L-Glutaminase Synthesis by Marine Halomonas meridiana Isolated from the Red Sea and Its Efficiency against Colorectal Cancer Cell Lines
Abstract
1. Introduction
2. Results
2.1. Isolation and Screening of L-Glutaminase-Producing Bacteria from the Red Sea
2.2. Molecular Identification via 16S rRNA Gene Sequencing and Phylogenetic Analysis
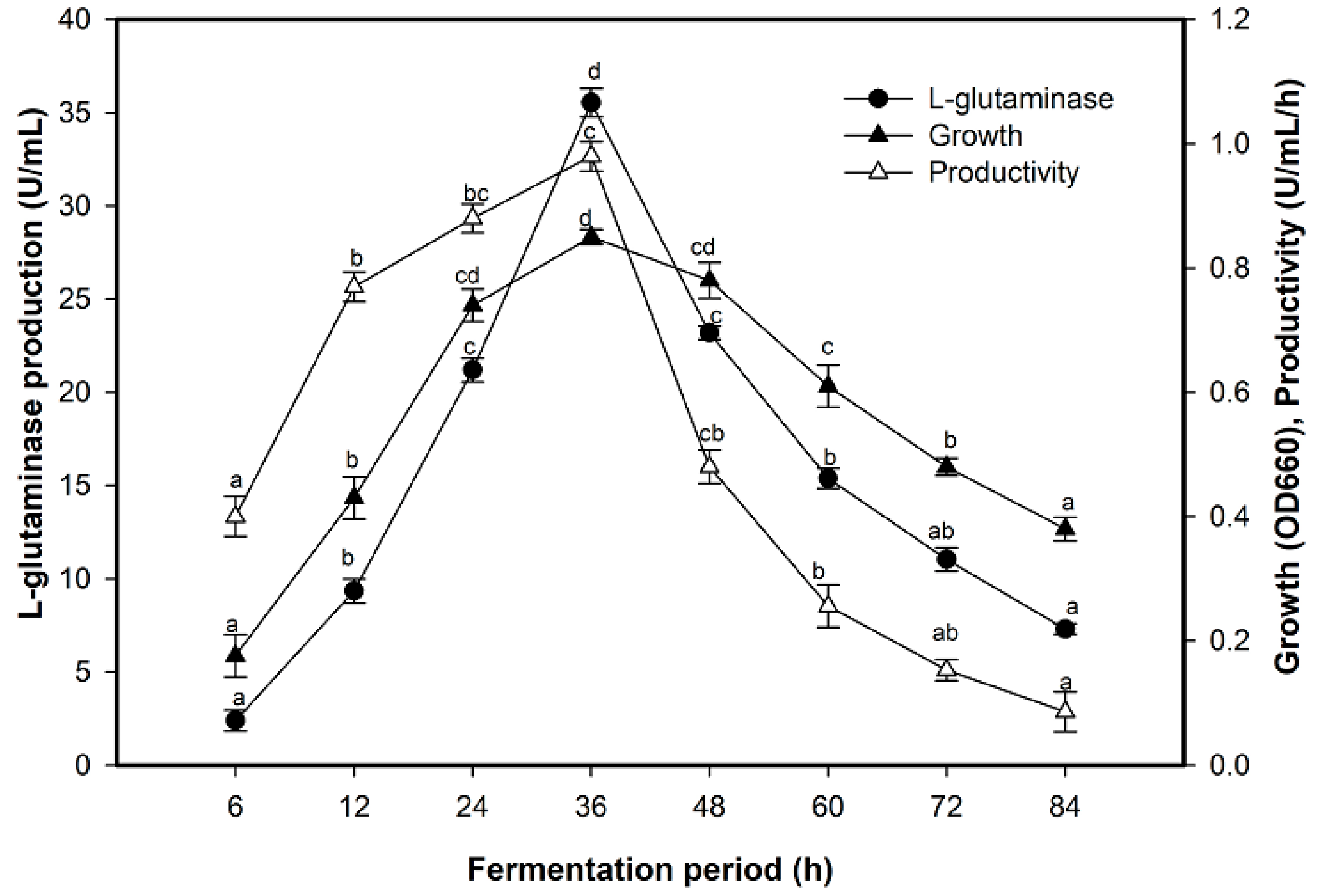
2.3. Effect of Fermentation Period on L-Glutaminase Production
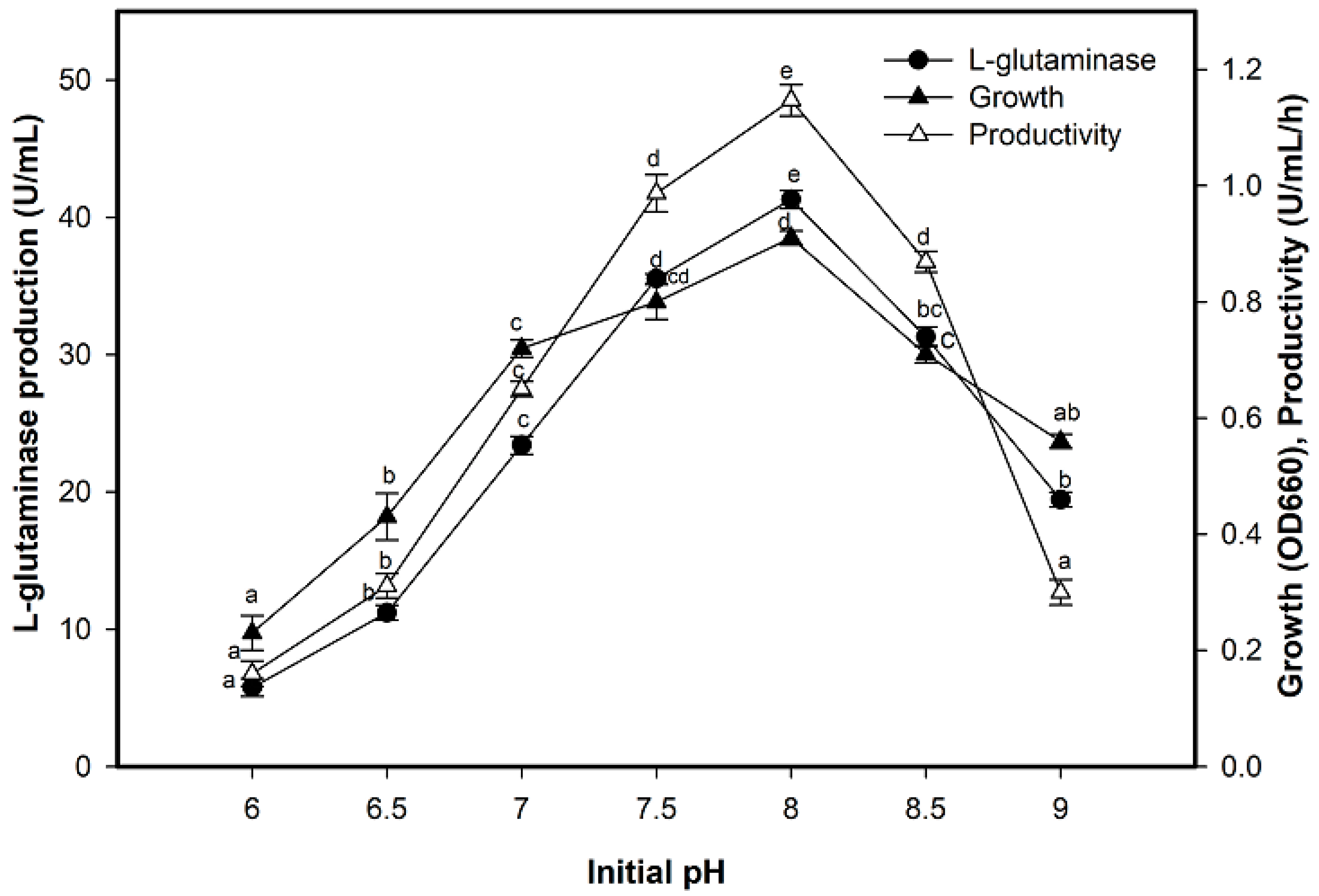
2.4. Effect of Initial pH on L-Glutaminase Production
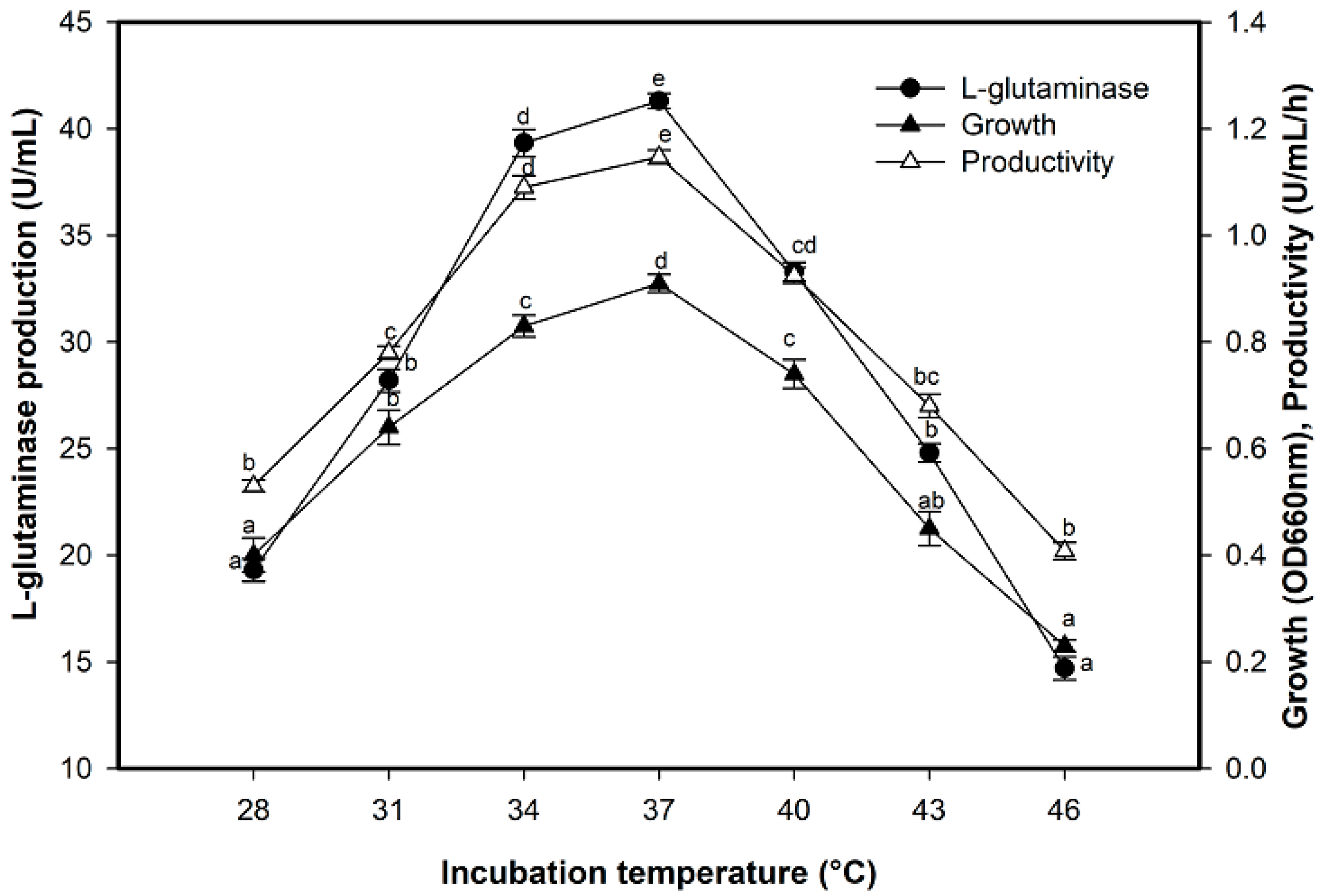
2.5. Effect of Incubation Temperature on L-Glutaminase Production
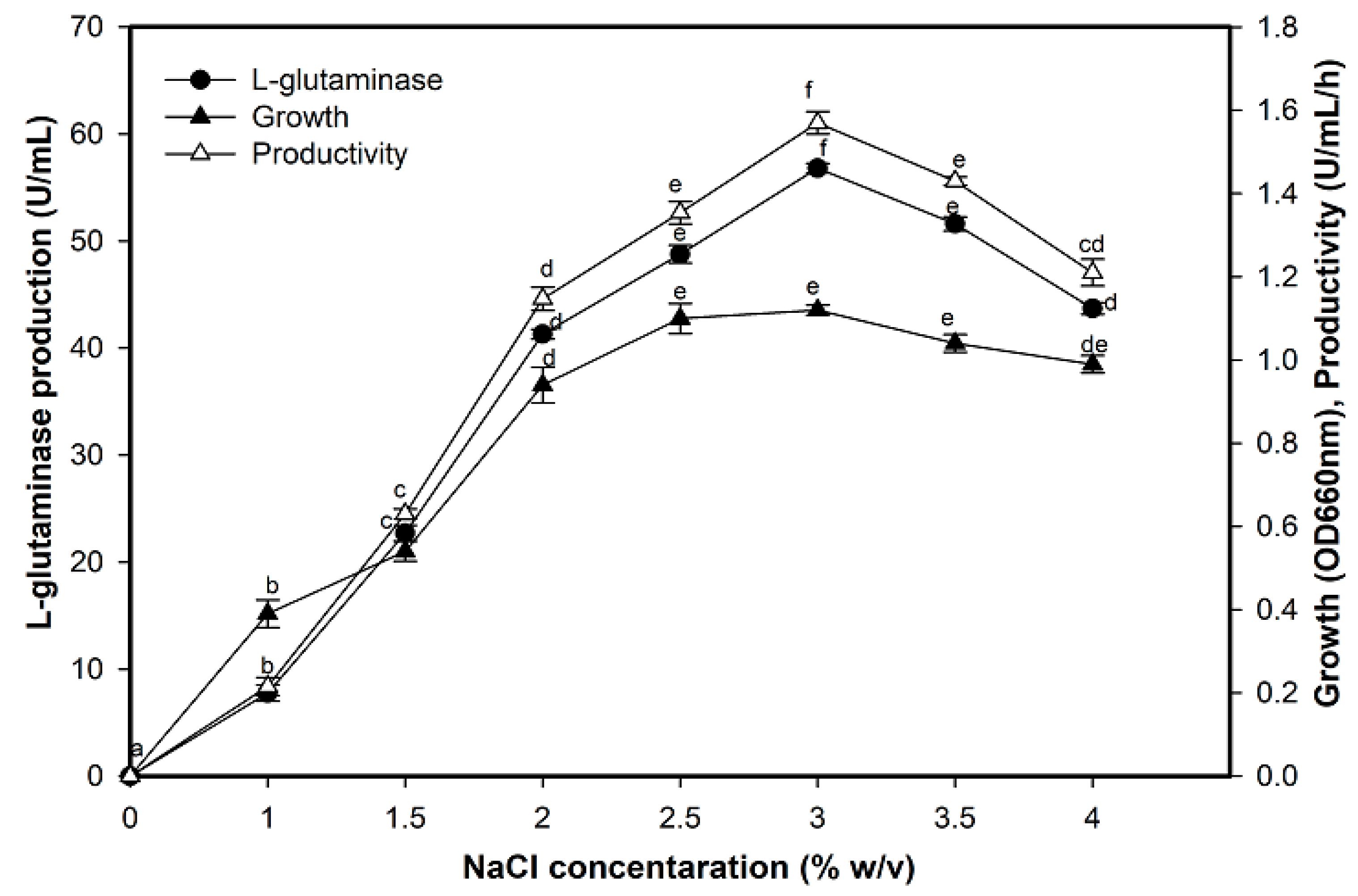
2.6. Effect of NaCl on L-Glutaminase Production
2.7. Effect of Carbon Sources on L-Glutaminase Production
2.8. Effect of Nitrogen Sources on the L-Glutaminase Production
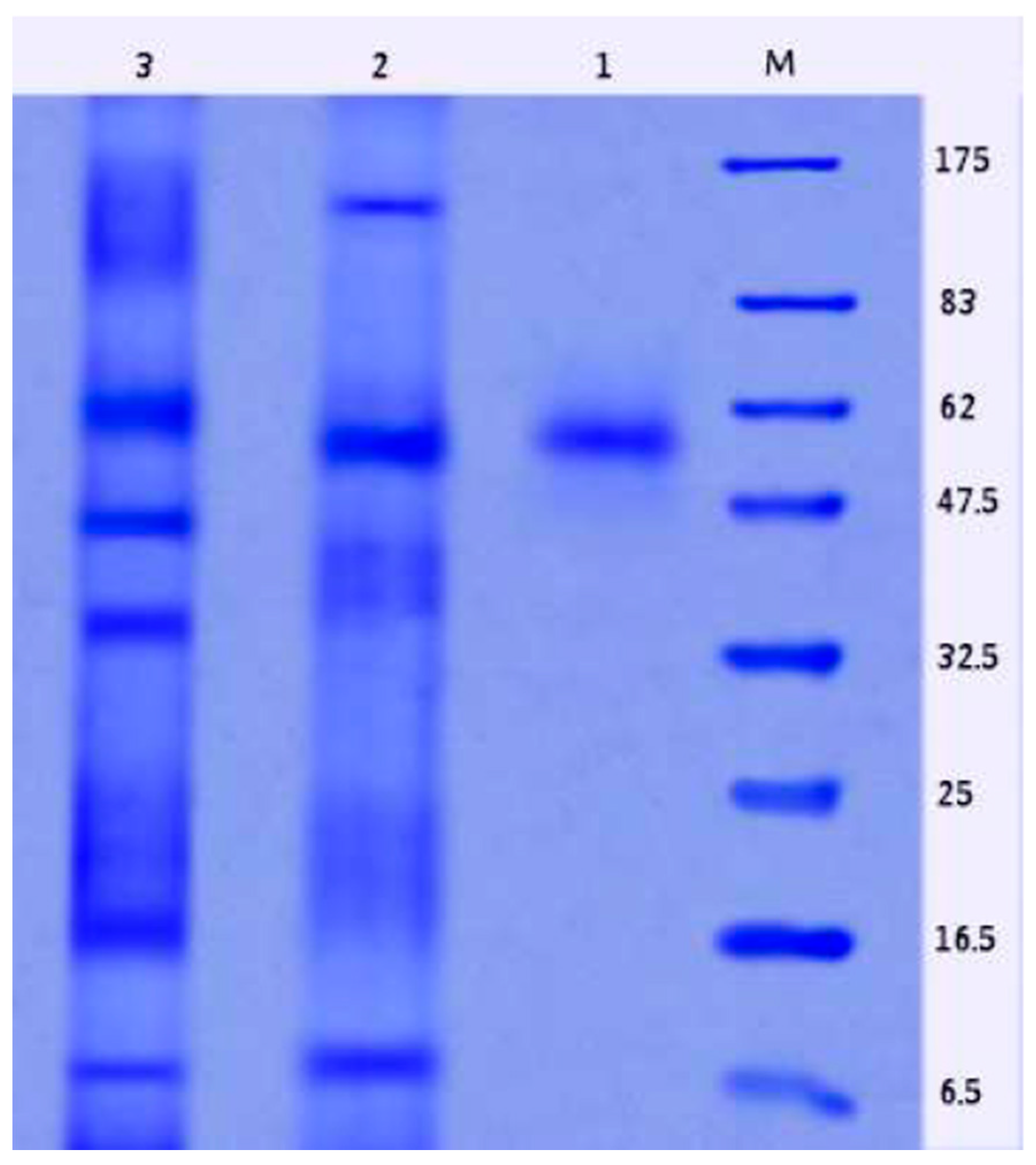
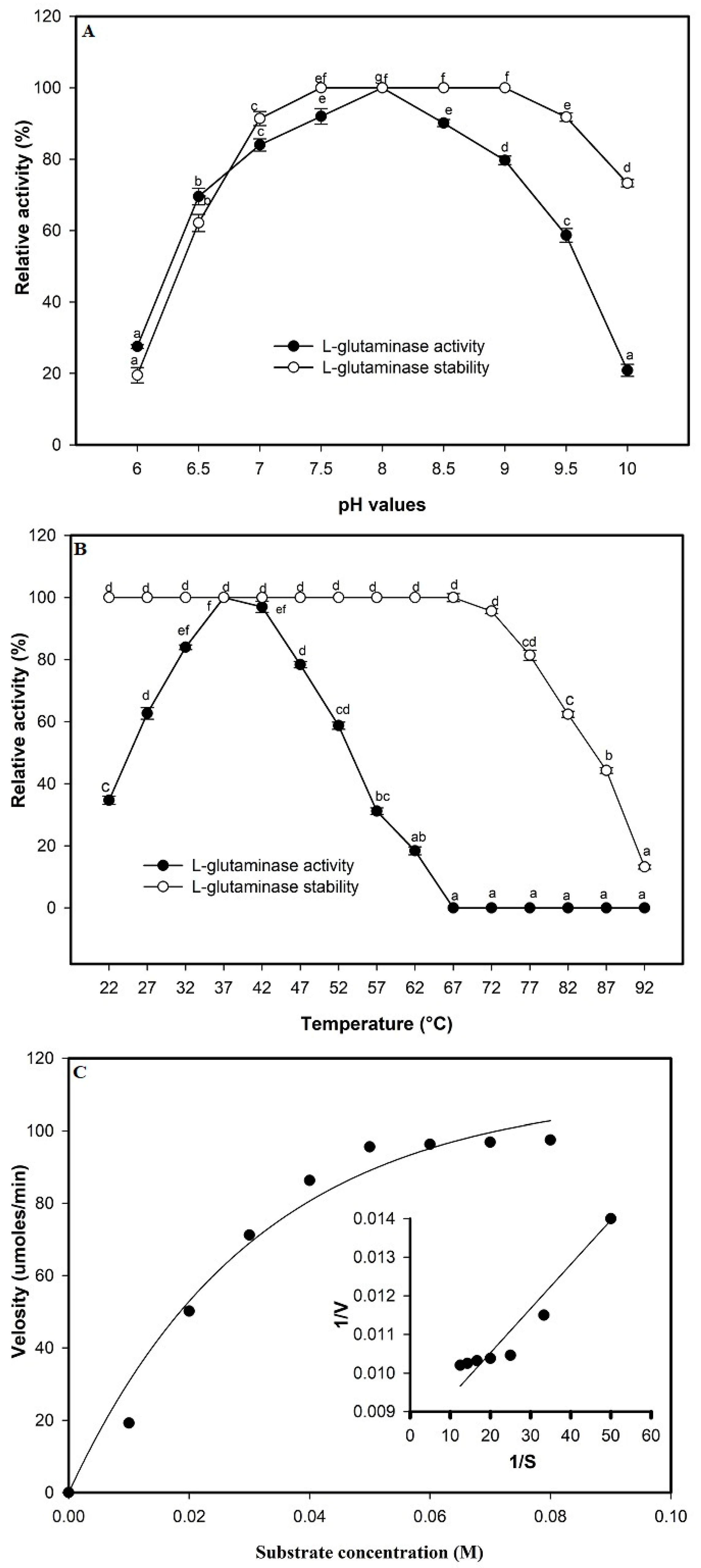
2.9. Properties of Purified L-Glutaminase
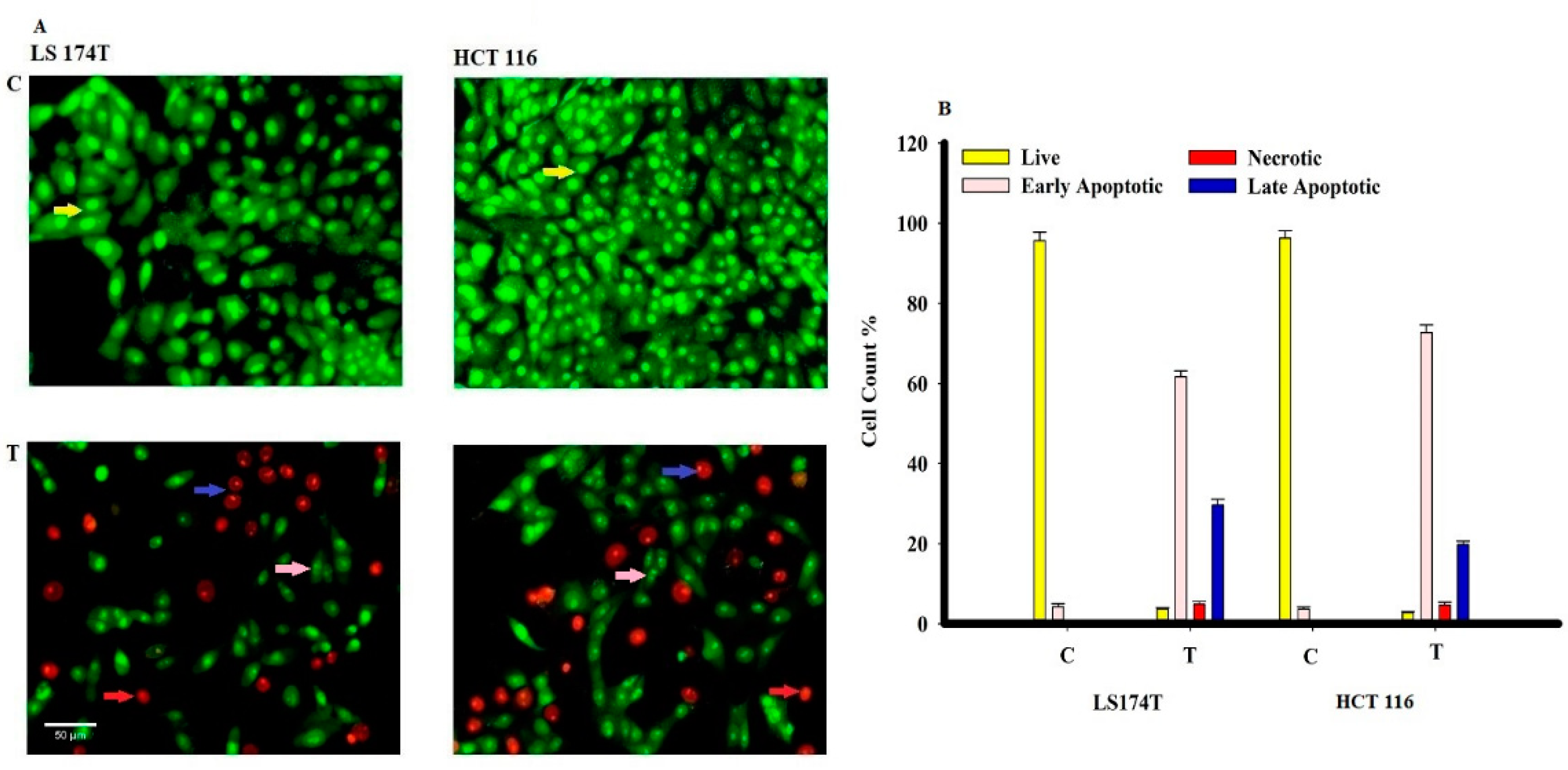
2.10. Cytotoxicity Assay of L-Glutaminase against Colorectal Cancer Cell Lines
2.11. Detection of Early and Late Apoptotic Cells
3. Discussion
4. Materials and Methods
4.1. Marine Sample Collection
4.2. Isolation and Selection of L-Glutaminase Producing Bacteria
4.3. Molecular Identification of the Promising Bacterial Isolate
4.4. Optimization of L-Glutaminase Production
4.5. L-Glutaminase Purification and Its Characterization
4.6. Analytical Methods
4.7. Anti-Colorectal Cancer Assay of L-Glutaminase
4.7.1. Cell Culture
4.7.2. Cytotoxicity Assay of L-Glutaminase
4.7.3. Detection of Early and Late Apoptotic Cells
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kiruthika, J.; Swathi, S. Purification and characterization of a novel broad spectrum anti-tumor L-glutaminase enzyme from marine Bacillus subtilis strain JK-79. Afr. J. Microb. Res. 2019, 13, 232–244. [Google Scholar]
