Shotgun Proteomics of Isolated Urinary Extracellular Vesicles for Investigating Respiratory Impedance in Healthy Preschoolers
Abstract
:1. Introduction
2. Results
2.1. Characteristics of Study Participants
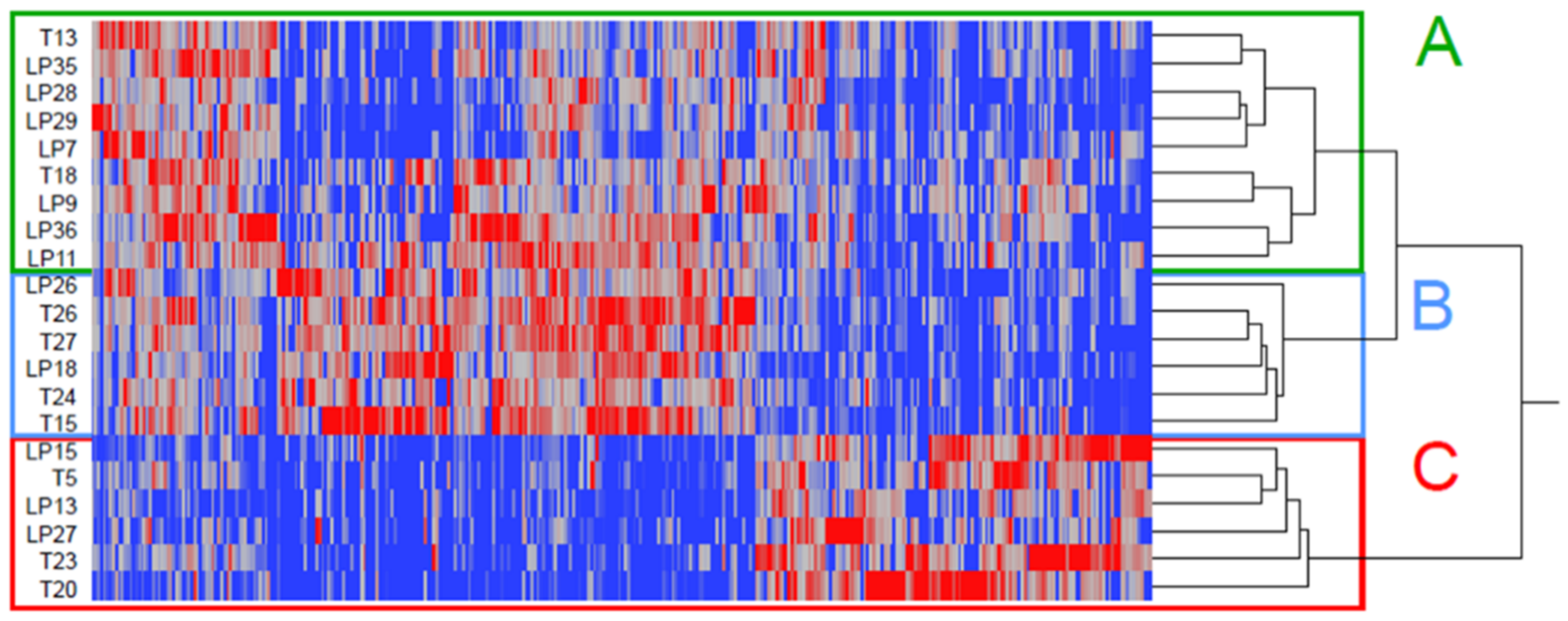
2.2. Isolation of Urinary Extracellular Vesicles and Proteomics Analysis
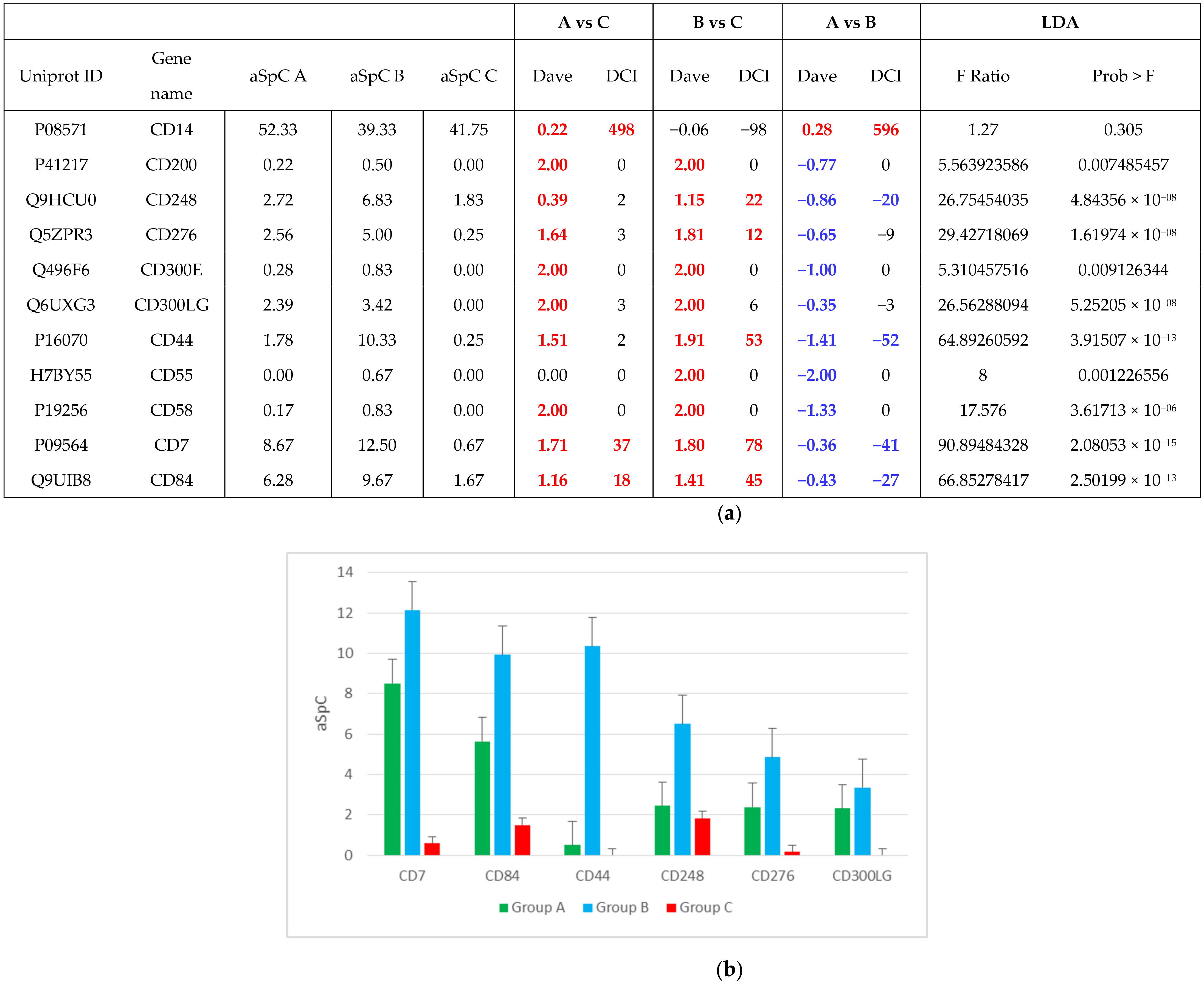
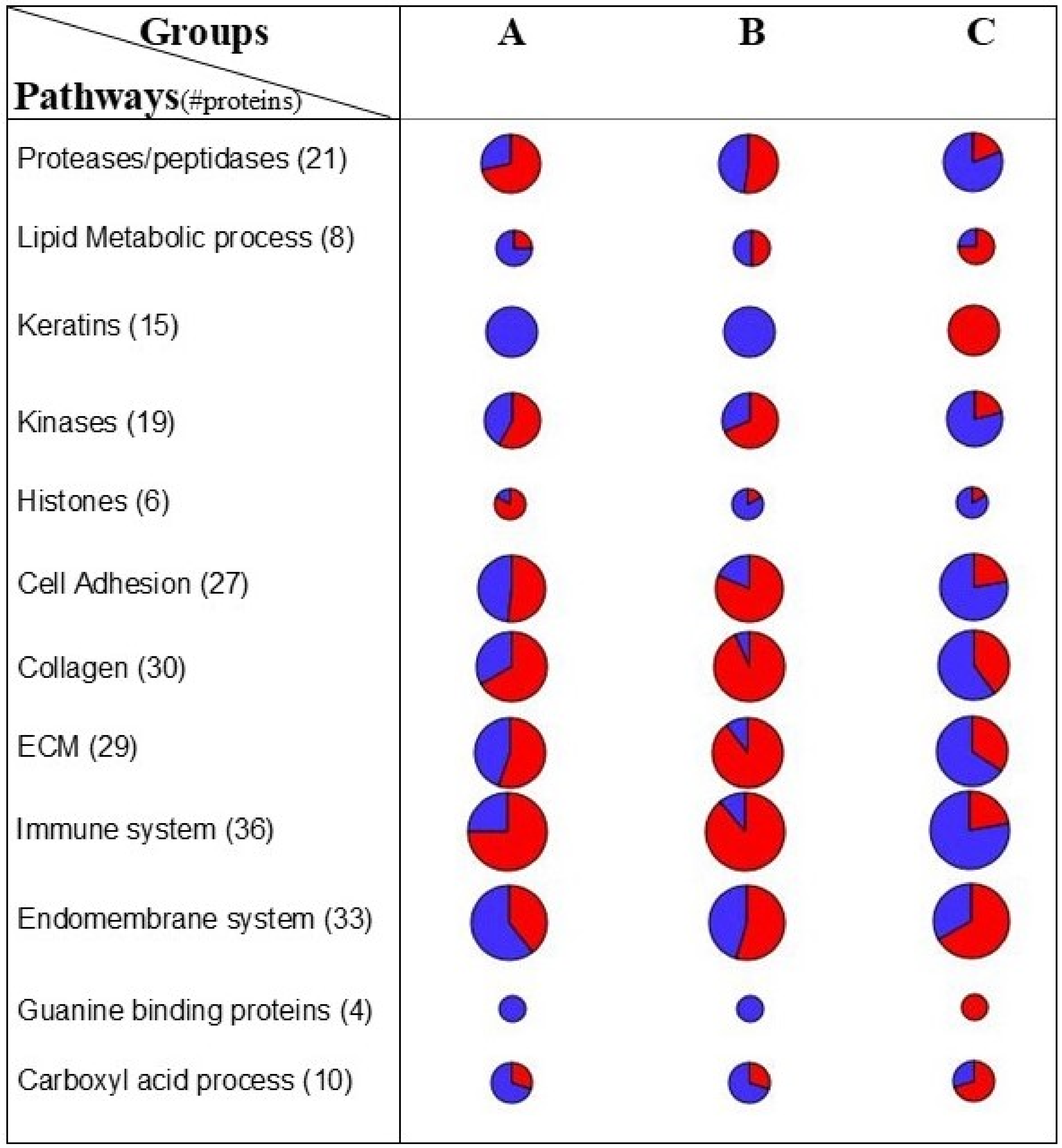
2.3. Linking FOT Parameters and Proteomics
3. Discussion
4. Materials and Methods
4.1. Participants and Study Design
4.2. Forced Oscillation Technique (FOT)
4.3. Isolation of Urinary Extracellular Vesicle
4.4. Nanosight Extracellular Vesicle Analysis
4.5. Proteomics Analysis of Urinary Extracellular Vesicle
4.6. Network Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Colin, A.A.; McEvoy, C.; Castile, R.G. Respiratory morbidity and lung function in preterm infants of 32 to 36 weeks’ gestational age. Pediatrics 2010, 126, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Er, I.; Gunlemez, A.; Uyan, Z.S.; Aydogan, M.; Oruc, M.; Isik, O.; Arisoy, A.E.; Turker, G.; Baydemir, C.; Gokalp, A.S. Evaluation of lung function on impulse oscillometry in preschool children born late preterm. Pediatr Int. 2016, 58, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Carraro, S.; Giordano, G.; Reniero, F.; Carpi, D.; Stocchero, M.; Sterk, P.J.; Baraldi, E. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy 2013, 68, 110–117. [Google Scholar] [CrossRef]
- Mauri, P.; Riccio, A.M.; Rossi, R.; Di Silvestre, D.; Benazzi, L.; De Ferrari, L.; Dal Negro, R.W.; Holgate, S.T.; Canonica, G.W. Proteomics of bronchial biopsies: Galectin-3 as a predictive biomarker of airway remodelling modulation in omalizumab-treated severe asthma patients. Immunol Lett. 2014, 162, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mauri, P.; Riccio, A.M.; Rossi, R.; Saccheri, F.; Bartezaghi, M.; Di Silvestre, D.; Rigoni, L.; Canonica, G.W. Proteomics analysis of asthma biomarkers: Proxima sub-study. Eur Respir. J. 2017, 50, 1163. [Google Scholar]
