Quantitative Detection of Chromium Pollution in Biochar Based on Matrix Effect Classification Regression Model
Abstract
:1. Introduction
2. Results and Discussion
2.1. Matrix Elemental Analysis
2.2. Spectral Classification of Matrix Effect
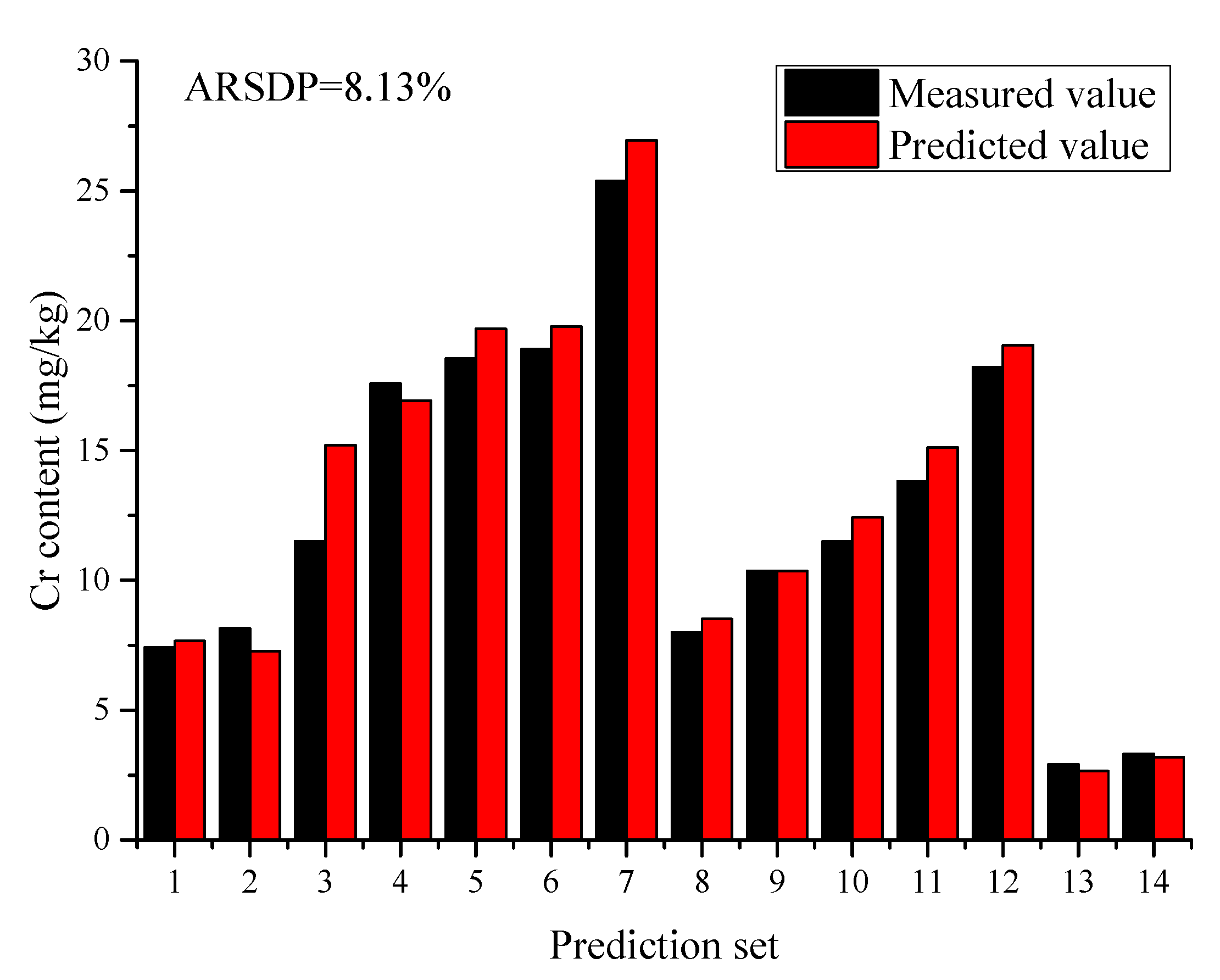
2.3. Classification Regression Model
3. Experimental Design and Methods
3.1. LIBS Device
3.2. Sample Preparation
3.3. Matrix Effect Classification Regression Modeling and Evaluation Criteria
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhang, Z.X.; Wu, J.; Chen, W.F. Review on Prepation and Application of Biochar. Adv. Mater. Res. 2014, 898, 456–460. [Google Scholar] [CrossRef]
- De Morais, C.P.; Barros, A.I.; Júnior, D.S.; Ribeiro, C.A.; Crespi, M.S.; Senesi, G.S.; Neto, J.A.G.; Ferreira, E.C. Calcium determination in biochar-based fertilizers by laser-induced breakdown spectroscopy using sodium as internal standard. Microchem. J. 2017, 134, 370–373. [Google Scholar] [CrossRef] [Green Version]
- Dong, M.; Wei, L.; Lu, J.; Li, W.; Lu, S.; Li, S.; Liu, C.; Yoo, J.H.; Li, W. A comparative model combining carbon atomic and molecular emissions based on partial least squares and support vector regression correction for carbon analysis in coal using LIBS. J. Anal. At. Spectrom. 2019, 34, 480–488. [Google Scholar] [CrossRef]
- Sheta, S.; Afgan, M.S.; Hou, Z.; Yao, S.-C.; Zhang, L.; Li, Z.; Wang, Z. Coal analysis by laser-induced breakdown spectroscopy: A tutorial review. J. Anal. At. Spectrom. 2019, 34, 1047–1082. [Google Scholar] [CrossRef]
- Andrade, D.F.; Filho, E.R.P. Direct Determination of Contaminants, Major and Minor Nutrients in Solid Fertilizers Using Laser-Induced Breakdown Spectroscopy (Libs). J. Agric. Food Chem. 2016, 64, 7890–7898. [Google Scholar] [CrossRef]
