Insight into Gentisic Acid Antidiabetic Potential Using In Vitro and In Silico Approaches
Abstract
:1. Introduction
2. Results and Discussion
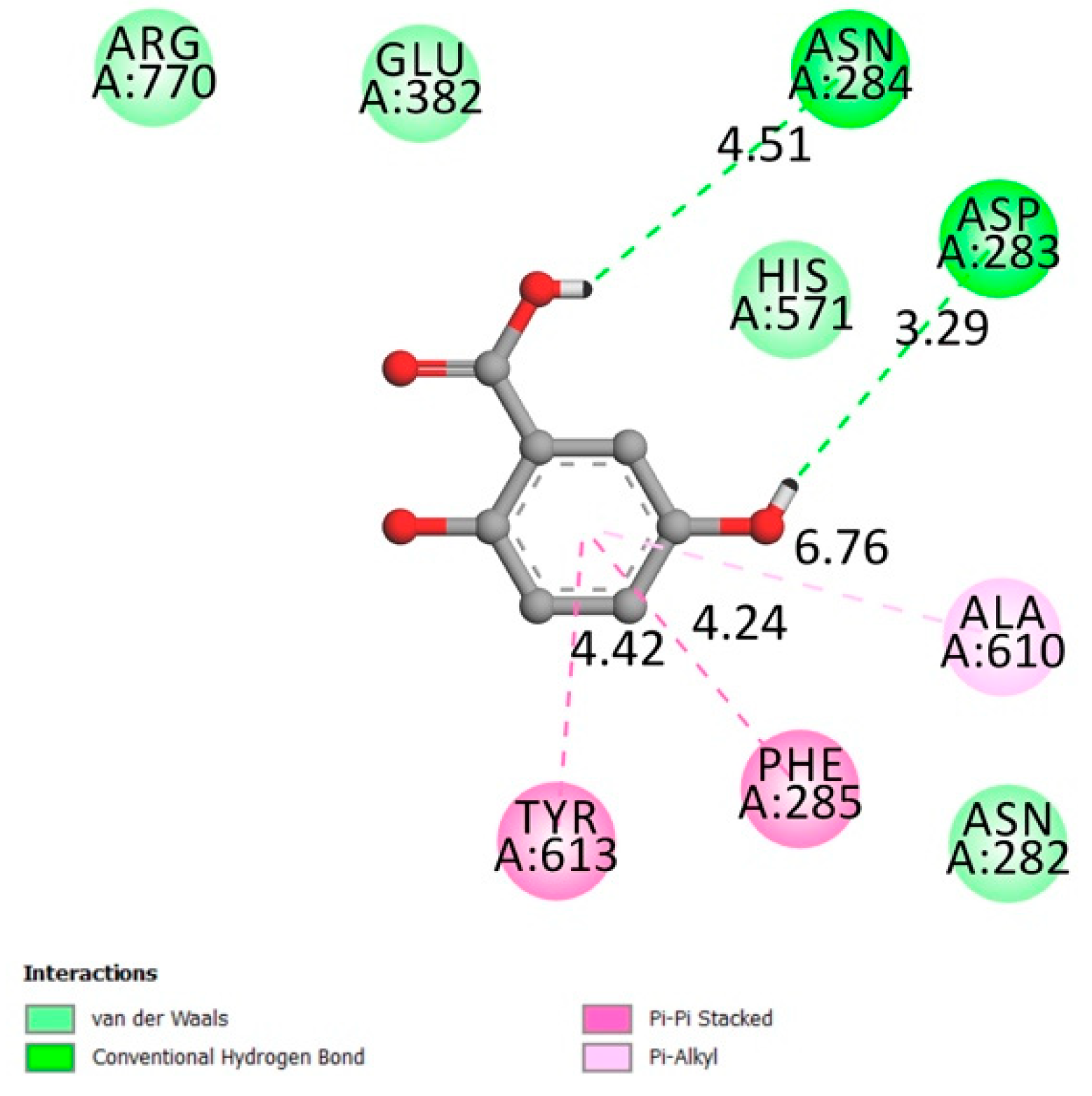
2.1. Molecular Docking
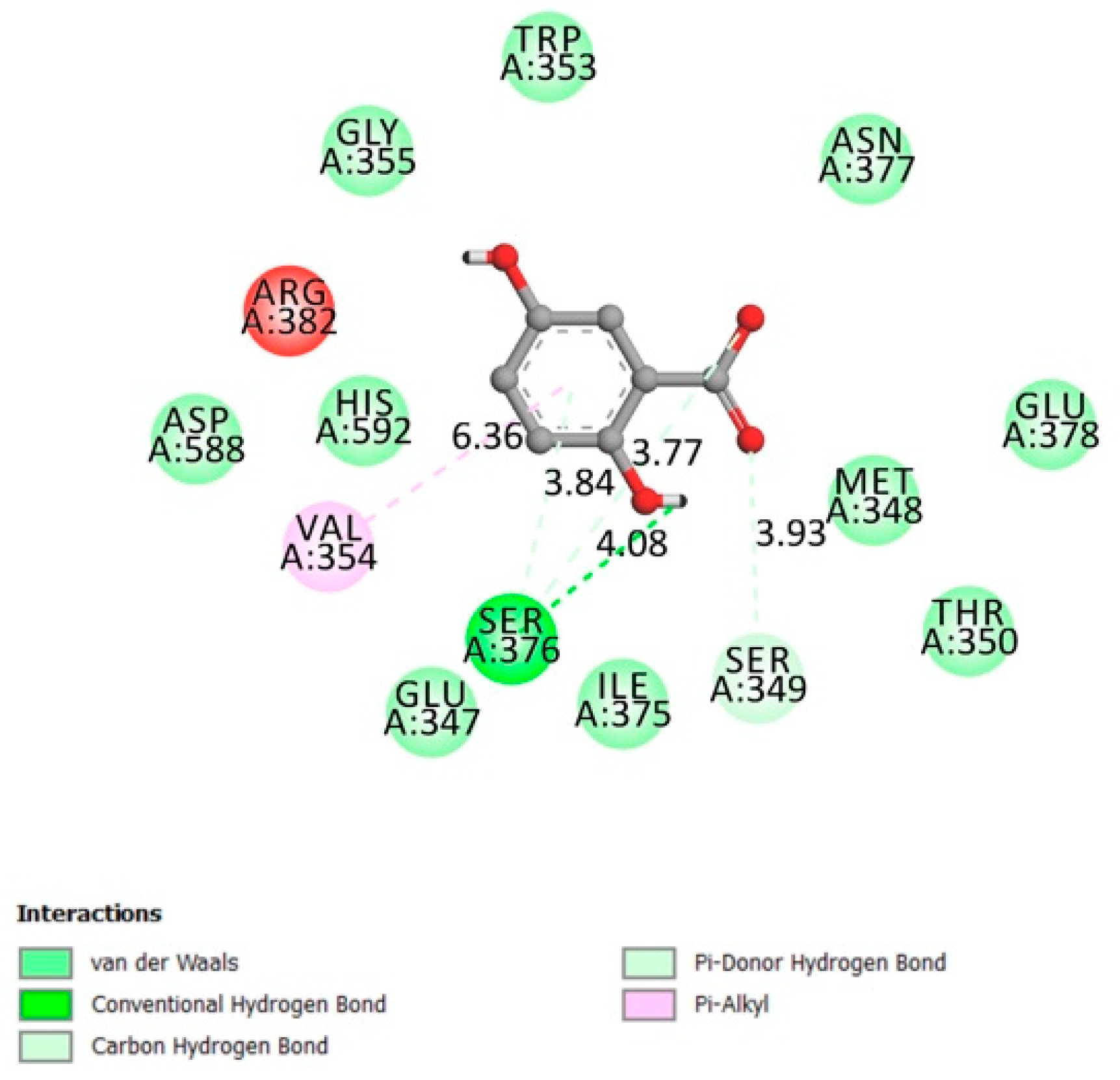
2.1.1. Protein Tyrosine Phosphatase 1B (PTP1B)
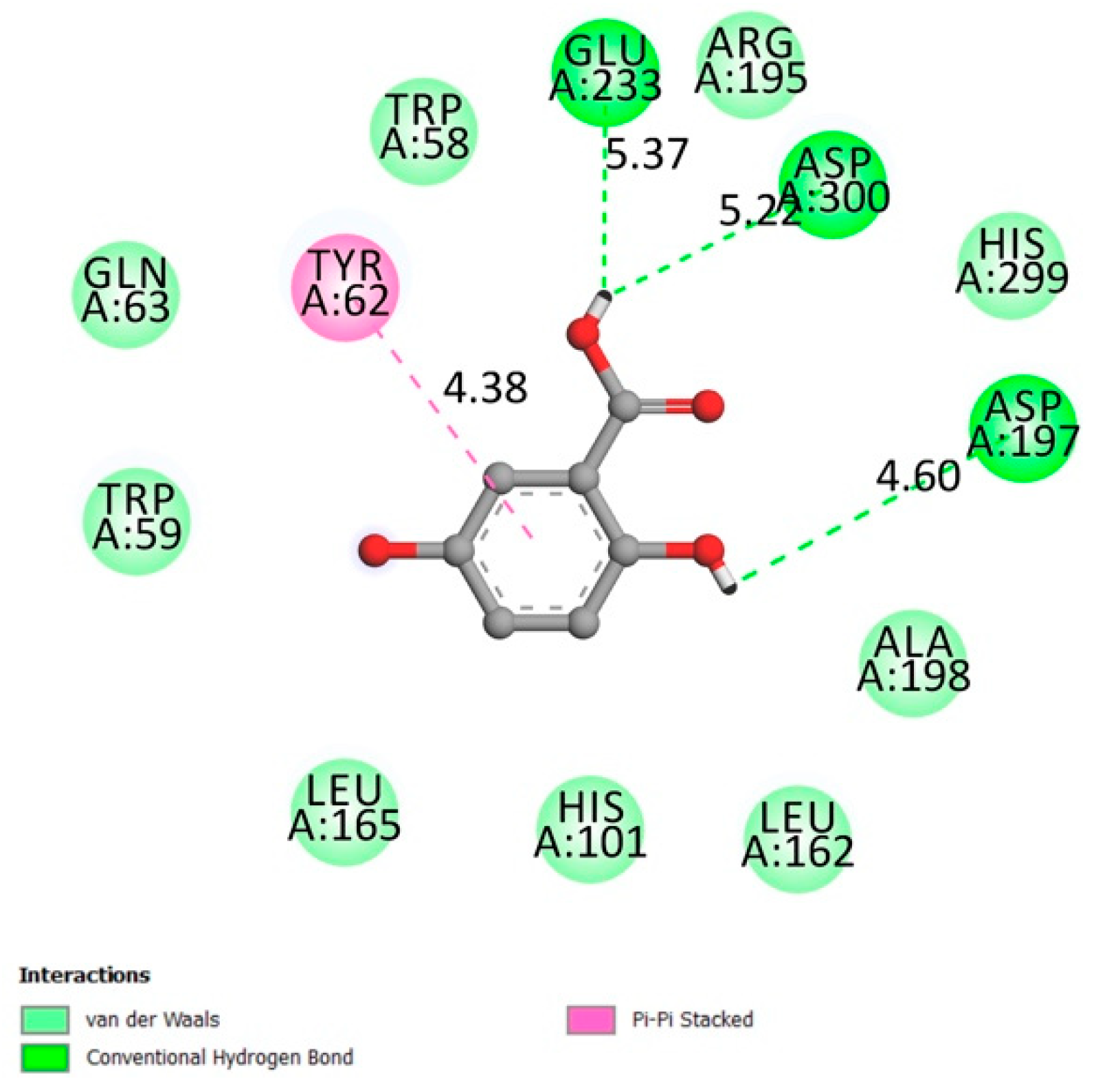
2.1.2. Dipeptidyl-Peptidase 4 (DPP4)
2.1.3. Free Fatty Acid Receptor 1 (FFAR1)
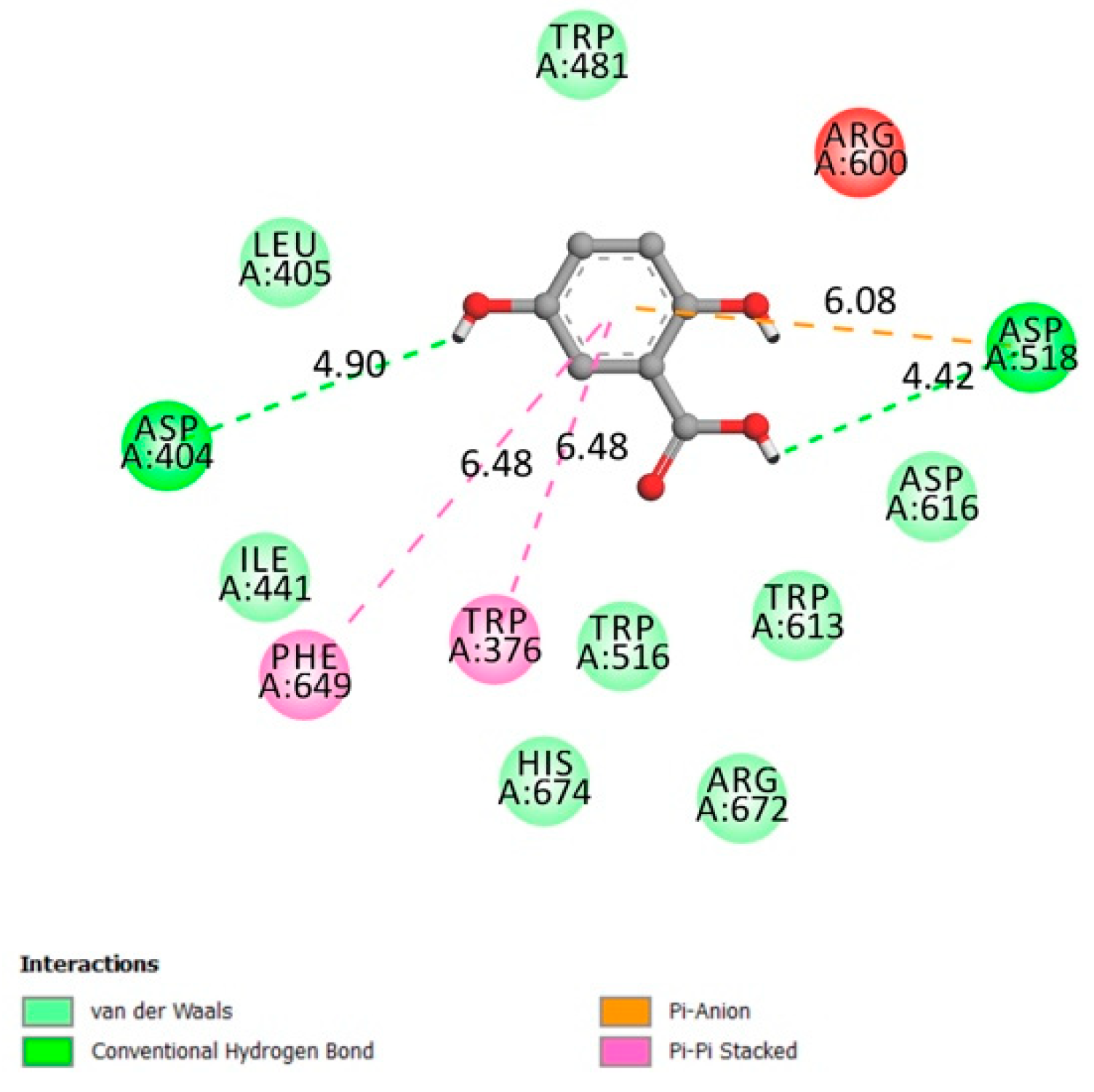
2.1.4. α-Amylase
2.1.5. Peroxisome Proliferator-Activated Receptor Gamma (PPARγ)
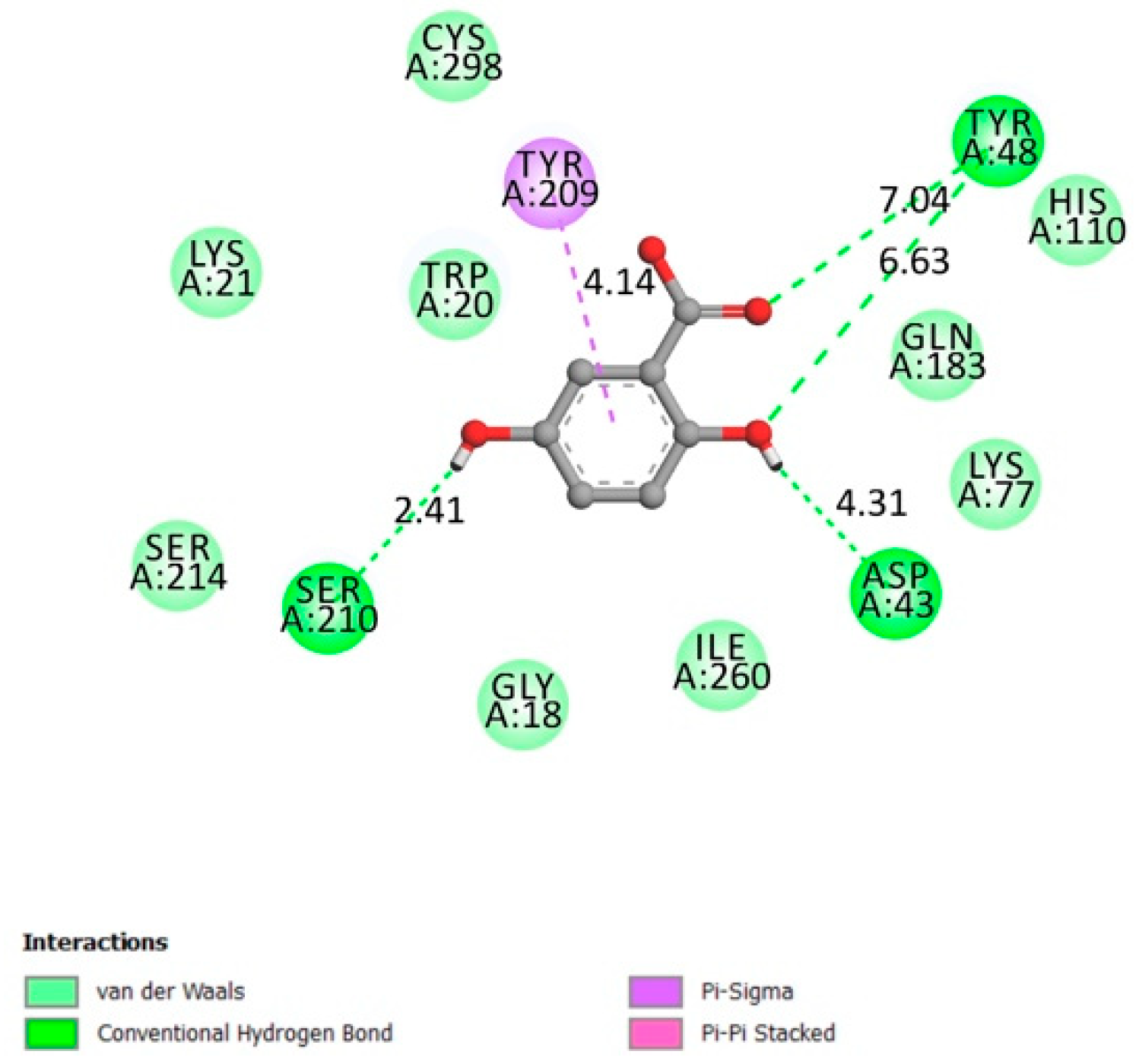
2.1.6. α-Glucosidase
2.1.7. Aldose Reductase
2.1.8. Glycogen Phosphorylase
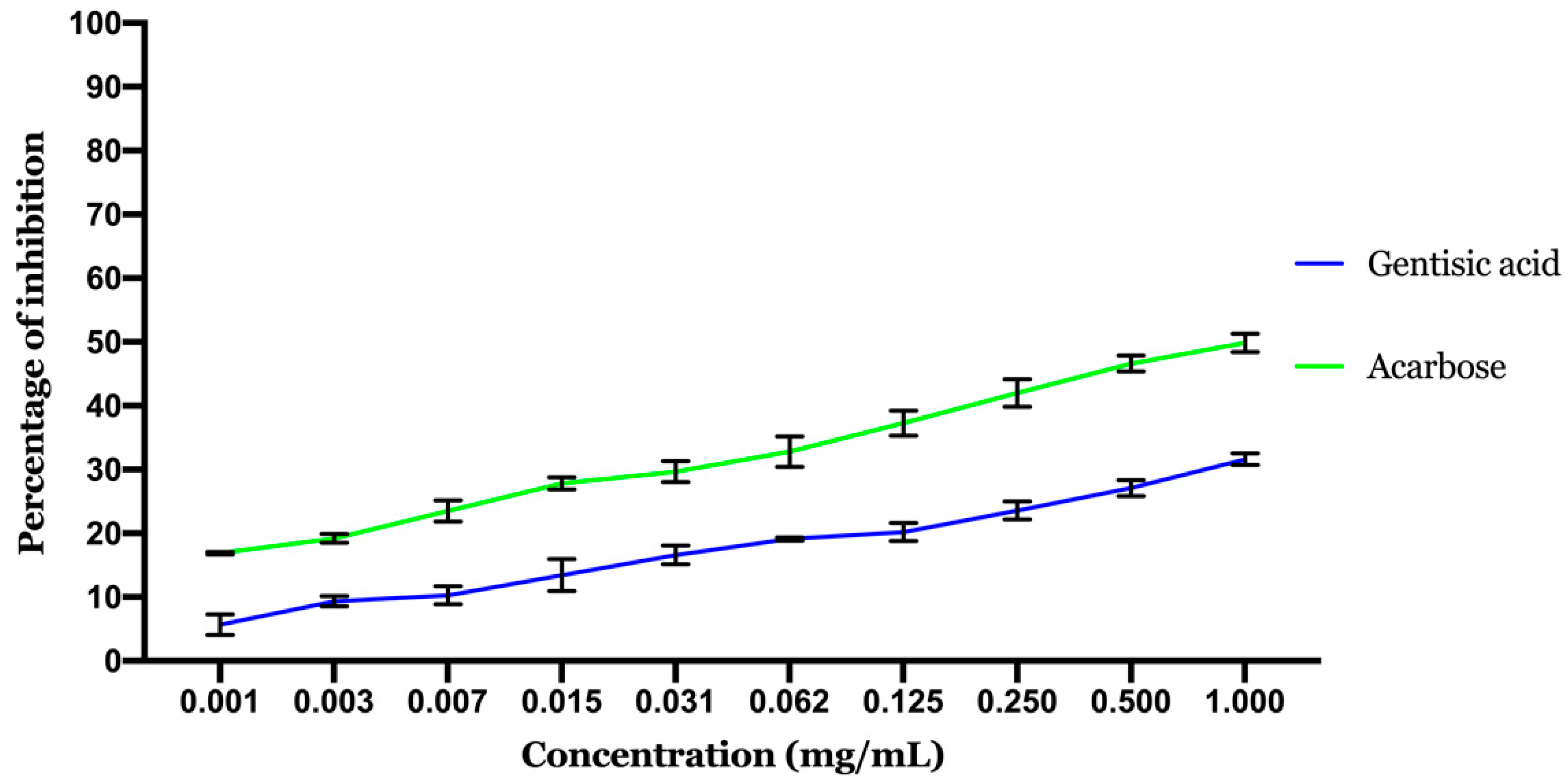
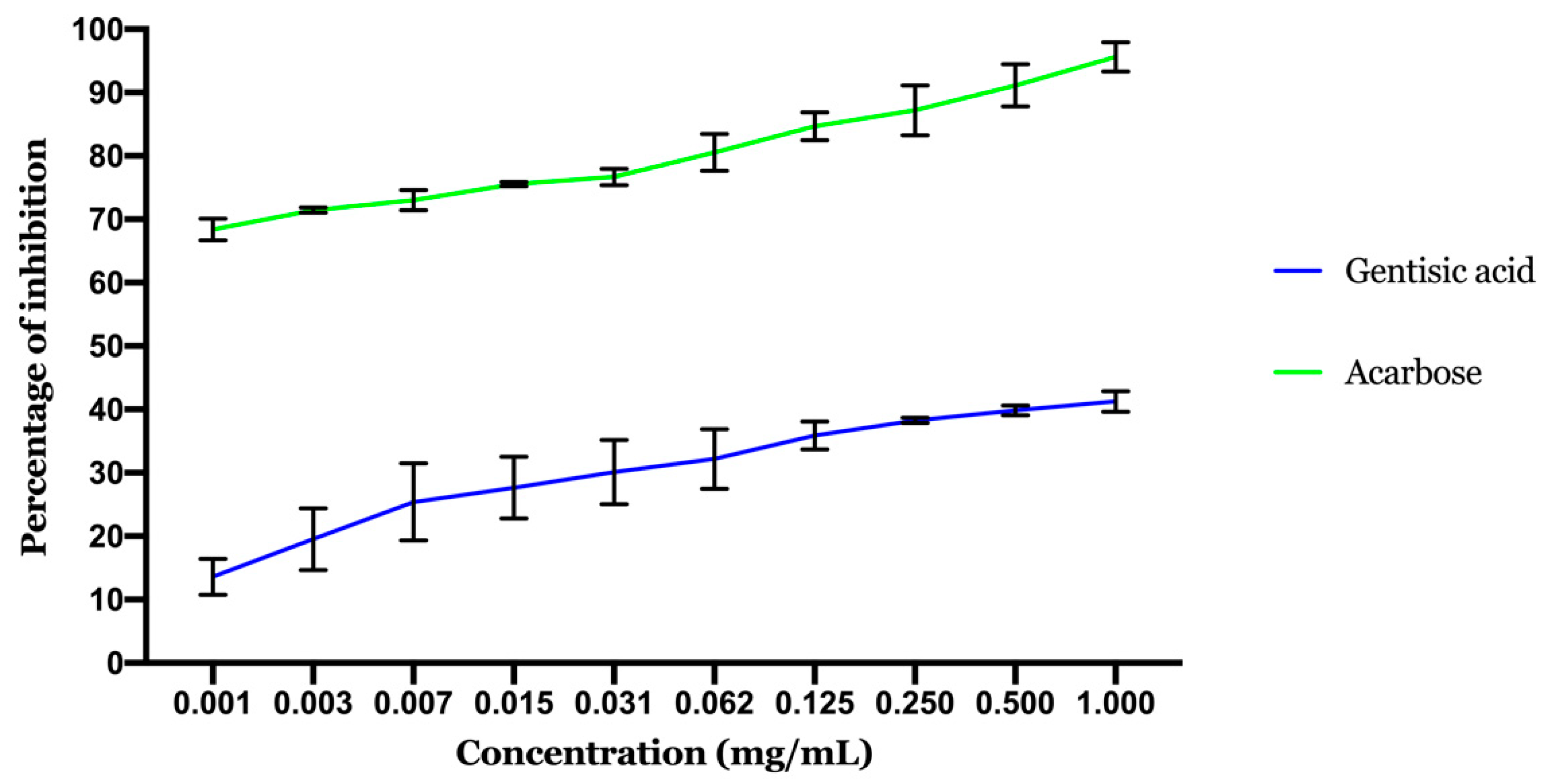
2.2. In Vitro Assays
2.2.1. Inhibitory Effect of α-Amylase
2.2.2. Inhibitory Effect of α-Glucosidase
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Molecular Docking
3.2.1. Ligand Preparation
3.2.2. Receptors’ Preparation
3.2.3. Docking Simulations
3.3. Enzymes In Vitro Inhibition
3.3.1. Assay of the α-Amylase Inhibitory Effect
3.3.2. Assay of the α-Glucosidase Inhibitory Effect
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Es-Safi, I.; Mechchate, H.; Amaghnouje, A.; Jawhari, F.Z.; Bari, A.; Cerruti, P.; Avella, M.; Andriy, A.; Andriy, D. Medicinal Plants Used to Treat Acute Digestive System Problems in the Region of Fez-Meknes in Morocco: An Ethnopharmacological Survey. Ethnobot. Res. Appl. 2020, 20. [Google Scholar] [CrossRef]
- Parveen, A.; Parveen, B.; Parveen, R.; Ahmad, S. Challenges and Guidelines for Clinical Trial of Herbal Drugs. J. Pharm. Bioallied Sci. 2015, 7, 329. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.-M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Süntar, I. Importance of Ethnopharmacological Studies in Drug Discovery: Role of Medicinal Plants. Phytochem. Rev. 2020, 19, 1199–1209. [Google Scholar] [CrossRef]