- Riccio, A.M.; Mauri, P.; De Ferrari, L.; Rossi, R.; Di Silvestre, D.; Bartezaghi, M.; Saccheri, F.; Canonica, G.W. Plasma Galectin-3 and urine proteomics predict FEV 1 improvement in omalizumab-treated patients with severe allergic asthma: Results from the PROXIMA sub-study. World Allergy Organ. J. 2020, 13, 100095–100105. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, K. Urinary proteomics and metabolomics in the diagnosis of pediatric disorders. Proteomics Clin Appl. 2015, 9, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.E.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteomics. 2009, 6, 267–283. [Google Scholar] [CrossRef]
- Becker, L.; Kheirandish-Gozal, L.; Peris, E.; Schoenfelt, K.Q.; Gozal, D. Contextualised urinary biomarker analysis facilitates diagnosis of paediatric obstructive sleep apnoea. Sleep Med. 2014, 15, 541–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Pol, E.; Hoekstra, A.G.; Sturk, A.; Otto, C.; van Leeuwen, T.G.; Nieuwland, R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J. Thromb Haemost. 2010, 8, 2596–2607. [Google Scholar] [CrossRef]
- Sereni, L.; Castiello, M.C.; Di Silvestre, D.; Della Valle, P.; Brombin, C.; Ferrua, F.; Cicalese, M.P.; Pozzi, L.; Migliavacca, M.; Bernardo, M.E.; et al. Lentiviral gene therapy corrects platelet phenotype and function in patients with Wiskott-Aldrich syndrome. J. Allergy Clin. Immunol 2019, 144, 825–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starodubtseva, N.L.; Kononikhin, A.S.; Bugrova, A.E.; Chagovets, V.; Indeykina, M.; Krokhina, K.N.; Nikitina, I.V.; Kostyukevich, Y.I.; Popov, I.A.; Larina, I.M.; et al. Investigation of urine proteome of preterm newborns with respiratory pathologies. J. Proteomics. 2016, 149, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.; De Palma, A.; Mauri, P. Emerging MS-based platforms for the characterization of tumor-derived exosomes isolated from human biofluids: Challenges and promises of MudPIT. Expert Rev. Proteomics. 2017, 14, 757–767. [Google Scholar] [CrossRef]