- Singh, V.K.; Kumar, V.; Sharma, J. Importance of laser-induced breakdown spectroscopy for hard tissues (bone, teeth) and other calcified tissue materials. Lasers Med Sci. 2014, 30, 1763–1778. [Google Scholar] [CrossRef]
- Jiang, T.; Guo, Z.; Ma, M.; Fang, L.; Yang, M.; Li, S.; Liu, J.; Zhao, N.; Huang, X.; Liu, W. Electrochemical Laser Induced Breakdown Spectroscopy for Enhanced Detection of Cd(II) without Interference in Rice on Layer-by-Layer Assembly of Gra-phene Oxides. Electrochim. Acta 2016, 216, 188–195. [Google Scholar] [CrossRef]
- Meng, D.; Zhao, N.; Wang, Y.; Ma, M.; Fang, L.; Gu, Y.; Jia, Y.; Liu, J. On-line/on-site analysis of heavy metals in water and soils by laser induced breakdown spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2017, 137, 39–45. [Google Scholar] [CrossRef]
- Peng, J.; He, Y.; Zhao, Z.; Jiang, J.; Zhou, F.; Liu, F.; Shen, T. Fast visualization of distribution of chromium in rice leaves by re-heating dual-pulse laser-induced breakdown spectroscopy and chemometric methods. Environ. Pollut. 2019, 252, 1125–1132. [Google Scholar] [CrossRef]
- Bunn, T.L.; Dietert, R.R.; Ladics, G.S.; Holsapple, M.P.; Bunn, R.R.D.T.L. Developmental Immunotoxicology Assessment in the Rat: Age, Gender, and Strain Comparisons after Exposure to Lead. Toxicol. Methods 2001, 11, 41–58. [Google Scholar] [CrossRef]
- Einsporn, S.; Bressling, J.; Koehler, A. Cellular Ellular Localization of Lead Using an Antibody-Based Detection System and Enzyme Activity Changes in the Gills and Digestive Gland of the Blue Mussel Mytilus Edulis. Environ. Toxicol. Chem. 2009, 28, 402–408. [Google Scholar] [CrossRef]