- Thomford, N.; Senthebane, D.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery. IJMS 2018, 19, 1578. [Google Scholar] [CrossRef] [Green Version]
- Kiokias, S.; Proestos, C.; Oreopoulou, V. Phenolic Acids of Plant Origin—A Review on Their Antioxidant Activity In Vitro (O/W Emulsion Systems) Along with Their in Vivo Health Biochemical Properties. Foods 2020, 9, 534. [Google Scholar] [CrossRef]
- Russell, W.; Duthie, G. Plant Secondary Metabolites and Gut Health: The Case for Phenolic Acids. Proc. Nutr. Soc. 2011, 70, 389–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morse, J.M. The Significance of Saturation. Qual. Health. Res. 1995, 5, 147–149. [Google Scholar] [CrossRef]
- Abedi, F.; Razavi, B.M.; Hosseinzadeh, H. A Review on Gentisic Acid as a Plant Derived Phenolic Acid and Metabolite of Aspirin: Comprehensive Pharmacology, Toxicology, and Some Pharmaceutical Aspects. Phytother. Res. 2020, 34, 729–741. [Google Scholar] [CrossRef]
- Shivanagoudra, S.R.; Perera, W.H.; Perez, J.L.; Athrey, G.; Sun, Y.; Wu, C.S.; Jayaprakasha, G.K.; Patil, B.S. In Vitro and in Silico Elucidation of Antidiabetic and Anti-Inflammatory Activities of Bioactive Compounds from Momordica charantia L. Bioorg. Med. Chem. 2019, 27, 3097–3109. [Google Scholar] [CrossRef]