- Sánchez-Gundín, J.; Fernández-Carballido, A.M.; Martínez-Valdivieso, L.; Barreda-Hernández, D.; Torres-Suárez, A.I. New trends in the therapeutic approach to metastati colorectal cancer. Int. J. Med. Sci. 2018, 15, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Alyabsi, M.; Alhumaid, A.; Allah-Bakhsh, H.; Alkelya, M.; Aziz, A.Z. Colorectal cancer in Saudi Arabia as the proof-of-principle model for implementing strategies of predictive, preventive, and personalized medicine in healthcare. EPMA 2020, 11, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Munro, M.J.; Wickremesekera, S.K.; Peng, L.; Tan, S.T.; Itinteang, T. Cancer stem cells in colorectal cancer: A review. J. Clin. Pathol. 2018, 71, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Dhankhar, R.; Gupta, V.; Kumar, S.; Kapoor, R.K.; Gulati, P. Microbial enzymes for deprivation of amino acid metabolism in malignant cells: Biological strategy for cancer treatment. Appl. Microb. Biotech. 2020, 104, 2857–2869. [Google Scholar] [CrossRef]
- Pandian, S.R.K.; Deepak, V.; Sivasubramaniam, S.D.; Nellaiah, H.; Sundar, K. Optimization and purification of anticancer enzyme Lglutaminase from Alcaligenes faecalis KLU102. Biologia 2014, 69, 1644–1651. [Google Scholar] [CrossRef]
- Singh, P.; Banik, R.M. Biochemical characterization and antitumor study of L-glutaminase from Bacillus cereus MTCC 1305. Appl. Biochem. Biotechnol. 2013, 171, 522–531. [Google Scholar] [CrossRef]
- Roberts, J.; MacAllister, T.W.; Sethuraman, N.; Inventors, M.E. Medical Enzymes AG, Assignee: Genetically Engineered Glutaminase and Its Use in Antiviral and Anticancer Therapy. U.S. Patent 6312939, 6 November 2001. [Google Scholar]
- Ye, M.; Liu, X.; Zhao, L. Production of a novel salt-tolerant L-glutaminase from Bacillus amyloliquefaciens using agro-industrial residues and its application in Chinese soy sauce fermentation. Biotechnology 2013, 12, 25–35. [Google Scholar] [CrossRef][Green Version]
- Amobonye, A.; Singh, S.; Pillai, S. Recent advances in microbial glutaminase production and applications—A concise review. Crit. Rev. Biotechnol. 2019, 39, 944–963. [Google Scholar] [CrossRef]
