Site-Specific Fluorescent Labeling of RNA Interior Positions
Abstract
:1. Introduction
2. Exploitation of Natural Posttranslational Modifications
2.1. Wybutosine
2.2. DHU
2.3. acp3U
2.4. s4U
2.5. Extensions beyond tRNA Labeling
3. Repurposing Enzymatic Transferase Reactions
3.1. SAM-Dependent Methyl Transferases (SAM-Mtases)
3.2. tRNA Guanine Transglycosylase (TGT)
4. Nucleic Acid-Assisted Labeling of Intact RNAs
4.1. RNA Acylation at DNA Induced Loops or Gaps
4.2. DNA Reactive Sequence Targeting of an Interior Adenosine
4.3. Evolving Ribozymes
5. Site-Specific Labeling Requiring De Novo RNA Preparation
5.1. Position-Selective Labeling of RNA (PLOR)
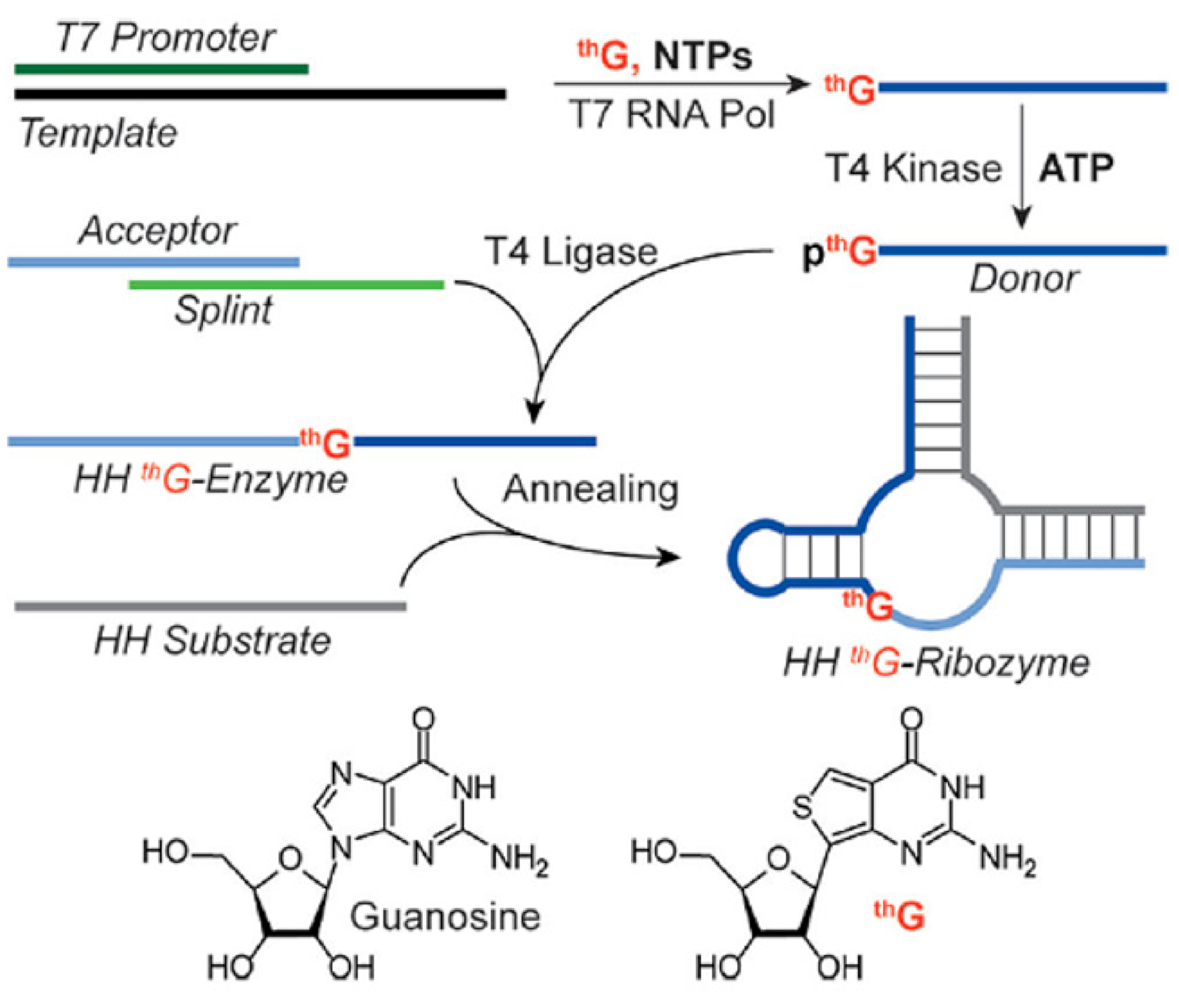
5.2. thG-Containing RNAs
5.3. Unnatural Base Pairs
6. Conclusions
Funding
Conflicts of Interest
References
- Sinkeldam, R.W.; Greco, N.J.; Tor, Y. Fluorescent analogs of biomolecular building blocks: Design, properties, and applications. Chem. Rev. 2010, 110, 2579–2619. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Chan, K.M.; Kool, E.T. Fluorescent nucleobases as tools for studying DNA and RNA. Nat. Chem. 2017, 9, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, Y.; Hudson, R.H. Base-modified fluorescent purine nucleosides and nucleotides for use in oligonucleotide probes. J. Photochem. Photobiol. C. Photochem. Rev. 2018, 36, 48–73. [Google Scholar] [CrossRef]
- Bood, M.; Sarangamath, S.; Wranne, M.S.; Grøtli, M.; Wilhelmsson, L.M. Fluorescent nucleobase analogues for base–base FRET in nucleic acids: Synthesis, photophysics and applications. Beilstein J. Org. Chem. 2018, 14, 114–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, B.Y.; Dziuba, D.; Benhida, R.; Demchenko, A.P.; Burger, A. Probing of Nucleic Acid Structures, Dynamics, and Interactions With Environment-Sensitive Fluorescent Labels. Front. Chem. 2020, 8, 112. [Google Scholar] [CrossRef] [Green Version]
- Klöcker, N.; Weissenboeck, F.P.; Rentmeister, A. Covalent labeling of nucleic acids. Chem. Soc. Rev. 2020, 49, 8749–8773. [Google Scholar] [CrossRef] [PubMed]
- Hanspach, G.; Trucks, S.; Hengesbach, M. Strategic labelling approaches for RNA single-molecule spectroscopy. RNA Biol. 2019, 16, 1119–1132. [Google Scholar] [CrossRef]
- Tomkuvienė, M.; Mickutė, M.; Vilkaitis, G.; Klimašauskas, S. Repurposing enzymatic transferase reactions for targeted labeling and analysis of DNA and RNA. Curr. Opin. Biotechnol. 2019, 55, 114–123. [Google Scholar] [CrossRef]
- George, J.T.; Srivatsan, S.G. Bioorthogonal chemistry-based RNA labeling technologies: Evolution and current state. Chem. Commun. 2020, 56, 12307–12318. [Google Scholar] [CrossRef]
- McCown, P.J.; Ruszkowska, A.; Kunkler, C.N.; Breger, K.; Hulewicz, J.P.; Wang, M.C.; Springer, N.A.; Brown, J.A. Naturally occurring modified ribonucleosides. Wiley Interdiscip Rev. RNA 2020, 11, e1595. [Google Scholar] [CrossRef]
- Itaya, T.; Kanai, T. Synthesis and structure of the hypermodified nucleoside of rat liver phenylalanine transfer ribonucleic Acid. Chem. Pharm. Bull. 2002, 50, 1318–1326. [Google Scholar] [CrossRef] [Green Version]