- Pearton, S.; Ren, F.; Wang, Y.-L.; Chu, B.H.; Chen, K.H.; Chang, C.Y.; Lim, W.; Lin, J. Recent Advances in Wide Bandgap Semiconductor Biological and Gas Sensors. MRS Proc. 2009, 1202, 1–59. [Google Scholar] [CrossRef]
- Douchiche, O.; Driouich, A.; Morvan, C. Spatial Regulation of Cell-Wall Structure in Response to Heavy Metal Stress: Cad-mium-Induced Alteration of the Methyl-Esterification Pattern of Homogalacturonans. Ann. Bot. 2010, 105, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Huang, E.P.; Marquis, C.P.; Gray, P.P. Development Ofsuper-Cho Protein-Free Medium Based on a Statistical Design. J. Chem. Technol. Biotechnol. 2007, 82, 431–441. [Google Scholar] [CrossRef]
- Kramer, K.; Fiedler, M.; Skerra, A.; Hock, B. A Generic Strategy for Subcloning Antibody Variable Regions from the Scfv Phage Display Vector Pcantab 5 E into Pask85 Permits the Economical Production of Fab Fragments and Leads to Improved Recom-binant Immunoglobulin Stability. Biosens. Bioelectron. 2002, 17, 305–313. [Google Scholar] [CrossRef]
- Li, H.; Liu, S.; Hassan, M.M.; Ali, S.; Ouyang, Q.; Chen, Q.; Wu, X.; Xu, Z. Rapid Quantitative Analysis of Hg2+ Residue in Dairy Products Using Sers Coupled with Aco-Bp-Adaboost Algorithm. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 223, 117281. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, M.G.; Lerose, R.; Cigliano, S.; Leone, A. Regulation by heavy metals and temperature of the human BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 2003, 541, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Rezk, R.; Galmed, A.; Abdelkreem, M.; Ghany, N.A.; Harith, M. Quantitative analysis of Cu and Co adsorbed on fish bones via laser-induced breakdown spectroscopy. Opt. Laser Technol. 2016, 83, 131–139. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, J.; Tian, Y.; Lu, B.; Hang, Y.; Chen, Q. Development of deep learning method for lead content prediction of lettuce leaf using hyperspectral images. Int. J. Remote Sens. 2020, 41, 2263–2276. [Google Scholar] [CrossRef]
