Clinical Amyloid Typing by Proteomics: Performance Evaluation and Data Sharing between Two Centres
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
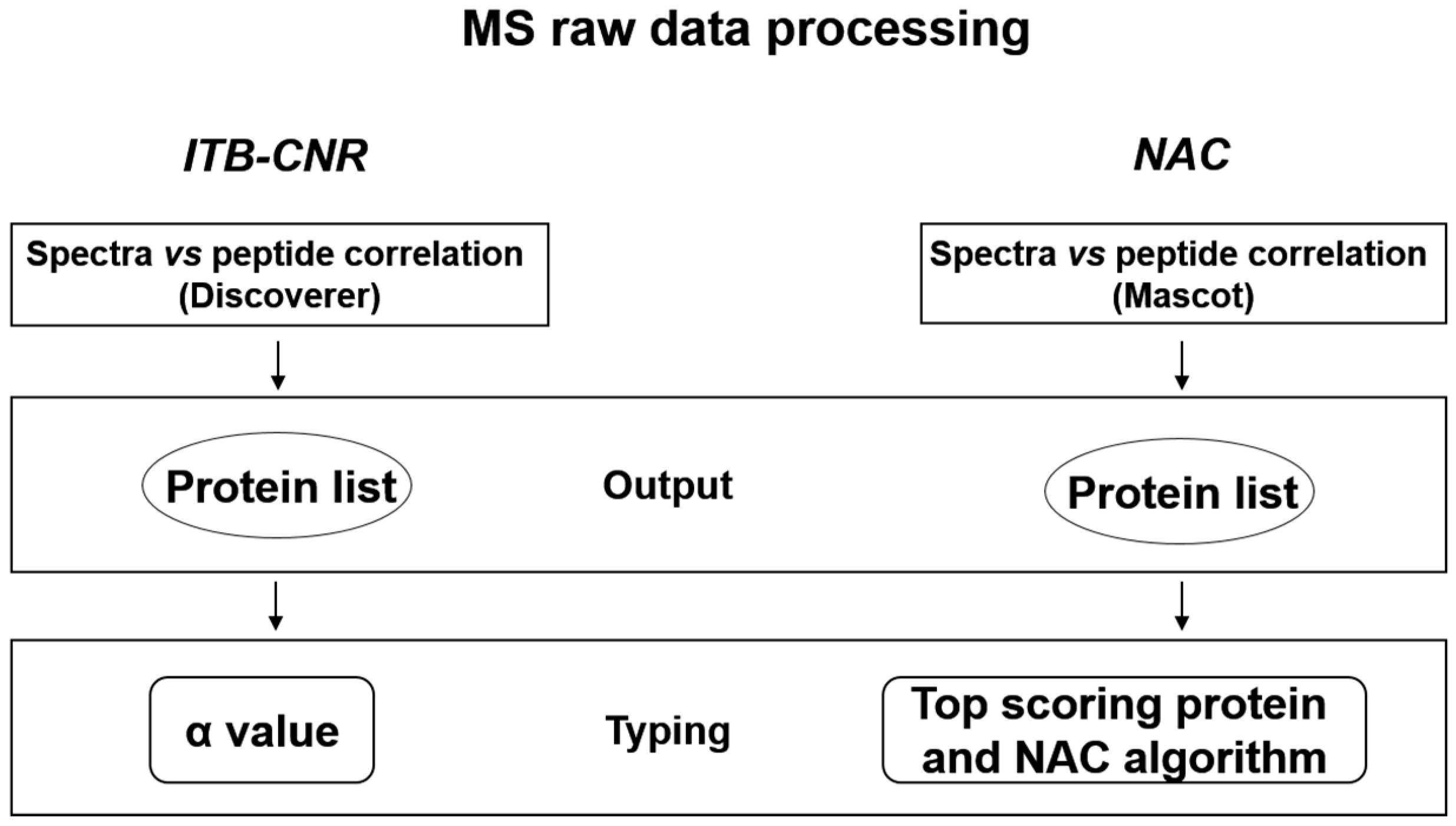
4.1. Proteomics Analysis at NAC
4.2. Proteomics Data Analysis at ITB-CNR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Pepys, M.B. Amyloidosis. Annu. Rev. Med. 2006, 57, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Pinney, J.H.; Whelan, C.J.; Petrie, A.; Dungu, J.; Banypersad, S.M.; Sattianayagam, P.; Wechalekar, A.; Gibbs, S.D.; Venner, C.P.; Wassef, N.; et al. Senile systemic amyloidosis: Clinical features at presentation and outcome. J. Am. Heart Assoc. 2013, 2, e000098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2018: Recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2018, 25, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puchtler, H.; Waldrop, F.S.; Meloan, S.N. A review of light, polarization and fluorescence microscopic methods for amyloid. Appl. Pathol. 1985, 3, 5–17. [Google Scholar] [PubMed]
- Linke, R.P. On typing amyloidosis using immunohistochemistry. Detailed illustrations, review and a note on mass spectrometry. Prog. Histochem. Cytochem. 2012, 47, 61–132. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, J.A.; Hunt, T.; Hawkins, P.N. Amyloid Typing: Experience from a Large Referral Centre. In Amyloid and Related Disorders; Picken, M., Dogan, A., Herrera, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 231–238. [Google Scholar]
- Theis, J.D.; Dasari, S.; Vrana, J.A.; Kurtin, P.J.; Dogan, A. Shotgun-proteomics-based clinical testing for diagnosis and classification of amyloidosis. J. Mass Spectrom. 