- de Crécy-Lagard, V.; Brochier-Armanet, C.; Urbonavicius, J.; Fernandez, B.; Phillips, G.; Lyons, B.; Noma, A.; Alvarez, S.; Droogmans, L.; Armengaud, J.; et al. Biosynthesis of wyosine derivatives in tRNA: An ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 2010, 27, 2062–2077. [Google Scholar]
- Eisinger, J.; Feuer, B.; Yamane, T. Luminescence and binding studies on tRNA-Phe. Proc. Natl. Acad. Sci. USA 1970, 65, 638–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beardsley, K.; Cantor, C.R. Studies of transfer RNA tertiary structure by singlet-singlet energy transfer. Proc. Natl. Acad. Sci. USA 1970, 65, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, S.R.; Tor, Y. tRNA(Phe) binds aminoglycoside antibiotics. Bioorg. Med. Chem. 1999, 9, 1979–1991. [Google Scholar] [CrossRef]
- Dwyer, B.G.; Johnson, E.; Cazares, E.; Holman, K.L.M.; Kirk, S.R. Ruthenium anticancer agent KP1019 binds more tightly than NAMI-A to tRNAPhe. J. Inorg. Biochem. 2018, 182, 177–183. [Google Scholar] [CrossRef]
- Paulsen, H.; Robertson, J.M.; Wintermeyer, W. Effect of ribosome binding and translocation on the anticodon of tRNAPhe as studied by wybutine fluorescence. Nucleic Acids Res. 1982, 10, 2651–2663. [Google Scholar] [CrossRef] [Green Version]
- Robertson, J.M.; Paulsen, H.; Wintermeyer, W. Pre-steady-state kinetics of ribosomal translocation. J. Mol. Biol. 1986, 192, 351–360. [Google Scholar] [CrossRef]
- RajBhandary, U.L.; Faulkner, R.D.; Stuart, A. Studies on Polynucleotides LXXIX. Yeast phenylalanine transfer ribonucleic acid: Products obtained by degradation with pancreatic ribonucleasE. J. Biol. Chem. 1968, 243, 575–583. [Google Scholar] [CrossRef]
- Thiebe, R.; Zachau, H.G. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur. J. Biochem. 1968, 5, 546–555. [Google Scholar] [CrossRef]
- MODOMICS a Database of RNA Modifications. Available online: https://iimcb.genesilico.pl/modomics/ (accessed on 12 February 2021).
- Finet, O.; Yague-Sanz, C.; Kruger, L.K.; Migeot, V.; Ernst, F.G.; Lafontaine, D.L.J.; Tran, P.; Wéry, M.; Morillon, A.; Hermand, D. Transcription-Wide Mapping of Dihydrouridine (d) Reveals That Mrna ihydrouridylation is Essential for Meiotic Chromosome Segregation. Available online: http://dx.doi.org/10.2139/ssrn.3569550 (accessed on 15 February 2021).
- Betteridge, T.; Liu, H.; Gamper, H.; Kirillov, S.; Cooperman, B.S.; Hou, Y.M. Fluorescent labeling of tRNAs for dynamics experiments. RNA 2007, 13, 1594–1601. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Betteridge, T.; Hou, Y.M. Fluorophore labeling to monitor tRNA dynamics. Methods Enzymol. 2009, 469, 69–93. [Google Scholar]
- Wintermeyer, W.; Zachau, H.G. Fluorescent derivatives of yeast tRNAPhe. Eur. J. Biochem. 1979, 98, 465–475. [Google Scholar] [CrossRef]
- Pan, D.; Qin, H.; Cooperman, B.S. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. RNA 2009, 15, 346–354. [Google Scholar] [CrossRef] [Green Version]
- Stevens, B.; Chen, C.; Farrell, I.; Zhang, H.; Kaur, J.; Broitman, S.L.; Smilansky, Z.; Cooperman, B.S.; Goldman, Y.E. FRET-based identification of mRNAs undergoing translation. PLoS ONE 2012, 7, 38344. [Google Scholar]
