Microbial Application to Improve Olive Mill Wastewater Phenolic Extracts
Abstract
1. Introduction
2. Results
2.1. Extraction of Phenols from OMW
2.1.1. OMW Analyses
2.1.2. Resins Adsorption
2.2. Enzymatic Tests
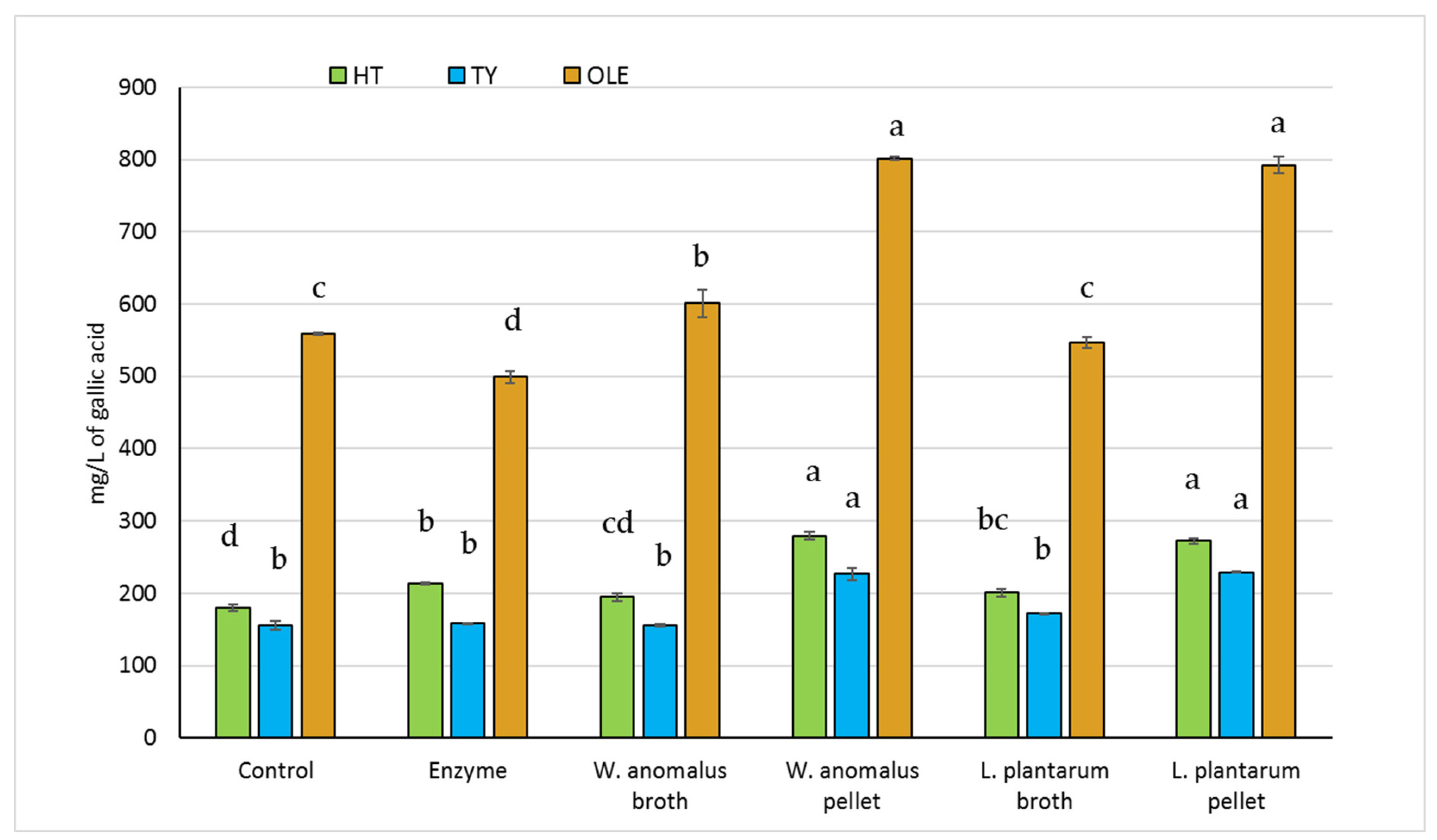
2.3. Bioconversion Efficiency by W. anomalus and L. plantarum
3. Discussion
4. Materials and Methods
4.1. Raw Material Treatment
4.2. Extraction of Phenols from OMW
4.3. Physicochemical Characterisation
4.4. HPLC Analysis
4.5. Microorganisms
4.6. Enzyme Production Test
4.7. Beta-Glucosidase Activity
4.8. Esterase Activity
4.9. Bioconversion of Phenolic Compounds
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
Sample Availability
References
- Owen, R.W.; Giacosa, A.; Hull, W.E.; Haubner, R.; Spiegelhalder, B.; Bartsch, H. The antioxidant/anticancer potential of phenolic compounds isolated from olive oil. Eur. J. Cancer. 2000, 36, 1235–1247. [Google Scholar] [CrossRef]
- Perona, J.S.; Cabello-Moruno, R.; Ruiz-Gutierrez, V. The role of virgin olive oil components in the modulation of endothelial function. J. Nutr. Biochem. 2006, 17, 429–445. [Google Scholar] [CrossRef]
- Visioli, F.; Caruso, D.; Grande, S.; Bosisio, R.; Villa, M.; Galli, G.; Sirtori, C.; Galli, C. Virgin olive oil study (VOLOS): Vasoprotective potential of extra virgin olive oil in mildly dyslipidemic patients. Eur. J. Nutr. 2005, 44, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Davalos, A.; López de las Hazas, M.C.; Crespo, M.C.; Tomé-Carneiro, J. An overview of the pharmacology of olive oil and its active ingredients. Br. J. Pharmacol. 2020, 177, 1316–1330. [Google Scholar] [CrossRef]
- Dermeche, S.; Nadour, M.; Larroche, C.; Moulti-Mati, F.; Michaud, P. Olive mill wastes: Biochemical characterizations and valorization strategies. Process Biochem. 2013, 48, 1532–1552. [Google Scholar] [CrossRef]
- Aggoun, M.; Arhab, R.; Cornu, A.; Portelli, J.; Barkat, M.; Graulet, B. Olive mill wastewater microconstituents composition according to olive variety and extraction process. Food Chem. 2016, 209, 72–80. [Google Scholar] [CrossRef]
- Fernández-Bolaños, J.; Rodríguez, G.; Rodríguez, R.; Guillén, R.; Jiménez, A. Extraction of interesting organic compounds from olive oil waste. Grasas Aceites 2006, 57, 95–106. [Google Scholar] [CrossRef]
- Alfano, A.; Corsuto, L.; Finamore, R.; Savarese, M.; Ferrara, F.; Falco, S.; Santabarbara, G.; De Rosa, M.; Schiraldi, C. Valorization of olive millwastewater by membrane processes to recover natural antioxidant compounds for cosmeceutical and nutraceutical applications or functional foods. Antioxidants 2018, 7, 72. [Google Scholar] [CrossRef]
- Fki, I.; Allouche, N.; Sayadi, S. The use of polyphenolic extract, purified hydroxytyrosol and 3,4-dihydroxyphenyl acetic acid from olive mill wastewater for the stabilization of refined oils: A potential alternative to synthetic antioxidants. Food Chem. 2005, 93, 197–204. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA). Scientific Opinion on the substantiation of health claims related to polyphenols in olive and protection of LDL particles from oxidative damage (ID 1333, 1638, 1639, 1696, 2865), maintenance of normal blood HDL-cholesterol concentrations (ID 1639), maintenance of normal blood pressure (ID 3781), anti-inflammatory properties (ID 1882), contributes to the upper respiratory tract health (ID 3468), can help to maintain a normal function of gastrointestinal tract (3779), and contributes to body defences against external agents (ID 3467) pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 2011, 9, 2033. Available online: www.efsa.europa.eu/efsajournal (accessed on 25 February 2021). [CrossRef]
- Hamza, M.; Khoufi, S.; Sayadi, S. Fungal enzymes as a powerful tool to release antioxidants from olive mill wastewater. Food Chem. 2012, 131, 1430–1436. [Google Scholar] [CrossRef]
- De Leonardis, A.; Aretini, A.; Alfano, G.; Macciola, V.; Ranalli, G. Isolation of a hydroxytyrosol-rich extract from olive leaves (Olea europaea L.) and evaluation of its antioxidant properties and bioactivity. Eur. Food Res. Technol. 2008, 226, 653–659. [Google Scholar] [CrossRef]
- Zagklis, D.P.; Vavouraki, A.I.; Kornaros, M.E.; Paraskeva, C.A. Purification of olive mill wastewater phenols through membrane filtration and resin adsorption/desorption. J. Hazard. Mater. 2015, 285, 69–76. [Google Scholar] [CrossRef]
- Bertin, L.; Ferri, F.; Scoma, A.; Marchetti, L.; Fava, F. Recovery of high added value natural polyphenols from actual olive mill wastewater through solid phase extraction. Chem. Eng. J. 2011, 171, 1287–1293. [Google Scholar] [CrossRef]
- Scoma, A.; Bertin, L.; Zanaroli, G.; Fraraccio, S.; Fava, F. A physicochemical–biotechnological approach for an integrated valorization of olive mill wastewater. Bioresource Technol. 2011, 102, 10273–10279. [Google Scholar] [CrossRef]
- Khoufi, S.; Hamza, M.; Sayadi, S. Enzymatic hydrolysis of olive wastewater for hydroxytyrosol enrichment. Bioresource Technol. 2011, 102, 9050–9058. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yong, Q.; Yu, S. Efficient bioconversion of oleuropein from olive leaf extract to antioxidant hydroxytyrosol by enzymatic hydrolysis and high temperature degradation. Biotechnol. Appl. Bioc. 2018, 65, 680–689. [Google Scholar] [CrossRef]
- Dammak, I.; Khoufi, S.; Sayadi, S. A performance comparison of olive oil mill wastewater enzymatic treatments. Food Bioprod. Process. 2016, 100, 61–71. [Google Scholar] [CrossRef]
- Liu, M.; Yong, Q.; Lian, Z.; Huang, C.; Yu, S. Continuous bioconversion of oleuropein from olive leaf extract to produce the bioactive product hydroxytyrosol using carrier-immobilized enzyme. Appl. Biochem. Biotech. 2020, 190, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, R.; Giorno, L.; Piacentini, E.; Mazzuca, S.; Drioli, E. Kinetic study of a biocatalytic membrane reactor containing immobilized b-glucosidase for the hydrolysis of oleuropein. J. Membr. Sci. 2009, 339, 215–223. [Google Scholar] [CrossRef]
- Kang, S.W.; Ko, E.H.; Lee, J.S.; Kim, S.W. Over-production of β-glucosidase by Aspergillus niger mutant from lignocellulosic biomass. Biotechnol. Lett. 1999, 21, 647–650. [Google Scholar] [CrossRef]