- Mullen, W.; Edwards, C.A.; Crozier, A. Absorption, Excretion and Metabolite Profiling of Methyl-, Glucuronyl-, Glucosyl- and Sulpho-Conjugates of Quercetin in Human Plasma and Urine after Ingestion of Onions. Br. J. Nutr. 2006, 96, 107. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Yang, C.; Gong, M.; Ke, Y.; Yuan, P.; Wang, X.; Li, M.; Zheng, X.; Feng, W. Antidiabetic Activity and Potential Mechanism of Amentoflavone in Diabetic Mice. Molecules 2019, 24, 2184. [Google Scholar] [CrossRef] [Green Version]
- Elchebly, M. Increased Insulin Sensitivity and Obesity Resistance in Mice Lacking the Protein Tyrosine Phosphatase-1B Gene. Science 1999, 283, 1544–1548. [Google Scholar] [CrossRef]
- Goldstein, B.J. Protein-Tyrosine Phosphatase 1B (PTP1B): A Novel Therapeutic Target for Type 2 Diabetes Mellitus, Obesity and Related States of Insulin Resistance. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2001, 1, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Klaman, L.D.; Boss, O.; Peroni, O.D.; Kim, J.K.; Martino, J.L.; Zabolotny, J.M.; Moghal, N.; Lubkin, M.; Kim, Y.-B.; Sharpe, A.H.; et al. Increased Energy Expenditure, Decreased Adiposity, and Tissue-Specific Insulin Sensitivity in Protein-Tyrosine Phosphatase 1B-Deficient Mice. Mol. Cell. Biol. 2000, 20, 5479–5489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Barford, D.; Flint, A.; Tonks, N. Structural Basis for Phosphotyrosine Peptide Recognition by Protein Tyrosine Phosphatase 1B. Science 1995, 268, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.O.; Ermolieff, J.; Jirousek, M.R. Protein Tyrosine Phosphatase 1B Inhibitors for Diabetes. Nat. Rev. Drug Discov. 2002, 1, 696–709. [Google Scholar] [CrossRef]
- Wiesmann, C.; Barr, K.J.; Kung, J.; Zhu, J.; Erlanson, D.A.; Shen, W.; Fahr, B.J.; Zhong, M.; Taylor, L.; Randal, M.; et al. Allosteric Inhibition of Protein Tyrosine Phosphatase 1B. Nat. Struct. Mol. Biol. 2004, 11, 730–737. [Google Scholar] [CrossRef]
