Lanthanide DO3A-Complexes Bearing Peptide Substrates: The Effect of Peptidic Side Chains on Metal Coordination and Relaxivity
Abstract
1. Introduction
2. Results and Discussion
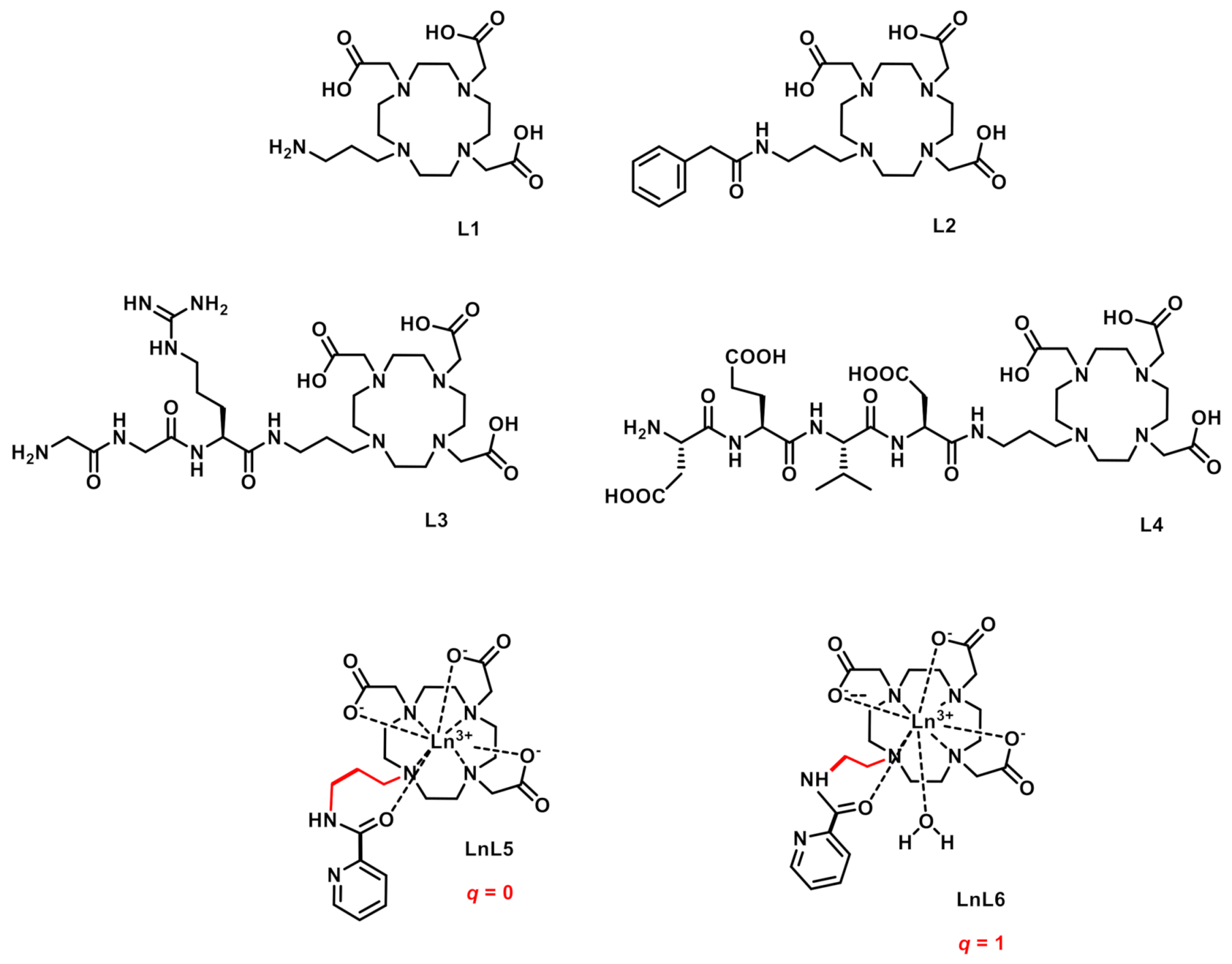
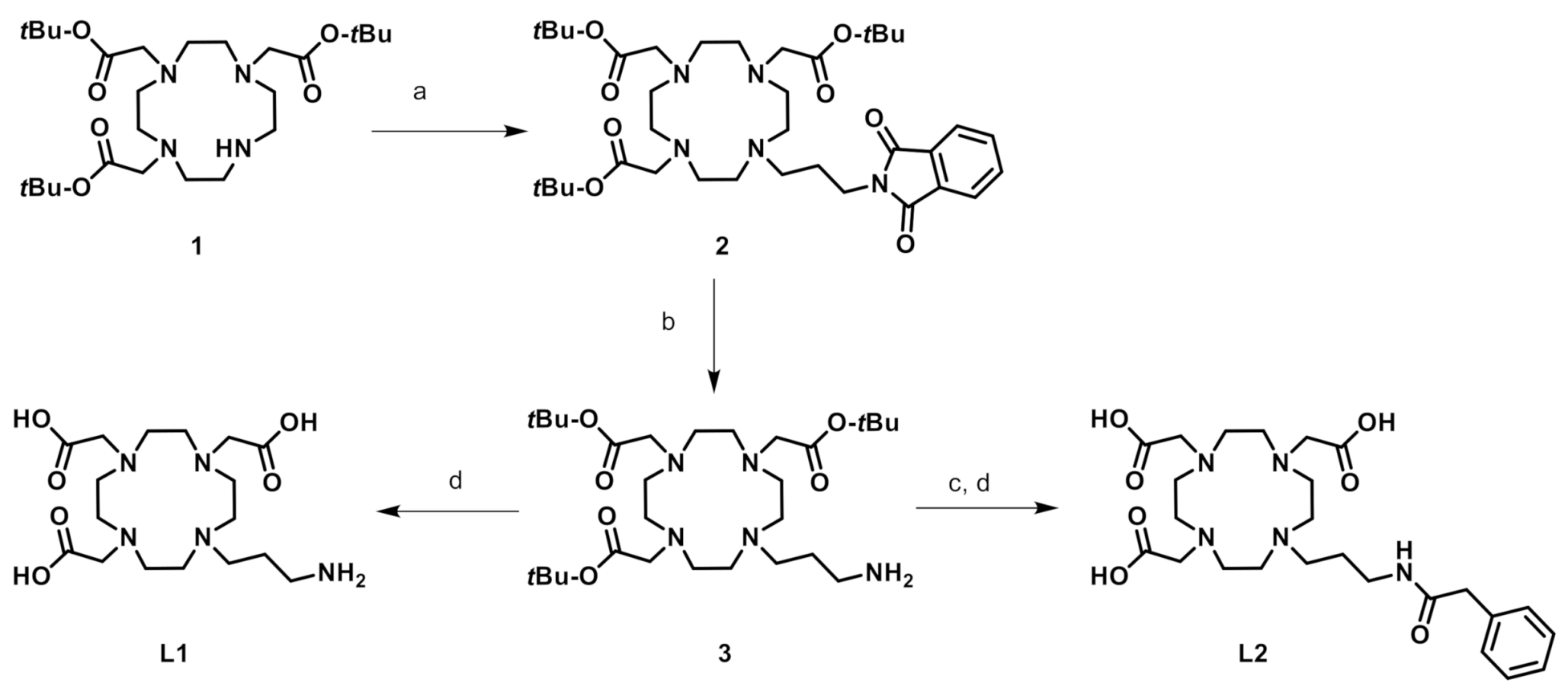
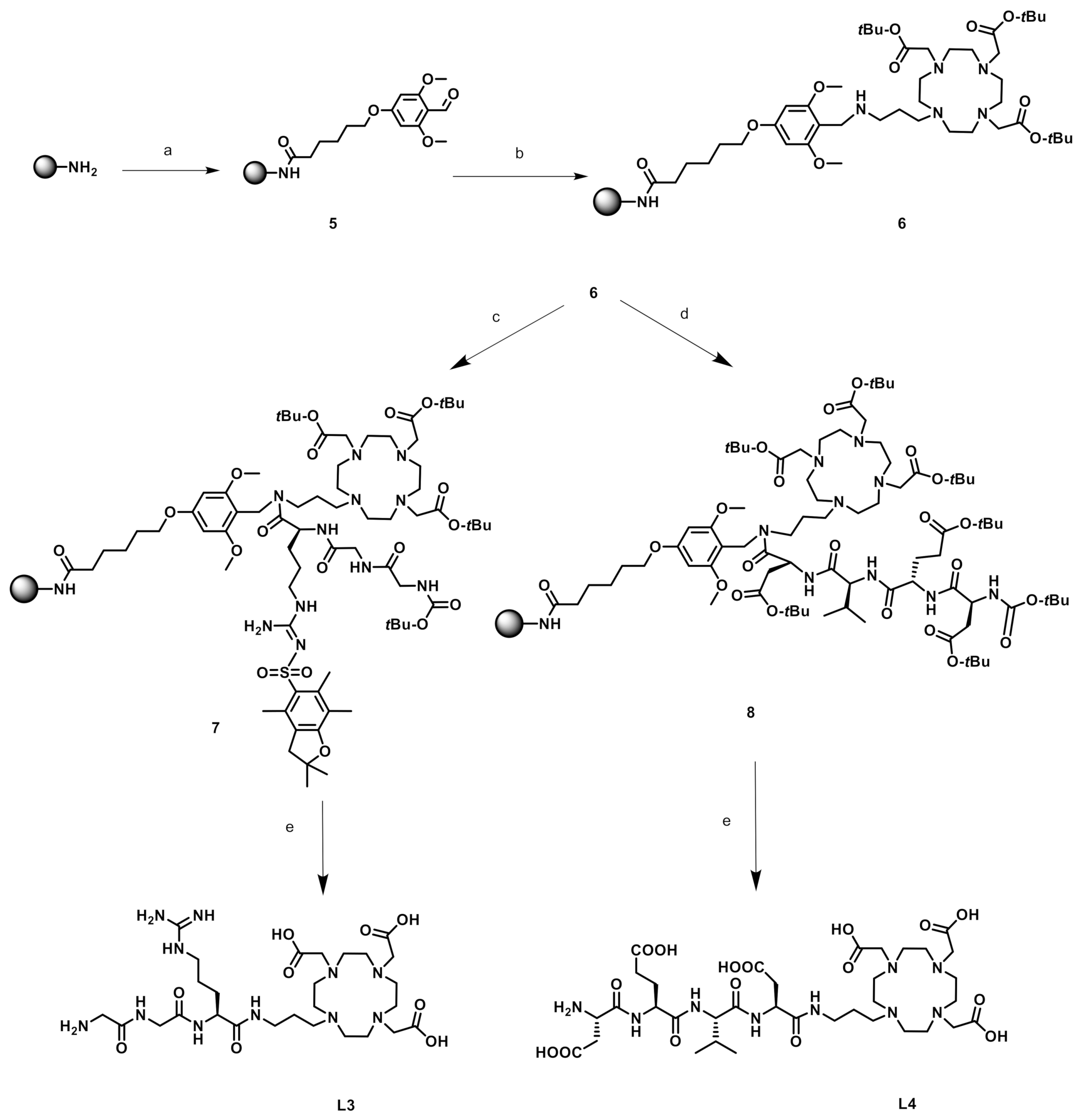
2.1. Syntheses of the Ligands and Complexes
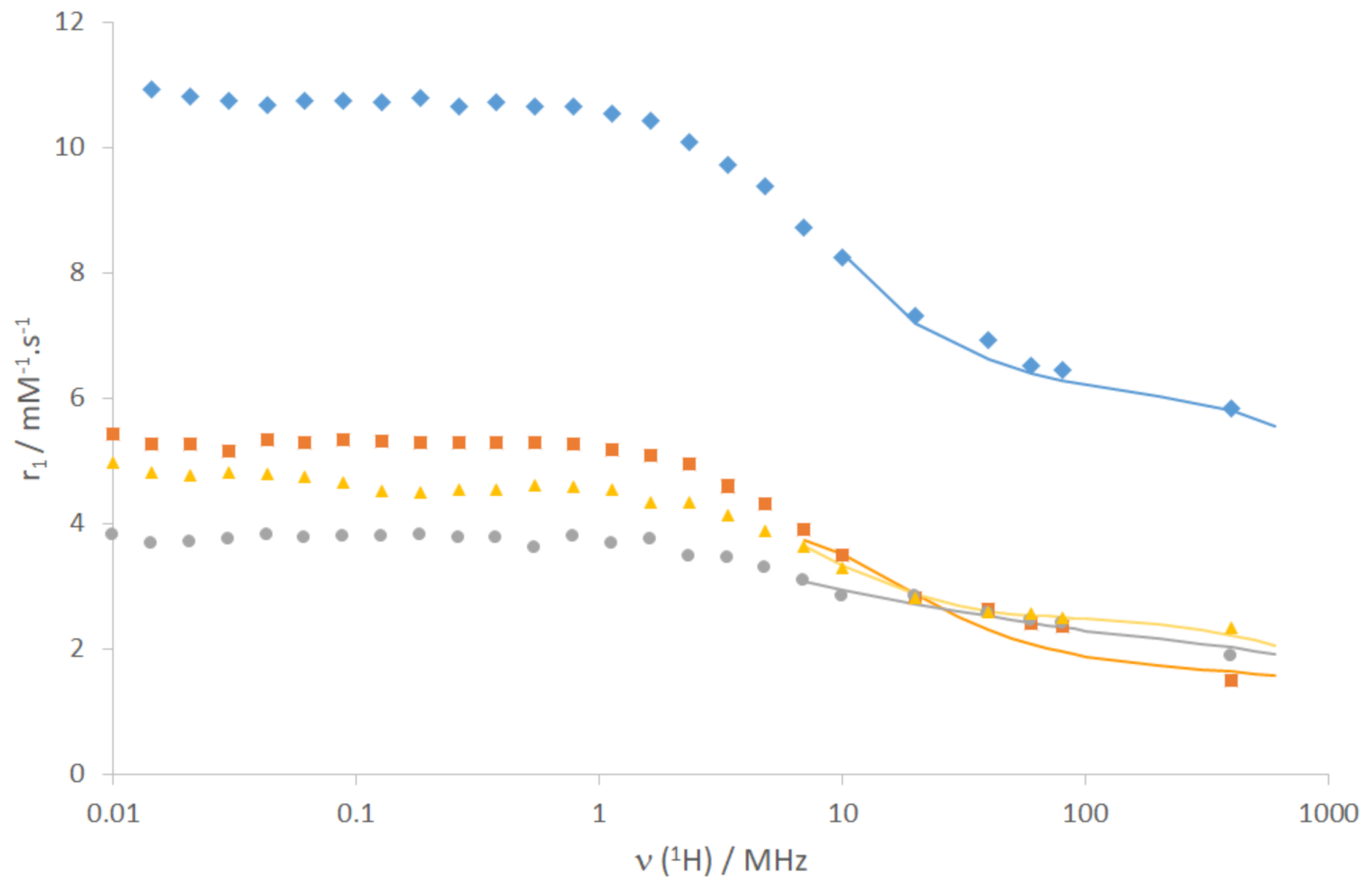
2.2. Relaxometric Characterization of the Gd3+ Complexes
- (a)
- As expected, EuL1 has the highest number of coordinated water molecules of the whole series (q ≈ 2), with a correspondingly high relaxivity for the Gd3+ analogue.
- (b)
- EuL2 has one coordinated water, implying that the coordination of the amide function still leaves space for one water molecule in the inner sphere. Despite their similar structure, this is in sharp contrast to LnL5, which has no inner-sphere water. Likely due to its higher flexibility around the methylene group, the benzyl moiety of LnL2 does not seem to hinder the access of the water