- Alexander, D.; Ellerby, R.; Hernandez, A.; Wu, F.; Amarasiriwardena, D. Investigation of simultaneous adsorption properties of Cd, Cu, Pb and Zn by pristine rice husks using ICP-AES and LA-ICP-MS analysis. Microchem. J. 2017, 135, 129–139. [Google Scholar] [CrossRef]
- Ayazi, Z.; Banihashemi, M. Determination of trace amount of silver in water samples by flame atomic absorption after preconcentration by ZnO nano sorbent. Sep. Sci. Technol. 2015, 51, 585–593. [Google Scholar] [CrossRef]
- Jing, F.; Yang, Z.; Chen, X.; Liu, W.; Guo, B.; Lin, G.; Huang, R.; Liu, W. Potentially hazardous element accumulation in rice tissues and their availability in soil systems after biochar amendments. J. Soils Sediments 2019, 19, 2957–2970. [Google Scholar] [CrossRef]
- Ito, S.; Tsukada, K. Matrix Effect and Correction by Standard Addition in Quantitative Liquid Chromatographic-Mass Spec-trometric Analysis of Diarrhetic Shellfish Poisoning Toxins. J. Chromatogr. 2001, 943, 39–46. [Google Scholar] [CrossRef]
- Choe, K.-Y.; Gajek, R. Determination of trace elements in human urine by ICP-MS using sodium chloride as a matrix-matching component in calibration. Anal. Methods 2016, 8, 6754–6763. [Google Scholar] [CrossRef]
- Unsal, Y.E.; Soylak, M.; Tuzen, M. Ultrasound-assisted ionic liquid-based dispersive liquid–liquid microextraction for preconcentration of patent blue V and its determination in food samples by UV–visible spectrophotometry. Environ. Monit. Assess. 2015, 187, 187–203. [Google Scholar] [CrossRef] [PubMed]