2013, 48, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, F.; Lavatelli, F.; Merlini, G.; Mauri, P. Clinical proteomics for diagnosis and typing of systemic amyloidosis. Proteom. Clin. Appl. 2013, 7, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Vrana, J.A.; Gamez, J.D.; Madden, B.J.; Theis, J.D.; Bergen, H.R., 3rd; Dogan, A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood 2009, 114, 4957–4959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, F.; Lavatelli, F.; Di Silvestre, D.; Valentini, V.; Rossi, R.; Palladini, G.; Obici, L.; Verga, L.; Mauri, P.; Merlini, G. Reliable typing of systemic amyloidosis through proteomic analysis of subcutaneous adipose tissue. Blood 2012, 119, 1844–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canetti, D.; Rendell, N.B.; Gilbertson, J.A.; Botcher, N.; Nocerino, P.; Blanco, A.; Di Vagno, L.; Rowczenio, D.; Verona, G.; Mangione, P.P.; et al. Diagnostic amyloid proteomics: Experience of the UK National Amyloidosis Centre. Clin. Chem. Lab. Med. 2020, 58, 948–957. [Google Scholar] [CrossRef]
- Chapman, J.R.; Liu, A.; Yi, S.S.; Hernandez, E.; Ritorto, M.S.; Jungbluth, A.A.; Pulitzer, M.; Dogan, A. Proteomic analysis shows that the main constituent of subepidermal localised cutaneous amyloidosis is not galectin-7. Amyloid 2020, 1–7. [Google Scholar] [CrossRef]
- Brambilla, F.; Lavatelli, F.; Di Silvestre, D.; Valentini, V.; Palladini, G.; Merlini, G.; Mauri, P. Shotgun protein profile of human adipose tissue and its changes in relation to systemic amyloidosis. J. Proteome. Res. 2013, 12, 5642–5655. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Tholey, A.; Krüger, S.; Schmidt, H.; Röcken, C. MALDI-Mass Spectrometry Imaging Identifies Vitronectin as a Common Constituent of Amyloid Deposits. J. Histochem. Cytochem. 2015, 63, 772–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollee, P.; Boros, S.; Loo, D.; Ruelcke, J.E.; Lakis, V.A.; Lê Cao, K.-A.; Renaut, P.; Hill, M.M. Implementation and evaluation of amyloidosis subtyping by laser-capture microdissection and tandem mass spectrometry. Clin. Proteom. 