- Röhrborn, D. DPP4 in Diabetes. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Matteucci, E.; Giampietro, O. Dipeptidyl Peptidase-4 (CD26): Knowing the Function before Inhibiting the Enzyme. CMC 2009, 16, 2943–2951. [Google Scholar] [CrossRef]
- Bjelke, J.R.; Christensen, J.; Branner, S.; Wagtmann, N.; Olsen, C.; Kanstrup, A.B.; Rasmussen, H.B. Tyrosine 547 Constitutes an Essential Part of the Catalytic Mechanism of Dipeptidyl Peptidase IV. J. Biol. Chem. 2004, 279, 34691–34697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, C.-H.; Tsai, C.-H.; Lin, C.-H.; Chou, C.-Y.; Chen, X. Identification of Hydrophobic Residues Critical for DPP-IV Dimerization. Biochemistry 2006, 45, 7006–7012. [Google Scholar] [CrossRef]
- Ren, X.-M.; Cao, L.-Y.; Zhang, J.; Qin, W.-P.; Yang, Y.; Wan, B.; Guo, L.-H. Investigation of the Binding Interaction of Fatty Acids with Human G Protein-Coupled Receptor 40 Using a Site-Specific Fluorescence Probe by Flow Cytometry. Biochemistry 2016, 55, 1989–1996. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Yano, J.; Hirozane, Y.; Kefala, G.; Gruswitz, F.; Snell, G.; Lane, W.; Ivetac, A.; Aertgeerts, K.; Nguyen, J.; et al. High-Resolution Structure of the Human GPR40 Receptor Bound to Allosteric Agonist TAK-875. Nature 2014, 513, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Morgan, N.G.; Dhayal, S. G-Protein Coupled Receptors Mediating Long Chain Fatty Acid Signalling in the Pancreatic Beta-Cell. Biochem. Pharmacol. 2009, 78, 1419–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sum, C.S.; Tikhonova, I.G.; Costanzi, S.; Gershengorn, M.C. Two Arginine-Glutamate Ionic Locks Near the Extracellular Surface of FFAR1 Gate Receptor Activation. J. Biol. Chem. 2009, 284, 3529–3536. [Google Scholar] [CrossRef] [Green Version]