molecule to Ln3+ as much as the pyridine. This difference highlights the difficulty of predicting the coordination mode, even in structurally analogous complexes. The hydration number q = 1 of GdL2 is in full accordance with its relaxivity, 70% higher than that of GdL5 [27].
- (c)
- EuL4 revealed to be non-hydrated, i.e., in addition to the amide coordination, either a carboxylate function from the peptide side chain (aspartate or glutamate) completes the coordination sphere to CN = 9, or the peptide is too “bulky” and prevents water access to Gd3+ (CN = 8). The relaxivity of GdL4 is remarkably high for a non-hydrated complex. Indeed, in the absence of hydration water, the relaxivity should be purely of outer-sphere origin, governed by the diffusion of bulk water molecules in the vicinity of the Gd3+. A rough estimation of an outer-sphere relaxivity gives a value of ca. 2.7 mM−1·s−1 at 20 MHz, 25 °C. For the outer-sphere mechanism, relaxivity is mainly determined by diffusion (of water molecules and the complex), and the distance of closest approach between water protons and Gd3+. Although the distance can vary depending on the geometry of the complex, this cannot plausibly account for such a high relaxivity. Much more likely, a second-sphere relaxivity mechanism operates due to the numerous carboxylate and amide functions of the peptid side-chain, which can retain water molecules in the vicinity of Gd3+ by hydrogen bonds.
- (d)
- For LnL3, the interpretation of the luminescence lifetimes and the estimation of q are more complex. In a first consideration, one can hypothesize a similar coordination mode to that of LnL2, involving the coordination of the macrocycle amines, the three macrocycle carboxylates and the amide to the Ln3+ ion. In this case, similarly to EuL2, Equation 1 indicates monohydration (q = 0.7(3)). However, this is not coherent with the relaxivities, which, for small molecular complexes like GdL2 and GdL3, are mainly dictated by rotation and thus molecular size. Given the bigger size of GdL3, if both GdL2 and GdL3 were monohydrated, GdL3 should have superior relaxivity. However, it is even lower than the relaxivity of the non-hydrated GdL4, suggesting that the number of water molecules coordinated could be rather close to 0. In LnL3, the amine functions of the peptide side chain can also potentially coordinate to the metal, which should then be taken into account in Equation 1. Considering the extra coordination of an amine in Equation 1, q = −0.4 is estimated for EuL3. Given the error on this value, the extra nitrogen coordination to Ln3+ leads to a q-value close to 0. (Though less likely due to steric reasons, an amide of the peptide chain might also coordinate; in this case, the estimated q would be 0.6). Overall, the combined analysis of the relaxivities and the luminescence lifetimes suggests that no water molecule is present in the first coordination sphere of LnL3, and the relaxivity of GdL3 is governed by a second sphere effect, similarly to GdL4.