2016, 13, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canetti, D.; Rendell, N.B.; Di Vagno, L.; Gilbertson, J.A.; Rowczenio, D.; Rezk, T.; Gillmore, J.D.; Hawkins, P.N.; Verona, G.; Mangione, P.P.; et al. Misidentification of transthyretin and immunoglobulin variants by proteomics due to methyl lysine formation in formalin-fixed paraffin-embedded amyloid tissue. Amyloid 2017, 24, 233–241. [Google Scholar] [CrossRef] [PubMed]
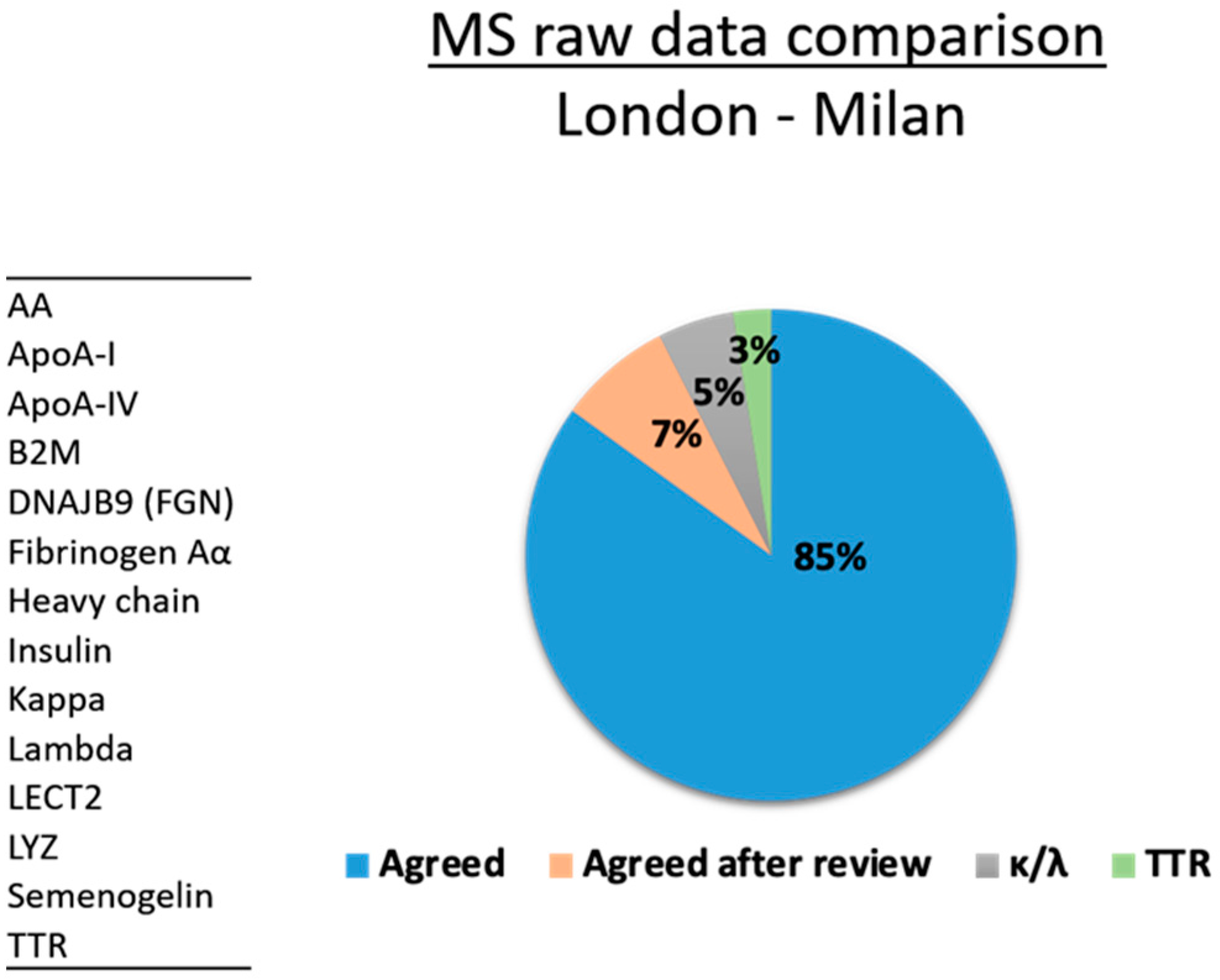
| Sample No. | NAC | ITB-CNR | Tissue Type | Agreed |
|---|---|---|---|---|
| 1 | LECT2 | LECT2 | renal | Y |
| 2 | LYZ | LYZ | renal | Y |
| 3 | Uncertain | LYZ/AL (λ)/Heavy chain | renal | Y |
| 4 | AL (λ) | AL (λ) | renal | Y |
| 5 | AA | AA | renal | Y |
| 6 | DNJB9 (FGN) | DNJB9 (FGN) | renal | Y |
| 7 | DNJB9 (FGN) | DNJB9 (FGN) | renal | Y |
| 8 | Fibrinogen Aα | Fibrinogen Aα | renal | Y |
| 9 | Heavy chain | Heavy chain | renal | Y |
| 10 | AL (κ) | AL (κ) | renal | Y |
| 11 | Fibrinogen Aα | Fibrinogen Aα | renal | Y |
| 12 | AL (λ) | AL (λ) | cardiac | Y |
| 13 | TTR | TTR | cardiac | Y |
| 14 | TTR | TTR | cardiac | Y |
| 15 | ApoA-IV | ApoA-IV | cardiac | Y |
| 16 | TTR | TTR | myocardial | Y |
| 17 | Insulin | Insulin | soft tissue | Y |
| 18 | AL (λ) | AL (λ) | soft tissue | Y |
| 19 | Heavy chain | Heavy chain | soft tissue | Y |
| 20 | AL (λ) | AL (λ) | lung parenchyma | Y |
| 21 | AL (κ) | AL (κ)/(λ) | lung parenchyma | N |
| 22 | B2M | B2M | liver | Y |
| 23 | LYZ | LYZ | liver | Y |
| 24 | AL (λ) | AL (λ) | salivary gland | Y |
| 25 | ApoA-I | TTR | bone marrow trephine | N |
| 26 | AA | AA | thyroid | Y |
| 27 | AL (λ) | AL (λ) | jejunal | Y |
| 28 | TTR | TTR | pancreas | Y |
| 29 | Semenogelin | Semenogelin | prostate | Y |
| 30 | AL (κ) | AL (κ) | vocal cord | Y |
| 31 | AL (κ) | AL (κ) | tongue | Y |
| 32 | AL (λ) | AL (λ) | urethral | Y |
| 33 | AL (κ) | AL (κ)/(λ) | skin | N |