- Albayrak, D.; Karakuş, E. A novel glutamine biosensor based on zinc oxide nanorod and glutaminase enzyme from Hypocria jecorina. Artif. Cells Nanomed. Biotechnol. 2016, 44, 92–97. [Google Scholar] [CrossRef]
- Binod, P.; Sindhu, R.; Madhavan, A.; Abraham, A.; Mathew, A.K.; Beevi, U.S.; Sukumaran, R.K.; Singh, S.P.; Pandey, A. Recent developments in L-glutaminase production and applications—An overview. Bioresour. Technol. 2017, 245, 1766–1774. [Google Scholar] [CrossRef]
- Cantor, J.R.; Panayiotou, V.; Agnello, G.; Georgiou, G.; Stone, E.M. Engineering reduced-immunogenicity enzymes for amino acid depletion therapy in cancer. Meth. Enzymol. 2012, 502, 291–319. [Google Scholar]
- De Berardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef]
- Mustafa, G.; AL-Juha, I.; Asmaa, A. Hussein. Characterization and antitumor activity study for a therapeutically important L-glutaminase purified from Staphylococcus aureus. Int. J. Psychosoc. Rehabil. 2020, 24, 3029–3037. [Google Scholar]
- Zolfaghar, M.; Amoozegar, M.A.; Khajeh, K.; Babavalian, H.; Tebyanian, H. Isolation and screening of extracellular anticancer enzymes from halophilic and halotolerant bacteria from different saline environments in Iran. Mol. Biol. Rep. 2019, 46, 3275–3286. [Google Scholar] [CrossRef]
- Shirazian, P.; Asad, S.; Amoozegar, M.A. The potential of halophilic and halotolerant bacteria for the production of antineoplastic enzymes: L-Asparaginase and L-glutaminase. EXCLI J. 2016, 15, 268–279. [Google Scholar] [PubMed]
- Nielsen, J.; Archer, J.; Essack, M.; Bajic, V.B.; Gojobori, T.; Mijakovic, I. Building a bio-based industry in the Middle East through harnessing the potential of the Red Sea biodiversity. Appl. Microbiol. Biotechnol. 2017, 101, 4837–4851. [Google Scholar] [CrossRef]