- Starodubtseva, N.L.; Kononikhin, A.S.; Bugrova, A.E.; Krokhina, K.N.; Nikitina, I.V.; Kostyukevich, Y.I.; Popov, I.A.; Frankevich, V.E.; Aleksandrova, N.V.; Ionov, O.V.; et al. Proteomic Analysis of the Urine for Diagnostics in Newborns. Bull. Exp. Biol. Med. 2016, 160, 867–870. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yayoi, Y.; Ohsawa, Y.; Koike, M.; Zhang, G.; Kominami, E.; Uchiyama, Y. Specific localization of lysosomal aminopeptidases in type II alveolar epithelial cells of the rat lung. Arch. Histol. Cytol. 2001, 64, 89–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohlmeier, S.; Nieminen, P.; Gao, J.; Kanerva, T.; Rönty, M.; Toljamo, T.; Bergmann, U.; Mazur, W.; Pulkkinen, V. Lung tissue proteomics identifies elevated transglutaminase 2 levels in stable chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 310, L1155–L1165. [Google Scholar] [CrossRef] [Green Version]
- Zha, W.; Su, M.; Huang, M.; Cai, J.; Du, Q. Administration of pigment epithelium-derived factor inhibits airway inflammation and remodeling in chronic OVA-induced mice via VEGF suppression. Allergy Asthma Immunol. Res. 2016, 8, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.C.; Chen, C.W.; Huang, Y.W.; Chao, L.; Chao, J.; Lin, Y.S.; Lin, C.F. Kallistatin protects against sepsis-related acute lung injury via inhibiting inflammation and apoptosis. Sci. Rep. 2015, 5, 12463. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Hagood, J.S.; Murphy-Ullrich, J.E. Thy-1 expression regulates the ability of rat lung fibroblasts to activate transforming growth factor-β in response to fibrogenic stimuli. Am. J. Pathol. 2004, 165, 659–669. [Google Scholar] [CrossRef]
- Liu, X.; Wong, S.S.; Taype, C.A.; Kim, J.; Shentu, T.P.; Espinoza, C.R.; Finley, J.C.; Bradley, J.E.; Head, B.P.; Patel, H.H.; et al. Thy-1 interaction with Fas in lipid rafts regulates fibroblast apoptosis and lung injury resolution. Lab. Invest. 2017, 97, 256–267. [Google Scholar] [CrossRef]