- J, S.G.; L, R.M. Hierarchical Clustering Via Joint between-within Distances: Extending Ward’s Minimum Variance Method. J. Classif. 2005, 22, 151–183. [Google Scholar]
- Du, X. Identifying Control and Management Plane Poison Message Failure by K-Nearest Neighbor Method. J. Netw. Syst. Manag. 2006, 14, 243–259. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Zhang, P.; Wang, X.; Cao, Y.; Han, L. Qualitative and quantitative correlation of physicochemical characteristics and lead sorption behaviors of crop residue-derived chars. Bioresour. Technol. 2018, 270, 545–553. [Google Scholar] [CrossRef]
- Medyńska-Juraszek, A.; Ćwieląg-Piasecka, I.; Jerzykiewicz, M.; Trynda, J. Wheat Straw Biochar as a Specific Sorbent of Cobalt in Soil. Materials 2020, 13, 2462. [Google Scholar] [CrossRef]
- Tang, Y.; Li, J.; Hao, Z.; Tang, S.; Zhu, Z.; Guo, L.; Li, X.; Zeng, X.; Duan, J.; Lu, Y. Multielemental self-absorption reduction in laser-induced breakdown spectroscopy by using microwave-assisted excitation. Opt. Express 2018, 26, 12121–12130. [Google Scholar] [CrossRef]
- Fu, X.; Duan, F.-J.; Huang, T.-T.; Ma, L.; Jiang, J.-J.; Li, Y.-C. A fast variable selection method for quantitative analysis of soils using laser-induced breakdown spectroscopy. J. Anal. At. Spectrom. 2017, 32, 1166–1176. [Google Scholar] [CrossRef]
- Duan, F.; Fu, X.; Jiang, J.; Huang, T.; Ma, L.; Zhang, C. Automatic variable selection method and a comparison for quantitative analysis in laser-induced breakdown spectroscopy. Spectrochim. Acta Part B At. Spectrosc. 2018, 143, 12–17. [Google Scholar] [CrossRef]