- Sum, C.S.; Tikhonova, I.G.; Neumann, S.; Engel, S.; Raaka, B.M.; Costanzi, S.; Gershengorn, M.C. Identification of Residues Important for Agonist Recognition and Activation in GPR40. J. Biol. Chem. 2007, 282, 29248–29255. [Google Scholar] [CrossRef] [Green Version]
- Suvd, D.; Fujimoto, Z.; Takase, K.; Matsumura, M.; Mizuno, H. Crystal Structure of Bacillus Stearothermophilus α-amylase: Possible Factors Determining the Thermostability. J. Biochem. 2001, 129, 461–468. [Google Scholar] [CrossRef]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Crystal Structures of the Psychrophilic α-amylase from Alteromonas Haloplanctis in Its Native Form and Complexed with an Inhibitor. Protein Sci. 1998, 7, 564–572. [Google Scholar] [CrossRef] [Green Version]
- Hsiu, J.; Fischer, E.H.; Stein, E.A. α-amylases As Calcium-Metalloenzymes. II. Calcium and the catalytic activity. Biochemistry 1964, 3, 61–66. [Google Scholar] [CrossRef]
- Ragunath, C.; Manuel, S.G.A.; Venkataraman, V.; Sait, H.B.R.; Kasinathan, C.; Ramasubbu, N. Probing the Role of Aromatic Residues at the Secondary Saccharide-Binding Sites of Human Salivary α-Amylase in Substrate Hydrolysis and Bacterial Binding. J. Mol. Biol. 2008, 384, 1232–1248. [Google Scholar] [CrossRef] [Green Version]
- Ramasubbu, N.; Ragunath, C.; Sundar, K.; Mishra, P.J.; Gyémánt, G.; Kandra, L. Structure-Function Relationships in Human Salivary α-Amylase: Role of Aromatic Residues. Biol. Sect. Cell. Mol. Biol. 2005, 60, 47–56. [Google Scholar]