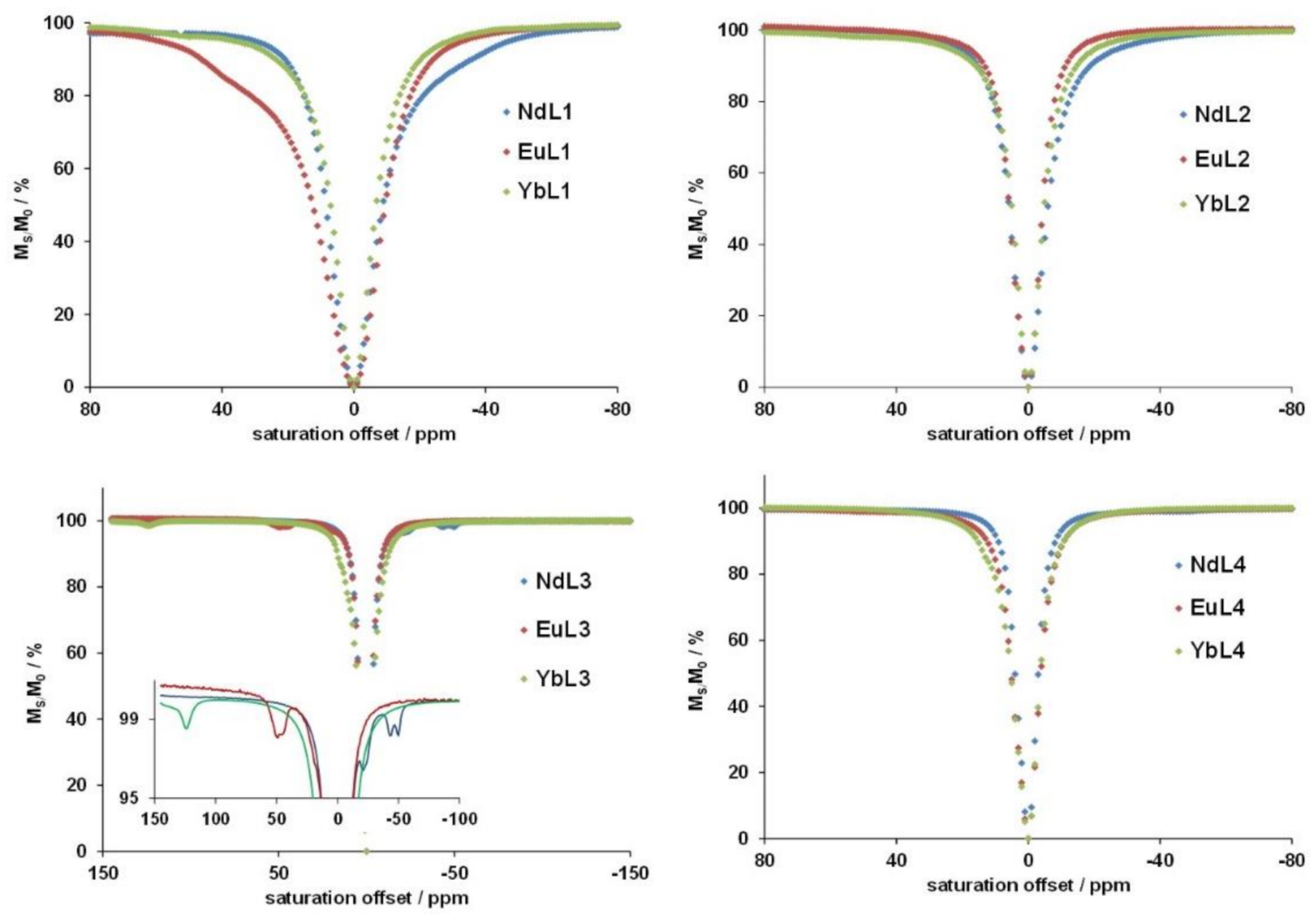
2.3. ParaCEST Properties of Eu3+, Yb3+ and Nd3+ Complexes
2.4. Enzymatic Cleavage
3. Materials and Methods
3.1. General Information
3.2. Synthesis
3.3. Sample Preparation
3.4. Relaxometric Measurements
3.5. Luminescence Lifetime Measurements
3.6. PARACEST and NMR
3.7. Enzymatic Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Soleimany, A.P.; Bhatia, S.N. Activity-based diagnostics: An emerging paradigm for disease detection and monitoring. Trends Mol. Med. 2020, 26, 450–468. [Google Scholar] [CrossRef] [PubMed]
- Wahsner, J.; Gale, E.M.; Rodríguez-Rodríguez, A.; Caravan, P. Chemistry of MRI contrast agents: Current challenges and new frontiers. Chem. Rev. 2018, 119, 957–1057. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, D.V.; Yoo, B.; Bernstein, A.S.; Pagel, M.D. Detecting enzyme activities with exogenous MRI contrast agents. Chem. Eur. J. 2014, 20, 9840–9850. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Meade, T.J. Molecular magnetic resonance imaging with Gd(III)-based contrast agents: Challenges and key advances. J. Am. Chem. Soc. 2019, 141, 17025–17041. [Google Scholar] [CrossRef]
- Kim, D.; Lee, Y.D.; Jo, S.; Kim, S.; Lee, T.S. Detection and imaging of cathepsin l in cancer cells using the aggregation of conjugated polymer dots and magnetic nanoparticles. Sens. Actuator B Chem. 2020, 307, 10. [Google Scholar] [CrossRef]
- Korchak, S.; Jagtap, A.P.; Glöggler, S. Signal-enhanced real-time magnetic resonance of enzymatic reactions at millitesla fields. Chem. Sci. 2021, 12, 314–319. [Google Scholar] [CrossRef]
- Wyskocka-Gajda, M.; Przypis, L.; Olesiejuk, M.; Krawczyk, T.; Kuznik, A.; Nawara, K.; Minoshima, M.; Sugihara, F.; Kikuchi, K.; Kuznik, N. A step towards gadolinium-free bioresponsive MRI contrast agent. Eur. J. Med. Chem. 2021, 211, 10. [Google Scholar] [CrossRef]
- Eills, J.; Cavallari, E.; Kircher, R.; Di Matteo, G.; Carrera, C.; Dagys, L.; Levitt, M.H.; Ivanov, K.L.; Aime, S.; Reineri, F.; et al. Singlet-contrast magnetic resonance imaging: Unlocking hyperpolarization with metabolism **. Angew. Chem.-Int. Edit. 2021, 60, 6791–6798. [Google Scholar] [CrossRef]
- Mason, S.D.; Joyce, J.A. Proteolytic networks in cancer. Trends Cell. Biol. 2011, 21, 228–237. [Google Scholar] [CrossRef]
- Vizovisek, M.; Ristanovic, D.; Menghini, S.; Christiansen, M.G.; Schuerle, S. The tumor proteolytic landscape: A challenging frontier in cancer diagnosis and therapy. Int. J. Mol. Sci. 2021, 22, 2514. [Google Scholar] [CrossRef]
- Merbach, A.; Helm, L.; Toth, E. The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging, 2nd ed.; John Wiley & Sons: Chichester, UK, 2013. [Google Scholar]