- Tu, C.; Santo, L.; Mishima, Y.; Raje, N.; Smilansky, Z.; Zoldan, J. Monitoring protein synthesis in single live cancer cells. Integr. Biol. 2016, 8, 645–653. [Google Scholar] [CrossRef]
- Kaur, J.; Raj, M.; Cooperman, B.S. Fluorescent labeling of tRNA dihydrouridine residues: Mechanism and distribution. RNA 2011, 17, 1393–1400. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Kirillov, S.V.; Cooperman, B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell 2007, 25, 519–525. [Google Scholar] [CrossRef] [Green Version]
- Grigoriadou, C.; Marzi, S.; Kirillov, S.; Gualerzi, C.O.; Cooperman, B.S. A quantitative kinetic scheme for 70 S translation initiation complex formation. J. Mol. Biol. 2007, 373, 562–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, M.Y.; Li, H.; Ghelfi, M.D.; Goldman, Y.E.; Cooperman, B.S. Ataluren and aminoglycosides stimulate read-through of nonsense codons by orthogonal mechanisms. Proc. Natl. Acad. Sci. USA 2021, 118, 2020599118. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Stevens, B.; Kaur, J.; Cabral, D.; Liu, H.; Wang, Y.; Zhang, H.; Rosenblum, G.; Smilansky, Z.; Goldman, Y.E.; et al. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol. Cell 2011, 42, 367–377. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhang, H.; Broitman, S.L.; Reiche, M.; Farrell, I.; Cooperman, B.S.; Goldman, Y.E. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat. Struct. Mol. Biol. 2013, 20, 582–588. [Google Scholar] [CrossRef]
- Rosenblum, G.; Chen, C.; Kaur, J.; Cui, X.; Zhang, H.; Asahara, H.; Chong, S.; Smilansky, Z.; Goldman, Y.E.; Cooperman, B.S. Quantifying elongation rhythm during full-length protein synthesis. J. Am. Chem. Soc. 2013, 135, 11322–11329. [Google Scholar] [CrossRef] [Green Version]
- Jamiolkowski, R.M.; Chen, C.; Cooperman, B.S.; Goldman, Y.E. tRNA Fluctuations Observed on Stalled Ribosomes Are Suppressed during Ongoing Protein Synthesis. Biophys. J. 2017, 113, 2326–2335. [Google Scholar] [CrossRef] [Green Version]
- Barhoom, S.; Kaur, J.; Cooperman, B.S.; Smorodinsky, N.I.; Smilansky, Z.; Ehrlich, M.; Elroy-Stein, O. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res. 2011, 39, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plochowietz, A.; Farrell, I.; Smilansky, Z.; Cooperman, B.S.; Kapanidis, A.N. In vivo single-RNA tracking shows that most tRNA diffuses freely in live bacteria. Nucleic Acids Res. 2017, 45, 926–937. [Google Scholar] [CrossRef] [Green Version]
- Dhakal, R.; Tong, C.; Anderson, S.; Kashina, A.S.; Cooperman, B.; Bau, H.H. Dynamics of intracellular stress-induced tRNA trafficking. Nucleic Acids Res. 2019, 47, 2002–2010. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Rai, A.; Hur, E.M.; Smilansky, Z.; Chang, K.T.; Min, K.T. DSCR1 is required for both axonal growth cone extension and steering. J. Cell Biol. 2016, 213, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilotte, J.; Chan, S.W.; Farnum, J.B.; Thomas, W.M.; Smilansky, Z.; Vanderklish, P.W. A heterogeneous tRNA granule structure exhibiting rapid, bi-directional neuritic transport. Eur. J. Cell Biol. 2018, 97, 168–179. [Google Scholar] [CrossRef]