- Chen, L.; Tang, R.Z.; Ruan, J.; Zhu, X.B.; Yang, Y. Up-regulation of THY1 attenuates interstitial pulmonary fibrosis and promotes lung fibroblast apoptosis during acute interstitial pneumonia by blockade of the WNT signaling pathway. Cell Cycle. 2019, 18, 670–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motokawa, I.; Endo, M.; Terada, K.; Horiguchi, H.; Miyata, K.; Kadomatsu, T.; Morinaga, J.; Sugizaki, T.; Ito, T.; Araki, K.; et al. Interstitial pneumonia induced by bleomycin treatment is exacerbated in Angptl2-deficient mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 311, L704–L713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tse-Ying, H.; Chih-Yuan, W.; Kuen-Yuan, C.; Li-Ting, H. Urinary Exosomal Thyroglobulin in Thyroid Cancer Patients With Post-ablative Therapy: A New Biomarker in Thyroid Cancer. Front. Endocrinol. 2020, 11, 382. [Google Scholar]
- Wang, S.; Kojima, K.; Mobley, J.A.; West, A.B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine. 2019, 45, 351. [Google Scholar] [CrossRef] [Green Version]
- Beydon, N.; Davis, S.D.; Lombardi, E.; Allen, J.L.; Arets, H.G.; Aurora, P.; Bisgaard, H.; Ducharme, F.M.; Eigen, H.; et al. American Thoracic Society/European Respiratory Society Working Group on Infant and Young Children Pulmonary Function Testing. An official American Thoracic Society/European Respiratory Society statement: Pulmonary function testing in preschool children. Am. J. Respir Crit. Care Med. 2007, 175, 1304–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calogero, C.; Simpson, S.J.; Lombardi, E.; Parri, N.; Cuomo, B.; Palumbo, M.; De Martino, M.; Shackleton, C.; Verheggen, M.; Gavidia, T.; et al. Respiratory impedance and bronchodilator responsiveness in healthy children aged 2–13 years. Pediatr. Pulmonol. 2013, 48, 707–715. [Google Scholar] [CrossRef]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot production of mesenchymal stem/stromal freeze-dried secretome for cell-free regenerative nanomedicine: A validated GMP-compliant process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [Green Version]
