Protective Effects of Cinnamaldehyde on the Inflammatory Response, Oxidative Stress, and Apoptosis in Liver of Salmonella typhimurium-Challenged Mice
Abstract
1. Introduction
2. Results
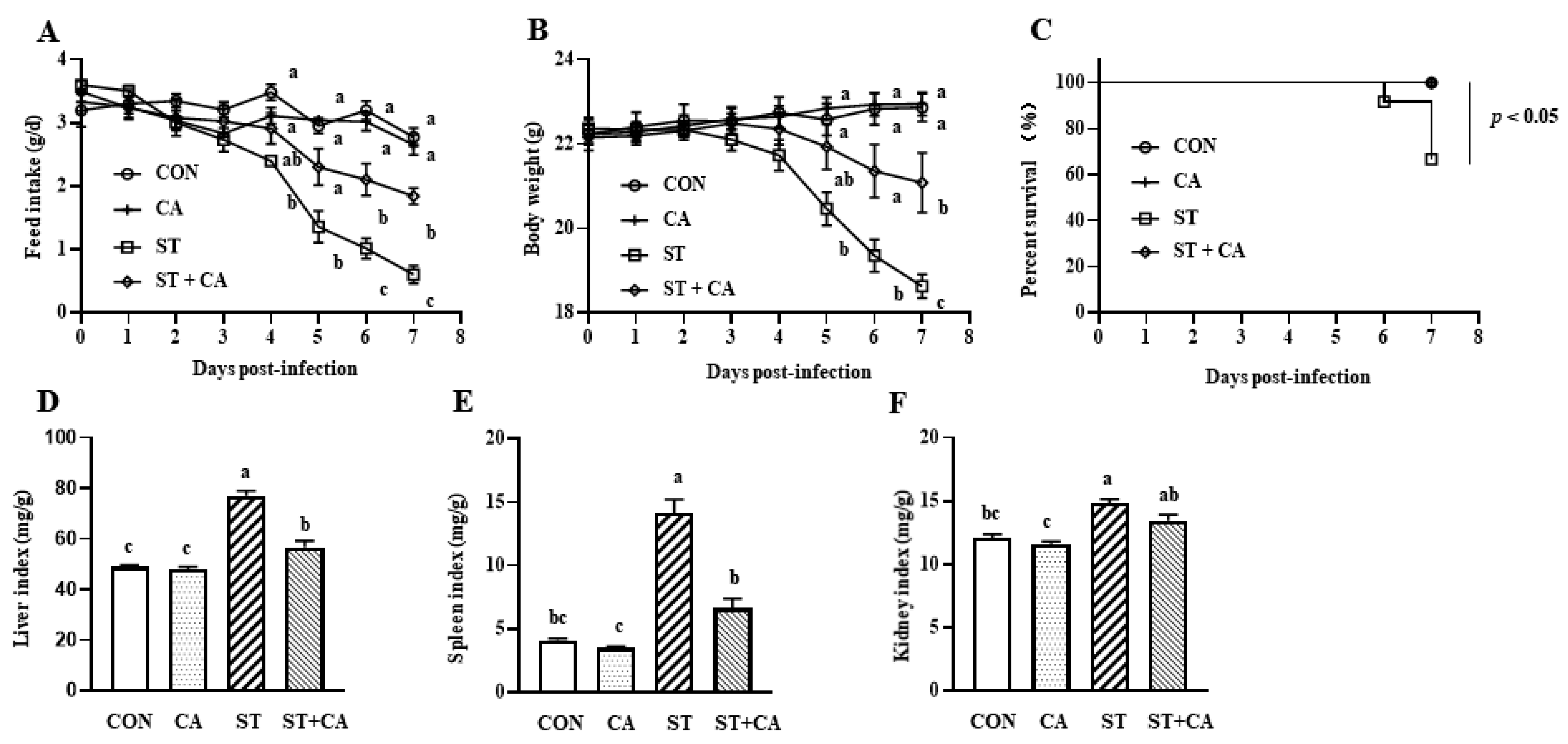