- Benítez-Cabello, A.; Calero-Delgado, B.; Rodríguez-Gómez, F.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. Biodiversity and multifunctional features of lactic acid bacteria isolated from table olive biofilms. Front. Microbiol. 2019, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Vaccalluzzo, A.; Pino, A.; De Angelis, M.; Bautista-Gallego, J.; Romeo, F.V.; Foti, P.; Caggia, C.; Randazzo, C.L. Effects of different stress parameters on growth and on oleuropein-degrading abilities of Lactiplantibacillus plantarum strains selected as tailored starter cultures for naturally table olives. Microorganisms 2020, 8, 1607. [Google Scholar] [CrossRef]
- Aponte, M.; Ungaro, F.; d’Angelo, I.; De Caro, C.; Russo, R.; Blaiotta, G.; Dal Piaz, F.; Calignano, A.; Miro, A. Improving in vivo conversion of oleuropein into hydroxytyrosol by oral granules containing probiotic Lactobacillus plantarum 299v and an Olea europaea standardized extract. Int. J. Pharmaceut. 2018, 543, 73–82. [Google Scholar] [CrossRef]
- Porru, C.; Rodríguez-Gόmez, F.; Benítez-Cabello, A.; Jiménez-Díaz, R.; Zara, G.; Budroni, M.; Mannazzu, I.; Arroyo-López, F.N. Genotyping, identification and multifunctional features of yeasts associated to Bosana naturally black table olive fermentations. Food Microbiol. 2018, 69, 33–42. [Google Scholar] [CrossRef]
- Bonatsou, S.; Karamouza, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z. Evaluating the probiotic potential and technological characteristics of yeasts implicated in cv. Kalamata natural black olive fermentation. Int. J. Food Microbiol. 2018, 271, 48–59. [Google Scholar] [CrossRef]
- Zullo, B.A.; Ciafardini, G. Evaluation of physiological properties of yeast strains isolated from olive oil and their in vitro probiotic trait. Food Microbiol. 2019, 78, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Padilla, B.; Gil, J.V.; Manzanares, P. Challenges of the non-conventional yeast Wickerhamomyces anomalus in winemaking. Fermentation 2018, 4, 68. [Google Scholar] [CrossRef]
- Belaqziz, M.; Tan, S.P.; El-Abbassi, A.; Kiai, H.; Hafidi, A.; O’Donovan, O.; McLoughlin, P. Assessment of the antioxidant and antibacterial activities of different olive processing wastewaters. PLoS ONE 2017, 12, e0182622. [Google Scholar] [CrossRef]
- Davies, L.C.; Vilhena, A.M.; Novais, J.M.; Martins-Dias, S. Olive mill wastewater characteristics: Modelling and statistical analysis. Grasas Aceites 2004, 55, 233–241. [Google Scholar] [CrossRef]
- Allouche, N.; Fki, I.; Sayadi, S. Toward a high yield recovery of antioxidants and purified hydroxytyrosol from olive mill wastewaters. J. Agric. Food Chem. 2004, 52, 267–273. [Google Scholar] [CrossRef]
- Obied, H.K.; Allen, M.S.; Bedgood, D.R.; Prenzler, P.D.; Robards, K.; Stockmann, R. Bioactivity and analysis of biophenols recovered from olive mill waste. J. Agric. Food Chem. 2005, 53, 823–837. [Google Scholar] [CrossRef]
- D’Antuono, I.; Kontogianni, V.G.; Kotsiou, K.; Linsalata, V.; Logrieco, A.F.; Tasioula-Margari, M.; Cardinali, A. Polyphenolic characterization of olive mill wastewaters, coming from Italian and Greek olive cultivars, after membrane technology. Food Res. Int. 2014, 65, 301–310. [Google Scholar] [CrossRef]