- Soren, J.P.; Paul, T.; Banerjee, A.; Mondal, K.C.; Das Mohapatra, P.K. Exploitation of agricultural waste as sole substrate for production of bacterial L-glutaminase under submerged fermentation and the proficient application of fermented hydrolysate as growth promoting agent for probiotic Organisms. Waste Biomass Valorization 2020, 11, 4245–4257. [Google Scholar] [CrossRef]
- Durthia, C.P.; Polaa, M.; Kolab, A.K.; Rajulapatia, S.B. Screening, optimization of culture conditions and scale-up for production of the L-glutaminase by novel isolated Bacillus sps. mutant endophyte using response surface methodology. Biocatal. Agric. Biotechnol. 2019, 18, 101077. [Google Scholar] [CrossRef]
- Qian, P.Y.; Wang, Y.; Lee, O.O.; Lau, S.C.K.; Yang, J.; Lafi, F.F.; Al-Suwailem, A.; Wong, T.Y.H. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 2011, 5, 507–518. [Google Scholar] [CrossRef]
- Putri, N.P.; Sari, C.N.; Gozan, M. Biosurfactant screening of Halomonas meridiana BK-AB4 for microbial enhanced oil recovery. Mater. Sci. Forum 2020, 988, 95–100. [Google Scholar] [CrossRef]
- Hussein, A.A.; Albarazanchi, S.I.; Alshanon, A.F. Evaluation of anticancer potential for L-glutaminase purified from Bacillus subtilis. Int. J. Pharm. Res. 2020, 12, 293–299. [Google Scholar]
- Jesuraj, S.A.V.; Sarker, M.D.M.R.; Ming, L.C.; Praya, S.M.J.; Ravikumar, M.; Wui, W.T. Enhancement of the production of L-glutaminase, an anticancer enzyme, from Aeromonas veronii by adaptive and induced mutation techniques. PLoS ONE 2017, 12, e0181745. [Google Scholar] [CrossRef]
- Iyer, P.; Singhal, R.S. Screening and selection of marine isolate for L-glutaminase production and media optimization using response surface methodology. Appl. Biochem. Biotechnol. 2009, 159, 233–250. [Google Scholar] [CrossRef] [PubMed]
- Jayabalan, R.; Jeeva, S.; Sasikumar, A.P.; Inbakandan, D.; Swaminathan, K.; Yun, S.E. Exracellular L-glutaminase production by marine Brevundimonas diminuta MTCC 8486. Int. J. Appl. Bioeng. 2010, 4, 19–24. [Google Scholar]
- Suresh, K.S.; Muthuvelayudham, R.; Viruthagiri, T. Medium optimization for production of L-glutaminase (EC 3.5.1.2) by Streptomyces griseus under submerged fermentation. IJSEA 2013, 2, 1–25. [Google Scholar]