- Koltun, B.; Ironi, S.; Gershoni-Emek, N.; Barrera, I.; Hleihil, M.; Nanguneri, S.; Sasmal, R.; Agasti, S.S.; Nair, D.; Rosenblum, K. Measuring mRNA translation in neuronal processes and somata by tRNA-FRET. Nucleic Acids Res. 2020, 48, 32. [Google Scholar] [CrossRef] [Green Version]
- Alroy, I.; Mansour, W.; Klepfish, M.; Sheinberger, Y. Expanding small-molecule target space to mRNA translation regulation. Drug Discov. Today 2020. [Google Scholar] [CrossRef] [PubMed]
- Barhoom, S.; Farrell, I.; Shai, B.; Dahary, D.; Cooperman, B.S.; Smilansky, Z.; Elroy-Stein, O.; Ehrlich, M. Dicodon monitoring of protein synthesis (DiCoMPS) reveals levels of synthesis of a viral protein in single cells. Nucleic Acids Res. 2013, 41, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Pampillo, M.; Guo, F.; Liu, S.; Cooperman, B.S.; Farrell, I.; Dahary, D.; Gan, B.S.; O’Gorman, D.B.; Smilansky, Z.; et al. Monitoring collagen synthesis in fibroblasts using fluorescently labeled tRNA pairs. J. Cell Physiol. 2014, 229, 1121–1129. [Google Scholar] [CrossRef]
- Volkov, I.L.; Lindén, M.; Aguirre Rivera, J.; Ieong, K.W.; Metelev, M.; Elf, J.; Johansson, M. tRNA tracking for direct measurements of protein synthesis kinetics in live cells. Nat. Chem. Biol. 2018, 14, 618–626. [Google Scholar] [CrossRef]
- Meyer, B.; Wurm, J.P.; Sharma, S.; Immer, C.; Pogoryelov, D.; Kötter, P.; Lafontaine, D.L.J.; Wöhnert, J.; Entian, K.-D. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 2016, 44, 4304–4316. [Google Scholar] [CrossRef] [Green Version]
- Umitsu, M.; Nishimasu, H.; Noma, A.; Suzuki, T.; Ishitani, R.; Nureki, O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc. Natl. Acad. Sci. USA 2009, 106, 15616–15621. [Google Scholar] [CrossRef] [Green Version]
- Meyer, B.; Wurm, J.P.; Kötter, P.; Leisegang, M.S.; Schilling, V.; Buchhaupt, M.; Held, M.; Bahr, U.; Karas, M.; Heckel, et.al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA. Nucleic Acids Res. 2011, 39, 1526–1537. [Google Scholar] [CrossRef]
- Fei, J.; Wang, J.; Sternberg, S.H.; MacDougall, D.D.; Elvekrog, M.M.; Pulukkunat, D.K.; Englander, M.T.; Gonzalez, R.L., Jr. A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. Methods Enzymol. 2010, 472, 221–259. [Google Scholar]
- Blanchard, S.C.; Kim, H.D.; Gonzalez, R.L., Jr.; Puglisi, J.D.; Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA 2004, 101, 12893–12898. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, S.C.; Gonzalez, R.L.; Kim, H.D.; Chu, S.; Puglisi, J.D. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004, 11, 1008–1014. [Google Scholar] [CrossRef]
- Wasserman, M.R.; Alejo, J.L.; Altman, R.B.; Blanchard, S.C. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 2016, 23, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Fei, J.; Gonzalez, R.L., Jr. The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc. Natl. Acad. Sci. USA 2014, 111, 12073–12078. [Google Scholar] [CrossRef] [Green Version]