- Ramasubbu, N.; Ragunath, C.; Mishra, P.J.; Thomas, L.M.; Gyemant, G.; Kandra, L. Human Salivary α-amylase Trp58 Situated at Subsite -2 Is Critical for Enzyme Activity. Eur. J. Biochem. 2004, 271, 2517–2529. [Google Scholar] [CrossRef]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARγ Signaling and Metabolism: The Good, the Bad and the Future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Pochetti, G.; Godio, C.; Mitro, N.; Caruso, D.; Galmozzi, A.; Scurati, S.; Loiodice, F.; Fracchiolla, G.; Tortorella, P.; Laghezza, A.; et al. Insights into the Mechanism of Partial Agonism: Crystal structures of the peroxisome proliferator-activated receptor γ ligand-binding domain in the complex with two enantiomeric ligands. J. Biol. Chem. 2007, 282, 17314–17324. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ghosh, S. Glucosidase. RCSB Protein Data Bank 2019. [Google Scholar] [CrossRef]
- Alqahtani, A.S.; Hidayathulla, S.; Rehman, M.T.; ElGamal, A.A.; Al-Massarani, S.; Razmovski-Naumovski, V.; Alqahtani, M.S.; El Dib, R.A.; AlAjmi, M.F. α-amylase and α-glucosidase Enzyme Inhibition and Antioxidant Potential of 3-Oxolupenal and Katononic Acid Isolated from Nuxia Oppositifolia. Biomolecules 2019, 10, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermans, M.M.; Kroos, M.A.; van Beeumen, J.; Oostra, B.A.; Reuser, A.J. Human Lysosomal α-glucosidase. Characterization of the Catalytic Site. J. Biol. Chem. 1991, 266, 13507–13512. [Google Scholar] [CrossRef]
- Tang, W.H.; Martin, K.A.; Hwa, J. Aldose Reductase, Oxidative Stress, and Diabetic Mellitus. Front. Pharmacol. 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Heather, L.C.; Clarke, K. Metabolism, Hypoxia and the Diabetic Heart. J. Mol. Cell. Cardiol. 2011, 50, 598–605. [Google Scholar] [CrossRef]