- Singh, P.; Shera, S.S.; Banik, J.; Banik, R.M. Optimization of cultural conditions using response surface methodology versus artificial neural network and modeling of L-glutaminase production by Bacillus cereus MTCC 1305. Bioresour. Technol. 2013, 137, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Sathish, T.; Kezia, D.; Bramhachari, P.V.; Prakasham, R.S. Multi-objective based superimposed optimization method for enhancement of L-glutaminase production by Bacillus subtilis RSP-GLU. KIJOMS 2018, 4, 50–60. [Google Scholar] [CrossRef]
- Kiruthika, J.; Murugesan, S. Studies on optimization of L-glutaminase production under submerged fermentation from marine Bacillus subtilis JK-79. Afr. J. Microbiol. Res. 2020, 14, 16–24. [Google Scholar]
- Prabhu, N.G.; Chandrasekaran, M. Impact of process parameters on l-glutaminase production by marine Vibrio costicola in solid state fermentation using polystyrene as an inert support. Process. Biochem. 1997, 32, 285–289. [Google Scholar] [CrossRef]
- Desai, S.S.; Chopra, S.J.; Hungund, B.S. Production, purification and characterization of L-glutaminase from Streptomyces sp. isolated from soil. J. Appl. Pharm. Sci. 2013, 6, 100–105. [Google Scholar] [CrossRef]
- Asy’ari, M.; Parwata, P.; Aditiawati, P.; Akhmaloka, A.; Hertadi, R. Isolation and identification of halostable lipase producing bacteria from the bledug kuwu mud crater located at Purwodadi-Grobogan, central Java, Indonesia. J. Pure Appl. Microbiol. 2014, 8, 3387–3396. [Google Scholar]
- Iyer, P.; Singhal, R.S. Production of glutaminase (E.C.3.2.1.5) from Zygosaccharomyces rouxii: Statistical optimization using response surface methodology. Bioresour. Technol. 2008, 99, 4300–4307. [Google Scholar] [CrossRef] [PubMed]
- Sabu, A.; Rajeev, S.; Chandrasekaran, C. Continuous production of extracellular L-glutaminase by Ca-alginate-immobilized marine Beauveria bassiana BTMF S-10 in Packed-Bed Reactor. Appl. Biochem. Biotechnol. 2002, 102, 71–79. [Google Scholar] [CrossRef]