- Tsai, A.; Puglisi, J.D.; Uemura, S. Probing the Translation Dynamics of Ribosomes Using Zero-Mode Waveguides. Prog. Mol. Biol. Transl. Sci. 2016, 139, 1–43. [Google Scholar] [PubMed]
- Bimai, O.; Arragain, S.; Golinelli-Pimpaneau, B. Structure-based mechanistic insights into catalysis by tRNA thiolation enzymes. Curr. Opin. Struct. Biol. 2020, 65, 69–78. [Google Scholar] [CrossRef]
- Lauhon, C.T.; Erwin, W.M.; Ton, G.N. Substrate specificity for 4-thiouridine modification in Escherichia coli. J. Biol. Chem. 2004, 279, 23022–23029. [Google Scholar] [CrossRef] [Green Version]
- Neumann, P.; Lakomek, K.; Naumann, P.T.; Erwin, W.M.; Lauhon, C.T.; Ficner, R. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014, 42, 6673–6685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Shang, J.; Qin, Z.; Tong, A.; Xiang, Y. Selective and sensitive fluorescence “turn-on” detection of 4-thiouridine in nucleic acids via oxidative amination. Chem. Commun. 2019, 55, 13096–13099. [Google Scholar] [CrossRef] [PubMed]
- Bou-Nader, C.; Pecqueur, L.; Barraud, P.; Fontecave, M.; Tisné, C.; Sacquin-Mora, S.; Hamdane, D. Conformational Stability Adaptation of a Double-Stranded RNA-Binding Domain to Transfer RNA Ligand. Biochemistry 2019, 58, 2463–2473. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Innovation by Evolution: Bringing New Chemistry to Life (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2019, 58, 14420–14426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motorin, Y.; Burhenne, J.; Teimer, R.; Koynov, K.; Willnow, S.; Weinhold, E.; Helm, M. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011, 39, 1943–1952. [Google Scholar] [CrossRef] [PubMed]
- Ovcharenko, A.; Weissenboeck, F.P.; Rentmeister, A. Tag-Free Internal RNA Labeling and Photocaging Based on mRNA Methyltransferases. Angew. Chem. Int. Ed. Engl. 2020. [Google Scholar] [CrossRef]
- Gu, X.; Santi, D.V. The T-arm of tRNA is a substrate for tRNA (m5U54)-methyltransferase. Biochemistry 1991, 30, 2999–3002. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.S.; Zoltek, M.A.; Simon, M.D. Reengineering a tRNA Methyltransferase To Covalently Capture New RNA Substrates. J. Am. Chem. Soc. 2019, 141, 17460–17465. [Google Scholar] [CrossRef]
- Busby, K.N.; Devaraj, N.K. Enzymatic covalent labeling of RNA with RNA transglycosylation at guanosine (RNA-TAG). Methods Enzym. 2020, 641, 373–399. [Google Scholar]
- Xiao, L.; Habibian, M.; Kool, E.T. Site-Selective RNA Functionalization via DNA-Induced Structure. J. Am. Chem. Soc. 2020, 142, 16357–16363. [Google Scholar] [CrossRef]
- Zhao, M.; Steffen, F.D.; Börner, R.; Schaffer, M.F.; Sigel, R.K.O.; Freisinger, E. Site-specific dual-color labeling of long RNAs for single-molecule spectroscopy. Nucleic Acids Res. 2018, 46, e13. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Börner, R.; Sigel, R.K.O.; Freisinger, E. Site-Specific Dual-Color Labeling of Long RNAs. Methods Mol Biol. 2020, 2106, 253–270. [Google Scholar]
- Li, Y.; Fin, A.; McCoy, L.; Tor, Y. Polymerase-Mediated Site-Specific Incorporation of a Synthetic Fluorescent Isomorphic G Surrogate into RNA. Angew. Chem. Int. Ed. 2017, 56, 1303–1307. [Google Scholar] [CrossRef] [Green Version]