- Sereni, L.; Castiello, M.C.; Marangoni, F.; Anselmo, A.; Di Silvestre, D.; Motta, S.; Draghici, E.; Mantero, S.; Thrasher, A.J.; Giliani, S.; et al. Autonomous role of Wiskott-Aldrich syndrome platelet deficiency in inducing autoimmunity and inflammation. J. Allergy Clin. Immunol. 2018, 142, 1272–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilario, M.; Kalousis, A. Approaches to dimensionality reduction in proteomic biomarker studies. Brief. Bioinform. 2008, 9, 102–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, F.; Lavatelli, F.; Di Silvestre, D.; Rossi, R.; Palladini, G.; Obici, L.; Verga, L.; Mauri, P.; Merlini, G. Reliable typing of systemic amyloidoses through proteomic analysis of subcutaneous adipose tissue. Blood 2012, 119, 1844–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawłowski, K.; Muszewska, A.; Lenart, A.; Szczepińska, T.; Godzik, A.; Grynberg, M. A widespread peroxiredoxin-like domain present in tumor suppression- and progression-implicated proteins. BMC Genomics. 2010, 11, 590. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Lundgren, D.H.; Hwang, S.I.; Wu, L.; Han, D.K. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics. 2010, 7, 39–53. [Google Scholar] [CrossRef]
- Zhang, B.; VerBerkmoes, N.C.; Langston, M.A.; Uberbacher, E.; Hettich, R.L.; Samatova, N.F. Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 2006, 5, 2909–2918. [Google Scholar] [CrossRef]
- Zhao, Y.; Karypis, G. Data clustering in life sciences. Mol. Biotechnol. 2005, 31, 55–80. [Google Scholar] [CrossRef]
- Roffia, V.; De Palma, A.; Lonati, C.; Silvestre, D.D.; Rossi, R.; Mantero, M.; Gatti, S.; Dondossola, D.; Valenza, F.; Mauri, P.; et al. Proteome Investigation of Rat Lungs subjected to Ex Vivo Perfusion (EVLP). Molecules. 2018, 23, 3061. [Google Scholar] [CrossRef] [Green Version]



| LP | T | p-Value | |
|---|---|---|---|
| n = 12 | n = 9 | ||
| Personal characteristics | |||
| Female (%) | 5 (41.67) | 2 (22.22) | 0.640 |
| Z-score Height, mean (SD) | −0.92 (1.08) | 0.21 (1.03) | 0.033 |
| Z-score BMI, mean (SD) | 0.64 (1.47) | 1.09 (1.09) | 0.337 |
| Age, years, mean (SD) | 3.67 (0.78) | 4.56 (0.73) | 0.015 |
| Birthweight, mean (SD), gr | 2123.33 (576.78) | 3012.22 (829.02) | 0.012 |
| Cesarean Delivery, n (%) | 10 (83.33) | 7 (77.78) | 1.000 |
| Maternal diseases in pregnancy, n (%) | 2 (16.67) | 0 (0.00) | 0.592 |