- Wang, T.; He, M.; Shen, T.; Liu, F.; He, Y.; Liu, X.; Qiu, Z. Multi-Element Analysis of Heavy Metal Content in Soils Using La-ser-Induced Breakdown Spectroscopy: A Case Study in Eastern China. Spectrochim. Acta Part B At. Spectrosc. 2018, 149, 300–312. [Google Scholar] [CrossRef]
- Duan, H.; Han, L.; Huang, G. Quantitative Analysis of Major Metals in Agricultural Biochar Using Laser-Induced Breakdown Spectroscopy with an Adaboost Artificial Neural Network Algorithm. Molecules 2019, 24, 3753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Li, A.; Wang, X.; Ding, C.; Qiu, S.; He, Y.; Lu, T.; He, F.; Zou, B.; Liu, R. The high-accuracy prediction of carbon content in semi-coke by laser-induced breakdown spectroscopy. J. Anal. At. Spectrom. 2020, 35, 984–992. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Li, G.; Du, Q.; Yang, F. Preparation of montmorillonite modified biochar with various temperatures and their mechanism for Zn ion removal. J. Hazard. Mater. 2020, 391, 121692. [Google Scholar] [CrossRef]
- Duan, H.; Ma, S.; Han, L.; Huang, G. A novel denoising method for laser-induced breakdown spectroscopy: Improved wavelet dual threshold function method and its application to quantitative modeling of Cu and Zn in Chinese animal manure composts. Microchem. J. 2017, 134, 262–269. [Google Scholar] [CrossRef]
- Taylor, P.J. Matrix effects: The Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry. Clin. Biochem. 2005, 38, 328–334. [Google Scholar] [CrossRef]
- Van Eeckhaut, A.; Lanckmans, K.; Sarre, S.; Smolders, I.; Michotte, Y. Validation of bioanalytical LC–MS/MS assays: Evaluation of matrix effects. J. Chromatogr. B 2009, 877, 2198–2207. [Google Scholar] [CrossRef]
- Xue, J.; Yang, Z.; Han, L.; Liu, Y.; Liu, Y.; Zhou, C. On-line measurement of proximates and lignocellulose components of corn stover using NIRS. Appl. Energy 2015, 137, 18–25. [Google Scholar] [CrossRef]