- Wakayama, M.; Yamagata, T.; Kamemura, A.; Bootim, N.; Yano, S.; Tachiki, T.; Yoshimune, K.; Moriguchi, M. Characterization of salt-tolerant glutaminase from Stenotrophomonas maltophilia NYW-81 and its application in Japanese soy sauce fermentation. J. Ind. Microbiol. Biotechnol. 2005, 32, 383–390. [Google Scholar] [CrossRef]
- Orabi, H.; El-Fakharany, E.; Abdelkhalek, E.; Sidkey, N. Production, optimization, purification, characterization, and anti-cancer application of extracellular L-glutaminase produced from the marine bacterial isolate. Prep. Biochem. Biotechnol. 2020, 50, 408–418. [Google Scholar] [CrossRef] [PubMed]
- More, S.S.; Swamy, R.; Mohan, N.; Navyashree, M.; Janardhan, B.; Niyonzima, F.N. Purification and characterization of anti-cancer L-glutaminase of Bacillus cereus strain LC13. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2016, 88, 695–705. [Google Scholar] [CrossRef]
- Sivakumar, K.; Sahu, M.K.; Manivel, P.R.; Kannan, L. Optimum conditions for L-glutaminase production by Actinomycete strain isolated from estuarine fish, Chanos chanos (Forskal, 1775). Ind. J. Exp. Biol. 2006, 44, 256–258. [Google Scholar]
- Han, B.Z.; Rombouts, F.M.; Robert Nout, M.J. Amino acid profiles of sufu, a Chinese fermented soybean food. J. Food Comp. Anal. 2004, 17, 689–698. [Google Scholar] [CrossRef]
- Suribabu, K.; Govardhan, T.L.; Hemalatha, K.P.J. Optimization of various nitrogen sources for the production of α-Amylase using Brevibacillus borstelensis R1 by submerged fermentation. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 791–800. [Google Scholar]
- Elshafei, A.M.; Hassan, M.M.; Ali, N.H.; Abouzeid, M.A.; Mahmoud, D.A.; Elghonemy, D.H. Purification, kinetic properties and antitumor activity of L-glutaminase from Penicillum brevicompactum NRC 829. Br. Microbiol. Res. J. 2014, 4, 97–115. [Google Scholar] [CrossRef]