- Wilson, D.K.; Bohren, K.M.; Gabbay, K.H.; Quiocho, F.A. An Unlikely Sugar Substrate Site in the 1.65 A Structure of the Human Aldose Reductase Holoenzyme Implicated in Diabetic Complications. Science 1992, 257, 81–84. [Google Scholar] [CrossRef]
- Livanova, N.B.; Chebotareva, N.A.; Eronina, T.B.; Kurganov, B.I. Pyridoxal 5′-Phosphate as a Catalytic and Conformational Cofactor of Muscle Glycogen Phosphorylase B. Biochemistry 2002, 67, 1089–1098. [Google Scholar] [CrossRef]
- Barford, D.; Johnson, L.N. The Allosteric Transition of Glycogen Phosphorylase. Nature 1989, 340, 609–616. [Google Scholar] [CrossRef]
- Goldsmith, E.; Sprang; Hamlin, R.; Xuong, N.; Fletterick, R. Domain Separation in the Activation of Glycogen Phosphorylase A. Science 1989, 245, 528–532. [Google Scholar] [CrossRef]
- Mechchate, H.; Es-Safi, I.; Bourhia, M.; Kyrylchuk, A.; El Moussaoui, A.; Conte, R.; Ullah, R.; Ezzeldin, E.; Mostafa, G.A.; Grafov, A.; et al. In-Vivo Antidiabetic Activity and In-Silico Mode of Action of LC/MS-MS Identified Flavonoids in Oleaster Leaves. Molecules 2020, 25, 5073. [Google Scholar] [CrossRef] [PubMed]
- Thyagaraju, B.M. Muralidhara Ferulic Acid Supplements Abrogate Oxidative Impairments in Liver and Testis in the Streptozotocin-Diabetic Rat. Zool. Sci. 2008, 25, 854–860. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Goyal, R.K. Cardioprotective Effects of Gallic Acid in Diabetes-Induced Myocardial Dysfunction in Rats. Pharmacogn. Res. 2011, 3, 239. [Google Scholar]
- Min, S.W.; Han, J.S. Polyopes Lancifolia Extract, a Potent α-Glucosidase Inhibitor, Alleviates Postprandial Hyperglycemia in Diabetic Mice. Prev. Nutr. Food Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, A.; Tamil, I.; Dineshkumar, B.; Nandhakumar, M.; Senthilkumar, M. In Vitro Study on α-Amylase Inhibitory Activity of an Indian Medicinal Plant, Phyllanthus Amarus. Indian J. Pharmacol. 2010, 42, 280. [Google Scholar] [CrossRef] [PubMed]