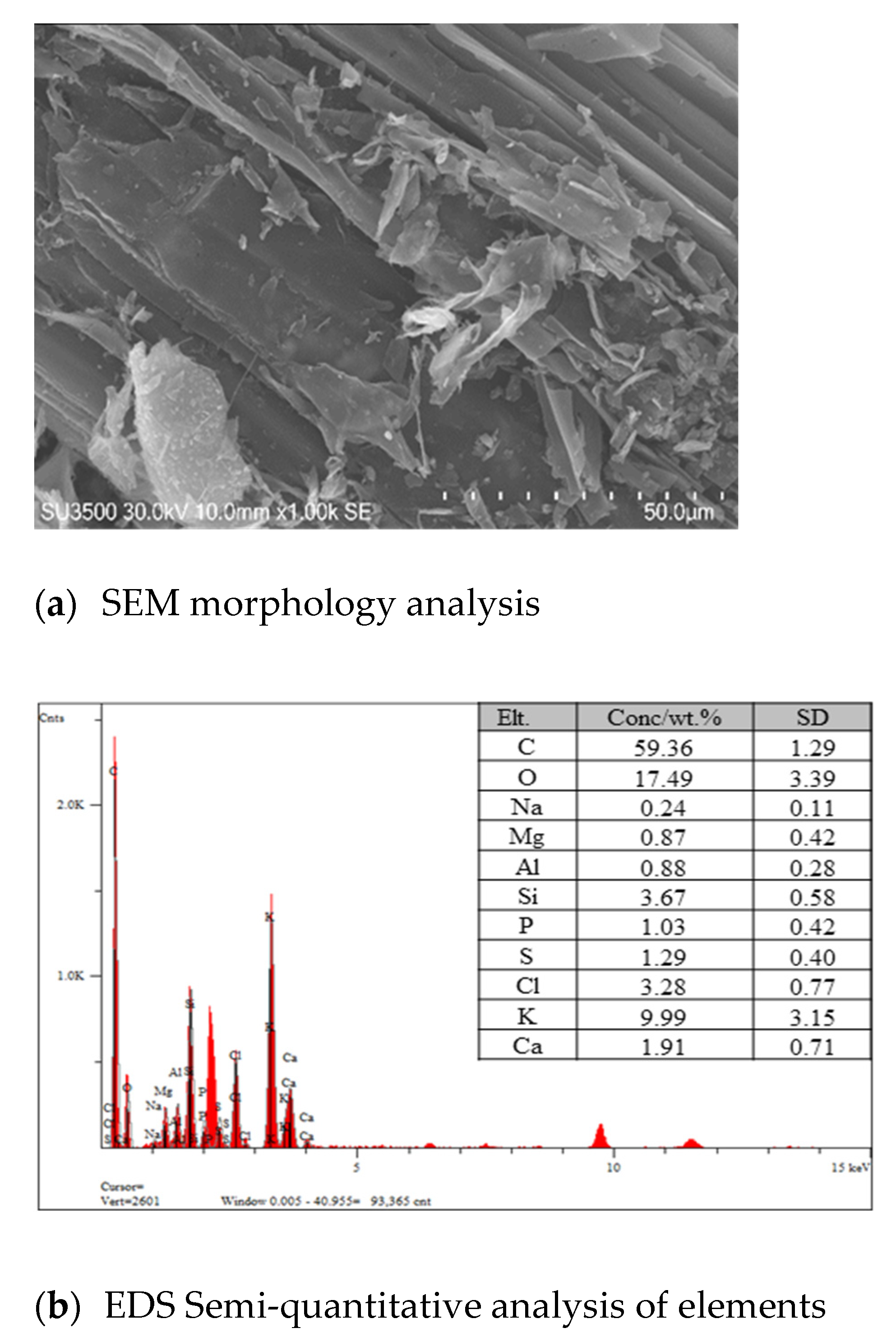
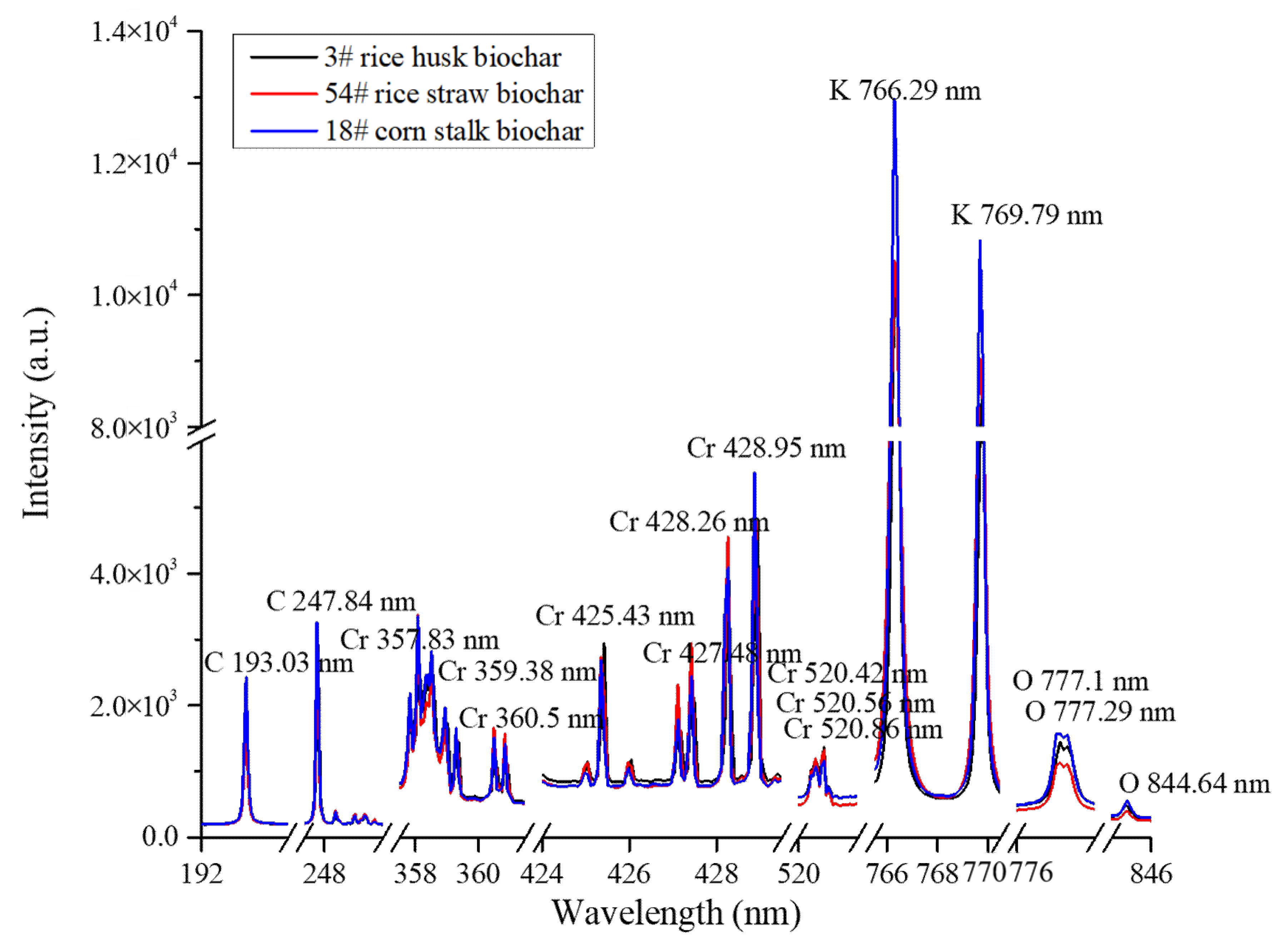
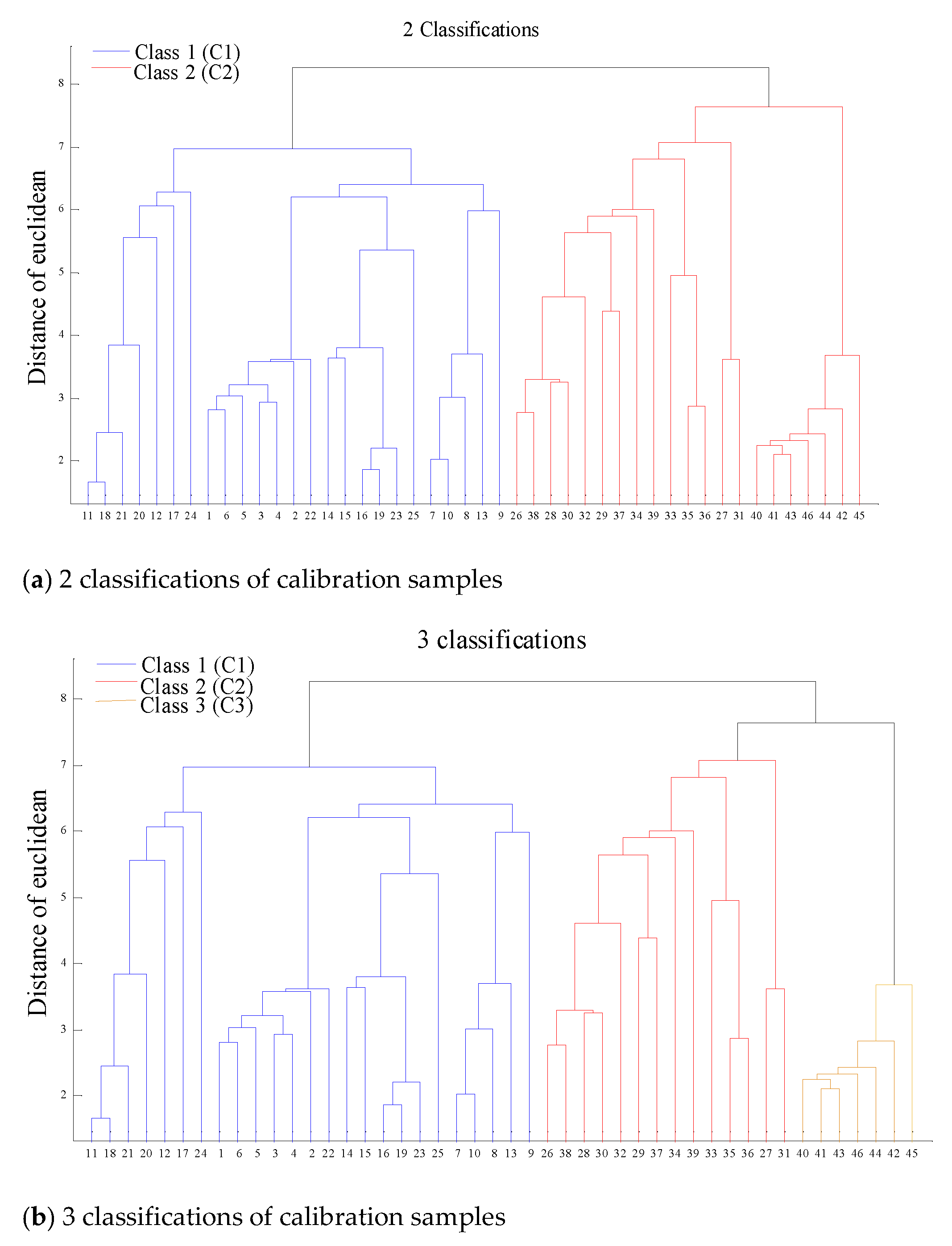
| Main Matrix | 2 classifications | 3 Classifications |
|---|---|---|
| C+O (76.85%) | C1: 1–25 (calibration), 47–53 (prediction) C2: 26–46 (calibration), 54–60(prediction) | c1: 1–25 (calibration), 47–53 (prediction) c2: 26–39 (calibration), 54–58 (prediction) c3: 40–46 (calibration), 59–60 (prediction) |
| Classifications | Classification | RMSECV (mg/kg) | RSDCV (%) | ARSDCV (%) |
|---|---|---|---|---|
| 1 | None | 2.38 | 18.53% | 18.53% |
| 2 | C1 | 1.99 | 12.70% | 18.11% |
| C2 | 2.23 | 23.51% | ||
| 3 | c1 | 1.99 | 12.70% | 12.32% |
| c2 | 1.38 | 11.02% | ||
| c3 | 0.45 | 13.24% |
| Particle | Element | Range (mg/kg) | RSDP/ARSDP (%) | Remarks | Ref. |
|---|---|---|---|---|---|
| Biochar | Cr | 2.92–25.38 | 8.13% | MEC-PLS 1 | In this work |
| Soil | Cr | 48–410 | 23.019 | MIPW-PLS 2 | Fu et al. 2017 [31] |
| Soil | Cr | 48–410 | 17.673 | FSC-MIPW-PLS 3 | Duan et al. 2018 [32] |
| Soil | Cr | 18.29–164.06 | 11.460 | Lasso 4 | Wang et al. 2018 [33] |
| Biochar | Cr | 5.05–19.15 | 17.41% | PLS | Duan et al. 2019 [34] |
| Derivation (#1~#15) | Content | Derivation (#16~#30) | Content | Derivation (#31~#45) | Content | Derivation (#46~#60) | Content |
|---|---|---|---|---|---|---|---|
| rice husk | 7.37 | corn stalk | 18.21 | rice straw | 10.67 | rice husk | 4.5 |
| rice husk | 7.47 | corn stalk | 18.63 | rice straw | 11.47 | rice husk | 7.42 |
| rice husk | 7.83 | corn stalk | 18.88 | rice husk | 11.73 | rice husk | 8.15 |
| rice husk | 8.11 | corn stalk | 18.9 | rice straw | 12.81 | rice husk | 11.51 |