- Egloff, D.; Oleinich, I.A.; Zhao, M.; König, S.L.; Sigel, R.K.; Freisinger, E. Sequence-Specific Post-Synthetic Oligonucleotide Labeling for Single-Molecule Fluorescence Applications. ACS Chem. Biol. 2016, 11, 2558–2567. [Google Scholar] [CrossRef] [PubMed]
- Ghaem Maghami, M.; Scheitl, C.P.M.; Höbartner, C. Direct in Vitro Selection of Trans.-Acting Ribozymes for Posttranscriptional, Site-Specific, and Covalent Fluorescent Labeling of RNA. J. Am. Chem. Soc. 2019, 141, 19546–19549. [Google Scholar] [CrossRef] [Green Version]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ghaem Maghami, M.; Dey, S.; Lenz, A.K.; Höbartner, C. Repurposing Antiviral Drugs for Orthogonal RNA-Catalyzed Labeling of RNA. Angew Chem. Int. Ed. Engl. 2020, 59, 9335–9339. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Holmstrom, E.; Yu, P.; Tan, K.; Zuo, X.; Nesbitt, D.J.; Sousa, R.; Stagno, J.R.; Wang, Y.X. Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA. Nat. Protoc. 2018, 13, 987–1005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, M.; Liu, Y. Optimization and characterization of position-selective labelling of RNA (PLOR) for diverse RNA and DNA sequences. RNA Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, M. Optimization of N-hydroxysuccinimide ester coupling with aminoallyl-modified RNA for fluorescent labeling. Bioengineered 2020, 11, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Feldman, A.W.; Romesberg, F.E. Expansion of the Genetic Alphabet: A Chemist’s Approach to Synthetic Biology. Acc. Chem. Res. 2018, 51, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Kimoto, M.; Hirao, I. Genetic alphabet expansion technology by creating unnatural base pairs. Chem. Soc. Rev. 2020, 49, 7602–7626. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, T.; Kimoto, M.; Kawai, R.; Yokoyama, S.; Hirao, I. Characterization of fluorescent, unnatural base pairs. Tetrahedron 2007, 63, 3528–3537. [Google Scholar] [CrossRef]
- Kimoto, M.; Mitsui, T.; Yamashige, R.; Sato, A.; Yokoyama, S.; Hirao, I. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 2010, 132, 15418–15426. [Google Scholar] [CrossRef]
- Someya, T.; Ando, A.; Kimoto, M.; Hirao, I. Site-specific labeling of RNA by combining genetic alphabet expansion transcription and copper-free click chemistry. Nucleic Acids Res. 2015, 43, 6665–6676. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cooperman, B.S. Site-Specific Fluorescent Labeling of RNA Interior Positions. Molecules 2021, 26, 1341. https://doi.org/10.3390/molecules26051341
Cooperman BS. Site-Specific Fluorescent Labeling of RNA Interior Positions. Molecules. 2021; 26(5):1341. https://doi.org/10.3390/molecules26051341
Chicago/Turabian StyleCooperman, Barry S. 2021. "Site-Specific Fluorescent Labeling of RNA Interior Positions" Molecules 26, no. 5: 1341. https://doi.org/10.3390/molecules26051341
APA StyleCooperman, B. S. (2021). Site-Specific Fluorescent Labeling of RNA Interior Positions. Molecules, 26(5), 1341. https://doi.org/10.3390/molecules26051341