- Reda, F.M. Kinetic properties of Streptomyces canarius L-glutaminase and its anticancer efficiency. Braz. J. Microbiol. 2015, 46, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Bu, X.D.; Li, N.; Tian, X.Q.; Huang, P.-L. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell 2011, 43, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gomez, C.; Campos-Sandoval, J.A.; Alonso, F.J.; Segura, J.A.; Manzanares, E.; Ruiz-Sanchez, P.; Gonzalez, M.E.; Marquez, J.; Mates, J.M. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem. J. 2005, 386, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, T.; Goto, I. Glutaminase and glutamine synthetase activities in human cirrhotic liver and hepatocellular carcinoma. Cancer Res. 1992, 52, 1192–1194. [Google Scholar]
- Boyan, B.D.; Sylvia, V.L.; Frambach, T.; Lohmann, C.H.; Dietl, J.; Dean, D.D.; Schwartz, Z. Estrogen-dependent rapid activation of protein kinase C in estrogen receptor-positive MCF-7 breast cancer cells and estrogen receptor-negative HCC38 cells is membrane-mediated and inhibited by tamoxifen. Endocrinology 2003, 144, 812–824. [Google Scholar] [CrossRef]
- Elbehairi, S.I.; Ismail, L.A.; Alfaifi, M.Y.; Elshaarawy, R.F.M.; Hafez, H.S. Chitosan nano-vehicles as biocompatible delivering tools for a new Ag(I) curcuminoid-Gboxin analog complex in cancer and inflammation therapy. Int. J. Biol. Macromol. 2020, 165, 2750–2764. [Google Scholar] [CrossRef]
- Lu, J.-J.; Perng, C.-L.; Lee, S.-Y.; Wan, C.-C. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol. 2000, 38, 2076–2080. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Al-Abd, A.M.; Lightfoot, D.A.; El-Shemy, H.A. Anti-cancer characteristics of mevinolin against three different solid tumor cell lines was not solely p53-dependent. J. Enzym. Inhib. Med. Chem. 2012, 27, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Alahdal, A.M.; Asfour, H.Z.; Ahmed, S.A.; Noor, A.O.; Al-Abd, A.M.; Elfaky, M.A.; Elhady, S.S. Anti-helicobacter, antitubercular and cytotoxic activities of scalaranes from the Red Sea sponge Hyrtios erectus. Molecules 2018, 23, 978. [Google Scholar] [CrossRef]