| 34 | AL (λ) | AL (λ) | fat aspirates | Y |
| 35 | No Amyloid Signature | FibAα/AL (κ)/no Amyloid Signature | fat aspirates | Y |
| 36 | AL (λ) | AL (λ) | fat aspirates | Y |
| 37 | AL (λ) | AL (λ) | fat aspirates | Y |
| 38 | TTR | TTR | fat aspirates | Y |
| 39 | TTR | TTR | fat aspirates | Y |
| 40 | No Amyloid Signature | AL(λ)/FibAα/DNAJB9/no Amyloid Signature | fat aspirates | Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canetti, D.; Brambilla, F.; Rendell, N.B.; Nocerino, P.; Gilbertson, J.A.; Di Silvestre, D.; Bergamaschi, A.; Lavatelli, F.; Merlini, G.; Gillmore, J.D.; et al. Clinical Amyloid Typing by Proteomics: Performance Evaluation and Data Sharing between Two Centres. Molecules 2021, 26, 1913. https://doi.org/10.3390/molecules26071913
Canetti D, Brambilla F, Rendell NB, Nocerino P, Gilbertson JA, Di Silvestre D, Bergamaschi A, Lavatelli F, Merlini G, Gillmore JD, et al. Clinical Amyloid Typing by Proteomics: Performance Evaluation and Data Sharing between Two Centres. Molecules. 2021; 26(7):1913. https://doi.org/10.3390/molecules26071913
Chicago/Turabian StyleCanetti, Diana, Francesca Brambilla, Nigel B. Rendell, Paola Nocerino, Janet A. Gilbertson, Dario Di Silvestre, Andrea Bergamaschi, Francesca Lavatelli, Giampaolo Merlini, Julian D. Gillmore, and et al. 2021. "Clinical Amyloid Typing by Proteomics: Performance Evaluation and Data Sharing between Two Centres" Molecules 26, no. 7: 1913. https://doi.org/10.3390/molecules26071913
APA StyleCanetti, D., Brambilla, F., Rendell, N. B., Nocerino, P., Gilbertson, J. A., Di Silvestre, D., Bergamaschi, A., Lavatelli, F., Merlini, G., Gillmore, J. D., Bellotti, V., Mauri, P., & Taylor, G. W. (2021). Clinical Amyloid Typing by Proteomics: Performance Evaluation and Data Sharing between Two Centres. Molecules, 26(7), 1913. https://doi.org/10.3390/molecules26071913