| Respiratory support, n (%) | 5 (41.67) | 3 (33.33) | 1.000 |
| Bronchiolitis 1st year of life, n (%) | 4 (33.33) | 4 (50.00) | 0.780 |
| Upper respiratory infection ever, n (%) | 5 (41.67) | 4 (44.44) | 1.000 |
| Pneumonia ever, n (%) | 0 (0.00) | 3 (33.33) | 0.126 |
| Environmental exposure | |||
| Proximity to high traffic road <200 m, n (%), | 10 (83.33) | 9 (100.00) | 0.592 |
| Current parental smoke exposure, n (%), | 4 (33.33) | 0 (0.00) | 0.173 |
| Current pet exposure, n (%), | 3 (25.00) | 2 (22.22) | 0.592 |
| Current mold exposure, n (%), | 2 (16.67) | 0 (0.00) | 0.592 |
| FOT parameters | |||
| Z-score | |||
| Rrs6 | 0.98 (1.49) | −0.27 (3.79) | 0.887 |
| Rrs8 | 0.75 (1.34) | −0.71 (3.86) | 0.619 |
| Rrs10 | 0.92 (1.35) | −0.57 (3.85) | 0.670 |
| Xrs6 | −1.36 (2.33) | −0.43 (2.23) | 0.522 |
| Xrs8 | −1.64 (2.05) | −0.86 (2.54) | 0.570 |
| Xrs10 | −1.55 (1.94) | −0.65 (2.22) | 0.522 |
| AX | 2.06 (2.26) | 0.80 (2.92) | 0.356 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrante, G.; Rossi, R.; Cilluffo, G.; Di Silvestre, D.; Brambilla, A.; De Palma, A.; Villa, C.; Malizia, V.; Gagliardo, R.; Torrente, Y.; et al. Shotgun Proteomics of Isolated Urinary Extracellular Vesicles for Investigating Respiratory Impedance in Healthy Preschoolers. Molecules 2021, 26, 1258. https://doi.org/10.3390/molecules26051258
Ferrante G, Rossi R, Cilluffo G, Di Silvestre D, Brambilla A, De Palma A, Villa C, Malizia V, Gagliardo R, Torrente Y, et al. Shotgun Proteomics of Isolated Urinary Extracellular Vesicles for Investigating Respiratory Impedance in Healthy Preschoolers. Molecules. 2021; 26(5):1258. https://doi.org/10.3390/molecules26051258
Chicago/Turabian StyleFerrante, Giuliana, Rossana Rossi, Giovanna Cilluffo, Dario Di Silvestre, Andrea Brambilla, Antonella De Palma, Chiara Villa, Velia Malizia, Rosalia Gagliardo, Yvan Torrente, and et al. 2021. "Shotgun Proteomics of Isolated Urinary Extracellular Vesicles for Investigating Respiratory Impedance in Healthy Preschoolers" Molecules 26, no. 5: 1258. https://doi.org/10.3390/molecules26051258