- Agalias, A.; Magiatis, P.; Skaltsounis, A.-L.; Mikros, E.; Tsar-Bopoulos, A.; Gikas, E.; Spanos, I.; Manios, T. A new process for the management of olive oil mill wastewater and recovery of natural antioxidants. J. Agric. Food Chem. 2007, 55, 2671–2676. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, J.; Carle, R.; Kammerer, D.R. Adsorption and ion exchange: Basic principles and their application in food processing. J. Agric. Food Chem. 2011, 59, 22–42. [Google Scholar] [CrossRef]
- Grohmann, K.; Manthey, J.A.; Cameron, R.G.; Buslig, B.S. Purification of citrus peel juice and molasses. J. Agric. Food Chem. 1999, 47, 4859–4860. [Google Scholar] [CrossRef]
- Kammerer, J.; Boschet, J.; Kammerer, D.R.; Carle, R. Enrichment and fractionation of major apple flavonoids, phenolic acids and dihydrochalcones using anion exchange resins. LWT-Food Sci. Technol. 2011, 44, 1079–1087. [Google Scholar] [CrossRef]
- Soto, M.L.; Conde, E.; González-López, N.; Conde, M.J.; Moure, A.; Sineiro, J.; Falqué, E.; Domínguez, H.; Núñez, M.J.; Parajó, J.C. Recovery and concentration of antioxidants from winery wastes. Molecules 2012, 17, 3008–3024. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, F.; Moreira, C.; Salgado, J.M.; Abrunhosa, L.; Venâncio, A.; Belo, I. Olive pomace valorization by Aspergillus species: Lipase production using solid-state fermentation. J. Sci. Food Agr. 2016, 96, 3583–3589. [Google Scholar] [CrossRef]
- Bhanja, T.; Kumari, A.; Banerjee, R. Enrichment of phenolics and free radical scavenging property of wheat koji prepared with two filamentous fungi. Bioresource Technol. 2009, 100, 2861–2866. [Google Scholar] [CrossRef]
- Matthews, A.; Grimaldi, A.; Walker, M.; Bartowsky, E.; Grbin, P.; Jiranek, V. Lactic Acid Bacteria as a potential source of enzymes for use in vinification. Appl. Environ. Microbiol. 2004, 5715–5731. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Gallego, J.; Rodriguez-Gomez, F.; Barrio, E.; Querol, A.; Garrido-Fernandez, A.; Arroyo-López, F.N. Exploring the yeast biodiversity of green table olive industrial fermentations for technological applications. Int. J. Food Microbiol. 2011, 147, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-López, F.N.; Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gomez, F.; Jiménez-Díaz, R.; García-García, P.; Querol, A.; Garrido-Fernández, A. Yeasts in table olive processing: Desirable or spoilage microorganisms? Int. J. Food Microbiol. 2012, 160, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Romero-Gil, V.; Bautista-Gallego, J.; Rodríguez-Gómez, F.; García-García, P.; Jiménez-Díaz, R.; Garrido-Fernández, A.; Arroyo-López, F.N. Evaluating the individual effects of temperature and salt on table olive related microorganisms. Food Microbiol. 2013, 33, 178–184. [Google Scholar] [CrossRef]
- Bonatsou, S.; Benitez, A.; Rodriguez-Gómez, F.; Panagou, E.Z.; Arroyo-Lopez, F.N. Selection of yeasts with multifunctional features for application as starters in natural black table olive processing. Food Microbiol. 2015, 46, 66–73. [Google Scholar] [CrossRef]
