matijošytė, I.; Arends, I.W.; de Vries, S.; Sheldon, R.A. Preparation and use of cross-linked enzyme aggregates (CLEAs) of laccases. J. Mol. Catal. B: Enzym. 2010, 62, 142–148. [Google Scholar] [CrossRef]
- Sheldon, R.A. Enzyme immobilization: The quest for optimum performance. Adv. Synth. Catal. 2007, 349, 1289–1307. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Q.; Dang, Y.; Shu, G. The effect of glutaraldehyde cross-linking on the enzyme activity of immobilized β-galactosidase on chitosan bead. Adv. J. Food Sci. Technol. 2013, 5, 932–935. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotech. 2004, 37, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Talekar, S.; Nadar, S.; Joshi, A.; Joshi, G. Pectin cross-linked enzyme aggregates (pectin-CLEAs) of glucoamylase. RSC Adv. 2014, 4, 59444–59453. [Google Scholar] [CrossRef]
- Hormigo, D.; García-Hidalgo, J.; Acebal, C.; De La Mata, I.; Arroyo, M. Preparation and characterization of cross-linked enzyme aggregates (CLEAs) of recombinant poly-3-hydroxybutyrate depolymerase from Streptomyces exfoliatus. Bioresour. Technol. 2012, 115, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Y.; Liu, W.; Wang, J.; Chen, H. A novel cross-linked enzyme aggregates (CLEAs) of papain and neu-trase-production, partial characterization and application. Int. J. Biol. Macromol. 2017, 95, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Tandjaoui, N.; Tassist, A.; Abouseoud, M.; Couvert, A.; Amrane, A. Preparation and characterization of cross-linked enzyme aggregates (CLEAs) of Brassica rapa peroxidase. Biocatal. Agric. Biotechnol. 2015, 4, 208–213. [Google Scholar] [CrossRef]
- Sangeetha, K.; Abraham, T.E. Preparation and characterization of cross-linked enzyme aggregates (CLEA) of Subtilisin for controlled release applications. Int. J. Biol. Macromol. 2008, 43, 314–319. [Google Scholar] [CrossRef]
- Tamborini, L.; Fernandes, P.; Paradisi, F.; Molinari, F. Flow bioreactors as complementary tools for biocatalytic process intensi-fication. Trends Biotechnol. 2018, 36, 73–88. [Google Scholar] [CrossRef]
- De Santis, P.; Meyer, L.-E.; Kara, S. The rise of continuous flow biocatalysis – fundamentals, very recent developments and future perspectives. React. Chem. Eng. 2020, 5, 2155–2184. [Google Scholar] [CrossRef]


| Run | Actual Level of Variables | |||
|---|---|---|---|---|
| Glutaraldehyde Concentration (mM) | Cross-Linking Time (h) | Enzyme Amount (U) | Activity Recovery (%) | |
| 1 | 45 | 1.00 | 0.50 | 62.32 |
| 2 | 35 | 1.00 | 0.50 | 72.17 |
| 3 | 40 | 1.50 | 0.45 | 85.80 |
| 4 | 30 | 1.50 | 0.45 | 54.67 |
| 5 | 45 | 1.00 | 0.40 | 70.88 |
| 6 | 40 | 1.50 | 0.35 | 63.45 |
| 7 | 40 | 1.50 | 0.45 | 79.71 |
| 8 | 45 | 2.00 | 0.50 | 67.86 |
| 9 | 35 | 2.00 | 0.50 | 74.92 |
| 10 | 40 | 1.50 | 0.45 | 73.30 |
| 11 | 35 | 2.00 | 0.40 | 40.61 |
| 12 | 40 | 0.50 | 0.45 | 40.83 |
| 13 | 50 | 1.50 | 0.45 | 67.86 |
| 14 | 35 | 1.00 | 0.40 | 62.32 |
| 15 | 40 | 1.50 | 0.45 | 89.41 |
| 16 | 40 | 1.50 | 0.45 | 79.55 |
| 17 | 45 | 2.00 | 0.40 | 52.42 |
| 18 | 40 | 2.50 | 0.45 | 35.98 |
| 19 | 40 | 1.50 | 0.55 | 85.69 |
| 20 | 40 | 1.50 | 0.45 | 77.88 |
| Storage Time (days) | Residual Activity (%) | |
|---|---|---|
| Free Enzyme | CLEAs | |
| 0 | 100 ± 0.4 | 100 ± 0.3 |
| 10 | 94.3 ± 0.7 | 98.9 ± 1.1 |
| 20 | 90.6 ± 1.5 | 97.5 ± 0.8 |
| 40 | 83.4 ± 2.1 | 94.2 ± 1.7 |
| 60 | 65.2 ± 1.4 | 87.4 ± 2.3 |
| 80 | 33.7 ± 0.9 | 79.8 ± 3.1 |
| 90 | 21.6 ± 3.4 | 74.5 ± 2.5 |
| Variables | Symbol | Unit | Coded Levels | ||||
|---|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | |||
| Glutaraldehyde Concentration | X1 | mM | 0.35 | 0.40 | 0.45 | 0.50 | 0.55 |
| Cross-Linking Time | X2 | h | 0.5 | 1.0 | 1.5 | 2.0 | 2.5 |
| Enzyme Amount | X3 | U | 30 | 35 | 40 | 45 | 50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yang, J.; Wang, K.; Duan, F.; Lu, L. Carrier-Free Immobilization of α-Galactosidase as Nano-Biocatalysts for Synthesizing Prebiotic α-Galacto-Oligosaccharides. Molecules 2021, 26, 1248. https://doi.org/10.3390/molecules26051248
Liu Y, Yang J, Wang K, Duan F, Lu L. Carrier-Free Immobilization of α-Galactosidase as Nano-Biocatalysts for Synthesizing Prebiotic α-Galacto-Oligosaccharides. Molecules. 2021; 26(5):1248. https://doi.org/10.3390/molecules26051248
Chicago/Turabian StyleLiu, Yan, Jingyi Yang, Ke Wang, Feiyu Duan, and Lili Lu. 2021. "Carrier-Free Immobilization of α-Galactosidase as Nano-Biocatalysts for Synthesizing Prebiotic α-Galacto-Oligosaccharides" Molecules 26, no. 5: 1248. https://doi.org/10.3390/molecules26051248
APA StyleLiu, Y., Yang, J., Wang, K., Duan, F., & Lu, L. (2021). Carrier-Free Immobilization of α-Galactosidase as Nano-Biocatalysts for Synthesizing Prebiotic α-Galacto-Oligosaccharides. Molecules, 26(5), 1248. https://doi.org/10.3390/molecules26051248