- Albright, F.; Stephenson, R.A.; Agarwal, N.; Teerlink, C.C.; Lawrence, W.T.; Farnham, J.M.; Albright, L.A. Prostate cancer risk prediction based on complete prostate cancer family history. J. Korean Med. Sci. 2015, 75, 390–398. [Google Scholar] [CrossRef] [PubMed]




| Purification Procedure | Enzyme Activity (U) | Protein (mg) | Specific Activity (U/mg) | Fold Purification | Yield (%) |
|---|---|---|---|---|---|
| Cell-Free Extract | 8633 | 764 | 11.29 | 1.0 | 100 |
| Ammonium Sulphate | 6754 | 295 | 22.89 | 2.03 | 78.23 |
| Sephadex G-200 Column | 3631 | 85.17 | 42.63 | 3.77 | 42.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mostafa, Y.S.; Alamri, S.A.; Alfaifi, M.Y.; Alrumman, S.A.; Elbehairi, S.E.I.; Taha, T.H.; Hashem, M. L-Glutaminase Synthesis by Marine Halomonas meridiana Isolated from the Red Sea and Its Efficiency against Colorectal Cancer Cell Lines. Molecules 2021, 26, 1963. https://doi.org/10.3390/molecules26071963
Mostafa YS, Alamri SA, Alfaifi MY, Alrumman SA, Elbehairi SEI, Taha TH, Hashem M. L-Glutaminase Synthesis by Marine Halomonas meridiana Isolated from the Red Sea and Its Efficiency against Colorectal Cancer Cell Lines. Molecules. 2021; 26(7):1963. https://doi.org/10.3390/molecules26071963
Chicago/Turabian StyleMostafa, Yasser S., Saad A. Alamri, Mohammad Y. Alfaifi, Sulaiman A. Alrumman, Serag Eldin I. Elbehairi, Tarek H. Taha, and Mohamed Hashem. 2021. "L-Glutaminase Synthesis by Marine Halomonas meridiana Isolated from the Red Sea and Its Efficiency against Colorectal Cancer Cell Lines" Molecules 26, no. 7: 1963. https://doi.org/10.3390/molecules26071963
APA StyleMostafa, Y. S., Alamri, S. A., Alfaifi, M. Y., Alrumman, S. A., Elbehairi, S. E. I., Taha, T. H., & Hashem, M. (2021). L-Glutaminase Synthesis by Marine Halomonas meridiana Isolated from the Red Sea and Its Efficiency against Colorectal Cancer Cell Lines. Molecules, 26(7), 1963. https://doi.org/10.3390/molecules26071963