- Pino, A.; Vaccalluzzo, A.; Solieri, L.; Romeo, F.V.; Todaro, A.; Caggia, C.; Arroyo-López, F.N.; Bautista-Gallego, J.; Randazzo, C.L. Effect of Sequential Inoculum of Beta-Glucosidase Positive and Probiotic Strains on Brine Fermentation to Obtain Low Salt Sicilian Table Olives. Front. Microbiol. 2019, 10, 174. [Google Scholar] [CrossRef]
- Bellumori, M.; Cecchi, L.; Innocenti, M.; Clodoveo, M.L.; Corbo, F.; Mulinacci, N. The EFSA Health Claim on Olive Oil Polyphenols: Acid Hydrolysis Validation and Total Hydroxytyrosol and Tyrosol Determination in Italian Virgin Olive Oils. Molecules 2019, 24, 2179. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, G.; Muzzalupo, I.; Muccilli, S.; Timpanaro, N.; Russo, M.P.; Guardo, M.; Rapisarda, P.; Romeo, F.V. New accessions of Italian table olives (Olea europaea): Characterization of genotypes and quality of brined products. Sci. Hortic. 2016, 213, 34–41. [Google Scholar] [CrossRef]
- Mackness, M.I.; Walker, C.H.; Rowlands, D.C.; Price, N.R. Esterase activity in homogenates of three strains of the rust red flour beetle Tribolium castaneum (Herbst). Comp. Biochem. Physiol. 1983, 74, 65–68. [Google Scholar] [CrossRef]

| Parameters | Means ± SD |
|---|---|
| pH | 4.80 ± 0.06 |
| Dry matter (g/100 mL) | 6.24 ± 0.02 |
| Hydroxytyrosol (HT) (mg/L) | 218.29 ± 4.94 |
| Tyrosol (TY) (mg/L) | 67.24 ± 5.57 |
| Oleuropein (OLE) (mg/L) | 207.49 ± 56.37 |
| Total phenols (mg/L) | 1343.26 ± 0.54 |
| Adsorbent Resin | Hydroxytyrosol (HT) | Tyrosol (TY) | Oleuropein (OLE) | Total Phenols |
|---|---|---|---|---|
| SP207 | 3239.7 ± 57.34 a | 2016.8 ± 49.36 a | 11,336.6 ± 199.65 a | 16,447.9 ± 16.31 a |
| XAD16 | 1381.7 ± 66.45 b | 1000.3 ± 120.75 b | 10,047.8 ± 120.29 ab | 13,144.6 ± 21.75 b |
| PAD900C | 294.3 ± 2.67 c | 90.0 ± 10.59 c | 4443.8 ± 176.32 c | 4880.3 ± 10.87 d |
| C18 | 191.1 ± 16.59 c | 50.4 ± 10.34 c | 8092.3 ± 218.15 b | 8763.6 ± 43.50 c |
| Samples | Beta-Glucosidase | Esterase |
|---|---|---|
| Lallzyme beta enzyme | 6979.9 ± 3.22 b | 316,713.9 ± 1032.48 c |
| W. anomalus | 7066.4 ± 5.36 b | 405,417.6 ± 516.24 b |
| L. plantarum | 2387.6 ± 3.22 c | 316,348.9 ± 1540.62 c |
| S. cerevisiae | 1437.9 ± 78.31 d | 399,577.0 ± 1548.72 b |
| A. niger | 314,313.8 ± 107.28 a | 4,506,229.2 ± 4646.15 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, F.V.; Granuzzo, G.; Foti, P.; Ballistreri, G.; Caggia, C.; Rapisarda, P. Microbial Application to Improve Olive Mill Wastewater Phenolic Extracts. Molecules 2021, 26, 1944. https://doi.org/10.3390/molecules26071944
Romeo FV, Granuzzo G, Foti P, Ballistreri G, Caggia C, Rapisarda P. Microbial Application to Improve Olive Mill Wastewater Phenolic Extracts. Molecules. 2021; 26(7):1944. https://doi.org/10.3390/molecules26071944
Chicago/Turabian StyleRomeo, Flora V., Gina Granuzzo, Paola Foti, Gabriele Ballistreri, Cinzia Caggia, and Paolo Rapisarda. 2021. "Microbial Application to Improve Olive Mill Wastewater Phenolic Extracts" Molecules 26, no. 7: 1944. https://doi.org/10.3390/molecules26071944
APA StyleRomeo, F. V., Granuzzo, G., Foti, P., Ballistreri, G., Caggia, C., & Rapisarda, P. (2021). Microbial Application to Improve Olive Mill Wastewater Phenolic Extracts. Molecules, 26(7), 1944. https://doi.org/10.3390/molecules26071944