- Vinogradov, E.; Sherry, A.D.; Lenkinski, R.E. CEST: From basic principles to applications, challenges and opportunities. J. Magn. Reson. 2013, 229, 155–172. [Google Scholar] [CrossRef]
- Woods, M.; Woessner, D.E.; Sherry, A.D. Paramagnetic lanthanide complexes as paracest agents for medical imaging. Chem. Soc. Rev. 2006, 35, 500–511. [Google Scholar] [CrossRef]
- Hingorani, D.V.; Bernstein, A.S.; Pagel, M.D. A review of responsive MRI contrast agents: 2005–2014. Contrast Media Mol. Imaging 2015, 10, 245–265. [Google Scholar] [CrossRef]
- Toth, E.; Bonnet, C.S. Responsive paracest contrast agents. Inorganics 2019, 7, 68. [Google Scholar] [CrossRef]
- Lilley, L.M.; Kamper, S.; Caldwell, M.; Chia, Z.K.; Ballweg, D.; Vistain, L.; Krimmel, J.; Mills, T.A.; MacRenaris, K.; Lee, P.; et al. Self-immolative activation of beta-galactosidase-responsive probes for in vivo mr imaging in mouse models. Angew. Chem. Int. Ed. 2020, 59, 388–394. [Google Scholar] [CrossRef]
- Sinharay, S.; Randtke, E.A.; Howison, C.M.; Ignatenko, N.A.; Pagel, M.D. Detection of enzyme activity and inhibition during studies in solution, in vitro and in vivo with catalycest MRI. Mol. Imaging Biol. 2018, 20, 240–248. [Google Scholar] [CrossRef]
- Ali, M.M.; Liu, G.; Shah, T.; Flask, C.A.; Pagel, M.D. Using two chemical exchange saturation transfer magnetic resonance imaging contrast agents for molecular imaging studies. Acc. Chem. Res. 2009, 42, 915–924. [Google Scholar] [CrossRef]
- Yoo, B.; Pagel, M.D. A paracest MRI contrast agent to detect enzyme activity. J. Am. Chem. Soc. 2006, 128, 14032–14033. [Google Scholar] [CrossRef]
- Yoo, B.; Sheth, V.R.; Howison, C.M.; Douglas, M.J.K.; Pineda, C.T.; Maine, E.A.; Baker, A.F.; Pagel, M.D. Detection of in vivo enzyme activity with catalycest MRI. Magn. Reson. Med. 2014, 71, 1221–1230. [Google Scholar] [CrossRef]
- Sinharay, S.; Howison, C.M.; Baker, A.F.; Pagel, M.D. Detecting in vivo urokinase plasminogen activator activity with a catalycest MRI contrast agent. NMR Biomed. 2017, 30, e3721. [Google Scholar] [CrossRef]
- Chauvin, T.; Durand, P.; Bernier, M.; Meudal, H.; Doan, B.T.; Noury, F.; Badet, B.; Beloeil, J.C.; Toth, E. Detection of enzymatic activity by paracest MRI: A general approach to target a large variety of enzymes. Angew. Chem. Int. Ed. 2008, 47, 4370–4372. [Google Scholar] [CrossRef]
- Chauvin, T.; Torres, S.; Rosseto, R.; Kotek, J.; Badet, B.; Durand, P.; Toth, E. Lanthanide(III) complexes that contain a self-immolative arm: Potential enzyme responsive contrast agents for magnetic resonance imaging. Chem. Eur. J. 2012, 18, 1408–1418. [Google Scholar] [CrossRef]