| rice husk | 8.22 | corn stalk | 18.98 | rice husk | 13.75 | corn stalk | 17.59 |
| rice husk | 8.53 | corn stalk | 19.15 | rice husk | 15.15 | corn stalk | 18.55 |
| rice husk | 10.59 | rice husk | 19.56 | rice straw | 16.47 | corn stalk | 18.91 |
| rice husk | 11.54 | corn stalk | 26.15 | corn stalk | 17.08 | corn stalk | 25.39 |
| rice husk | 12.11 | corn stalk | 26.34 | corn stalk | 20.15 | rice straw | 7.99 |
| rice husk | 12.52 | corn stalk | 28.51 | rice straw | 2.79 | rice straw | 10.36 |
| corn stalk | 15.27 | rice straw | 7.9 | rice straw | 2.9 | rice straw | 11.5 |
| corn stalk | 15.61 | rice straw | 8.37 | rice straw | 3.04 | rice straw | 13.81 |
| rice husk | 17.28 | rice straw | 9.32 | rice straw | 3.28 | corn stalk | 18.22 |
| corn stalk | 17.94 | rice straw | 9.9 | rice straw | 3.3 | rice straw | 2.92 |
| corn stalk | 18.03 | rice straw | 10.6 | rice straw | 3.98 | rice straw | 3.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Zhu, R.; Zhang, L.; Zhang, R.; Huang, G.; Duan, H. Quantitative Detection of Chromium Pollution in Biochar Based on Matrix Effect Classification Regression Model. Molecules 2021, 26, 2069. https://doi.org/10.3390/molecules26072069
Guo M, Zhu R, Zhang L, Zhang R, Huang G, Duan H. Quantitative Detection of Chromium Pollution in Biochar Based on Matrix Effect Classification Regression Model. Molecules. 2021; 26(7):2069. https://doi.org/10.3390/molecules26072069
Chicago/Turabian StyleGuo, Mei, Rongguang Zhu, Lixin Zhang, Ruoyu Zhang, Guangqun Huang, and Hongwei Duan. 2021. "Quantitative Detection of Chromium Pollution in Biochar Based on Matrix Effect Classification Regression Model" Molecules 26, no. 7: 2069. https://doi.org/10.3390/molecules26072069