- Pistia-Brueggeman, G.; Hollingsworth, R.I. A Preparation and Screening Strategy for Glycosidase Inhibitors. Tetrahedron 2001, 57, 8773–8778. [Google Scholar] [CrossRef]

| Affinity (kcal/mol) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Molecule | PTP1B | DPP4 | FFAR1 | α-amylase | PPAR Gamma | α-glucosidase | Aldose Reductase | Glycogen Phosphorylase |
| Gentisic acid | −6.1 | −6.7 | None | −5.6 | None | −6.1 | −6.9 | −6.3 |
| Amentoflavone | −8.8 | −10.5 | None | −11.3 | None | −9.5 | −10.0 | −10.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mechchate, H.; Es-safi, I.; Mohamed Al kamaly, O.; Bousta, D. Insight into Gentisic Acid Antidiabetic Potential Using In Vitro and In Silico Approaches. Molecules 2021, 26, 1932. https://doi.org/10.3390/molecules26071932
Mechchate H, Es-safi I, Mohamed Al kamaly O, Bousta D. Insight into Gentisic Acid Antidiabetic Potential Using In Vitro and In Silico Approaches. Molecules. 2021; 26(7):1932. https://doi.org/10.3390/molecules26071932
Chicago/Turabian StyleMechchate, Hamza, Imane Es-safi, Omkulthom Mohamed Al kamaly, and Dalila Bousta. 2021. "Insight into Gentisic Acid Antidiabetic Potential Using In Vitro and In Silico Approaches" Molecules 26, no. 7: 1932. https://doi.org/10.3390/molecules26071932
APA StyleMechchate, H., Es-safi, I., Mohamed Al kamaly, O., & Bousta, D. (2021). Insight into Gentisic Acid Antidiabetic Potential Using In Vitro and In Silico Approaches. Molecules, 26(7), 1932. https://doi.org/10.3390/molecules26071932