- He, J.F.; Bonnet, C.S.; Eliseeva, S.V.; Lacerda, S.; Chauvin, T.; Retailleau, P.; Szeremeta, F.; Badet, B.; Petoud, S.; Toth, E.; et al. Prototypes of lanthanide(III) agents responsive to enzymatic activities in three complementary imaging modalities: Visible/near-infrared luminescence, paracest-, and T1-MRI. J. Am. Chem. Soc. 2016, 138, 2913–2916. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhang, R.; Zhang, T.; Xing, D. H2O2-responsive biodegradable nanomedicine for cancer-selective dual-modal imaging guided precise photodynamic therapy. Biomaterials 2019, 207, 39–48. [Google Scholar] [CrossRef]
- Redy, O.; Kisin-Finfer, E.; Sella, E.; Shabat, D. A simple fret-based modular design for diagnostic probes. Org. Biomol. Chem. 2012, 10, 710–715. [Google Scholar] [CrossRef]
- Congreve, A.; Parker, D.; Gianolio, E.; Botta, M. Steric control of lanthanide hydration state: Fast water exchange at gadolinium ion in a mono-amide dota complex. Dalton Trans. 2004, 9, 1441–1445. [Google Scholar] [CrossRef]
- Yoo, B.; Pagel, M.D. A facile synthesis of a-amino-dota as a versatile molecular imaging probe. Tet. Lett. 2006, 47, 7327–7330. [Google Scholar] [CrossRef]
- Yang, Y. Side Reactions in Peptide Synthesis. In Peptide Racemization; Academic Press: Cambridge, MA, USA, 2016; Chapter 11; pp. 257–292. [Google Scholar]
- Yoo, B.; Sheth, V.R.; Pagel, M.D. An amine-derivatized, dota-loaded polymeric support for Fmoc solid phase peptide synthesis. Tet. Lett. 2009, 50, 4459–4462. [Google Scholar] [CrossRef]
- Supkowski, R.M.; Horrocks, D., Jr. Displacement of inner-sphere water molecules from Eu(III) analogues of Gd(III) MRI contrast agents by carbonate and phosphonate anions: Dissociation constants from luminescence data in the rapid-exchange limit. Inorg. Chem. 1999, 38, 5616–5619. [Google Scholar] [CrossRef]
- Toth, E.; Ni Dhubhghaill, O.M.; Besson, G.; Helm, L.; Merbach, A.E. Coordination equilibrium—A clue for fast water exchange on potential magnetic resonance imaging contrast agents? Magn. Reson. Chem. 1999, 37, 701–708. [Google Scholar] [CrossRef]
- Lebduskova, P.; Hermann, P.; Helm, L.; Toth, E.; Kotek, J.; Binnemans, K.; Rudovsky, J.; Lukes, I.; Merbach, A.E. Gadolinium(III) complexes of mono- and diethyl esters of monophosphonic acid analogue of dota as potential MRI contrast agents: Solution structures and relaxometric studies. Dalton Trans. 2007, 4, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Krchová, T.; Kotek, J.; Jirák, D.; Havlíčková, J.; Císařová, I.; Hermann, P. Lanthanide(III) complexes of aminoethyl-do3a as paracest contrast agents based on decoordination of the weakly bound amino group. Dalton Trans. 2013, 42, 15735–15747. [Google Scholar] [CrossRef] [PubMed]
- Aime, S.; Barge, A.; Castelli, D.D.; Fedeli, F.; Mortillaro, A.; Nielsen, F.U.; Terreno, E. Paramagnetic lanthanide(III) complexes as ph-sensitive chemical exchange saturation transfer (CEST) contrast agents for MRI applications. Magn. Reson. Med. 2002, 47, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Terreno, E.; Delli Castelli, D.; Cravotto, G.; Milone, L.; Aime, S. Ln(III)-dotamgiy complexes: A versatile series to assess the determinants of the efficacy of paramagnetic chemical exchange saturation transfer agents for magnetic resonance imaging applications. Investig. Radiol. 2004, 39, 235–243. [Google Scholar] [CrossRef]
- Aime, S.; Botta, M.; Fasano, M.; Marques, M.P.M.; Geraldes, C.F.G.C.; Pubanz, D.; Merbach, A.E. Conformational and coordination equilibria on dota complexes of lanthanide metal ions in aqueous solution studied by 1h nmr spectroscopy. Inorg. Chem. 1997, 36, 2059–2068. [Google Scholar] [CrossRef]

| τH2O (ms) 1 | τD2O (ms) 1 | q2 | r1 (mM−1·s−1) 3 | |
|---|---|---|---|---|
| LnL1 | 0.422 (4) | 1.54 (1) | 1.6 (3) | 8.26 |
| LnL2 | 0.506 (3) | 1.431 (9) | 1.0 (3) | 4.02 |
| LnL3 | 0.638 (6) | 1.72 (1) | −0.4 (4) 4 | 3.27 |
| LnL4 | 0.925 (8) | 1.64 (1) | 0.1 (3) | 3.65 |
| GdL1 | GdL2 | GdL3 | GdL4 | |
|---|---|---|---|---|
| q | 2 | 1 | 0 | 0 |
| qSS | 0 | 0 | 1 | 1 |
| kex298 (106 s−1) | 11 a | 111 b | - | - |
| ΔH≠ (kJ·mol−1) | 33.6 a | 21.0 b | - | - |
| kex298 SS (106 s−1) | - | - | 2000 c | 2000 c |
| ΔH≠ SS (kJ·mol−1) | - | - | 30 c | 30 c |
| ER (kJ·mol−1) | 15 (3) | 68 (7) | 12 (6) | 15 (4) |
| τR298 (ps) | 95 (5) | 55 (5) | 160(10) | 260 (20) |
| rGdH (Å) | 3.1 | 3.1 | 3.8 d | 3.8 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laine, S.; Morfin, J.-F.; Galibert, M.; Aucagne, V.; Bonnet, C.S.; Tóth, É. Lanthanide DO3A-Complexes Bearing Peptide Substrates: The Effect of Peptidic Side Chains on Metal Coordination and Relaxivity. Molecules 2021, 26, 2176. https://doi.org/10.3390/molecules26082176
Laine S, Morfin J-F, Galibert M, Aucagne V, Bonnet CS, Tóth É. Lanthanide DO3A-Complexes Bearing Peptide Substrates: The Effect of Peptidic Side Chains on Metal Coordination and Relaxivity. Molecules. 2021; 26(8):2176. https://doi.org/10.3390/molecules26082176
Chicago/Turabian StyleLaine, Sophie, Jean-François Morfin, Mathieu Galibert, Vincent Aucagne, Célia S. Bonnet, and Éva Tóth. 2021. "Lanthanide DO3A-Complexes Bearing Peptide Substrates: The Effect of Peptidic Side Chains on Metal Coordination and Relaxivity" Molecules 26, no. 8: 2176. https://doi.org/10.3390/molecules26082176
APA StyleLaine, S., Morfin, J.-F., Galibert, M., Aucagne, V., Bonnet, C. S., & Tóth, É. (2021). Lanthanide DO3A-Complexes Bearing Peptide Substrates: The Effect of Peptidic Side Chains on Metal Coordination and Relaxivity. Molecules, 26(8), 2176. https://doi.org/10.3390/molecules26082176