2.1. The Protective Effect of Cinnamaldehyde on the Feed Intake, Body Weight Change, and Organ Index of Salmonella typhimurium-Infected Mice
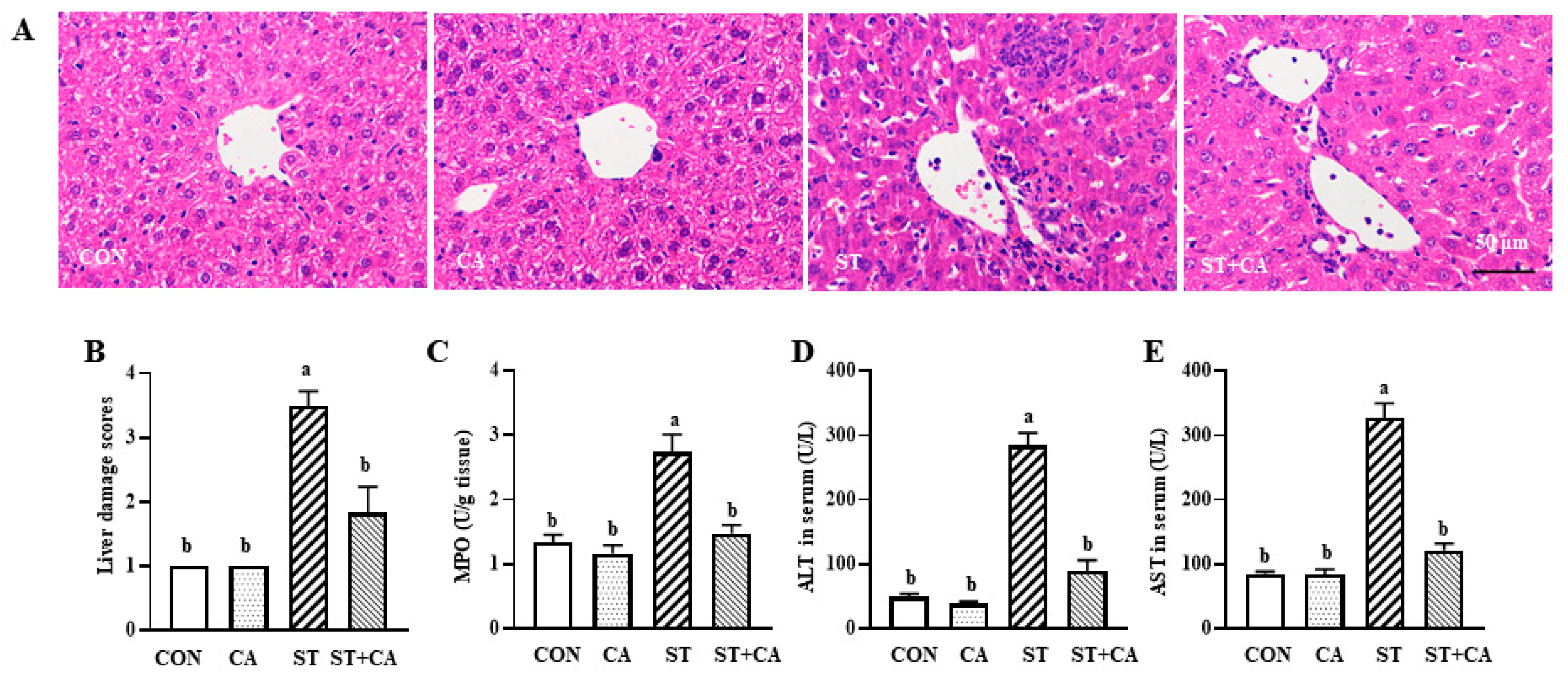
2.2. Cinnamaldehyde Alleviated the Salmonella typhimurium-Induced Liver Damage
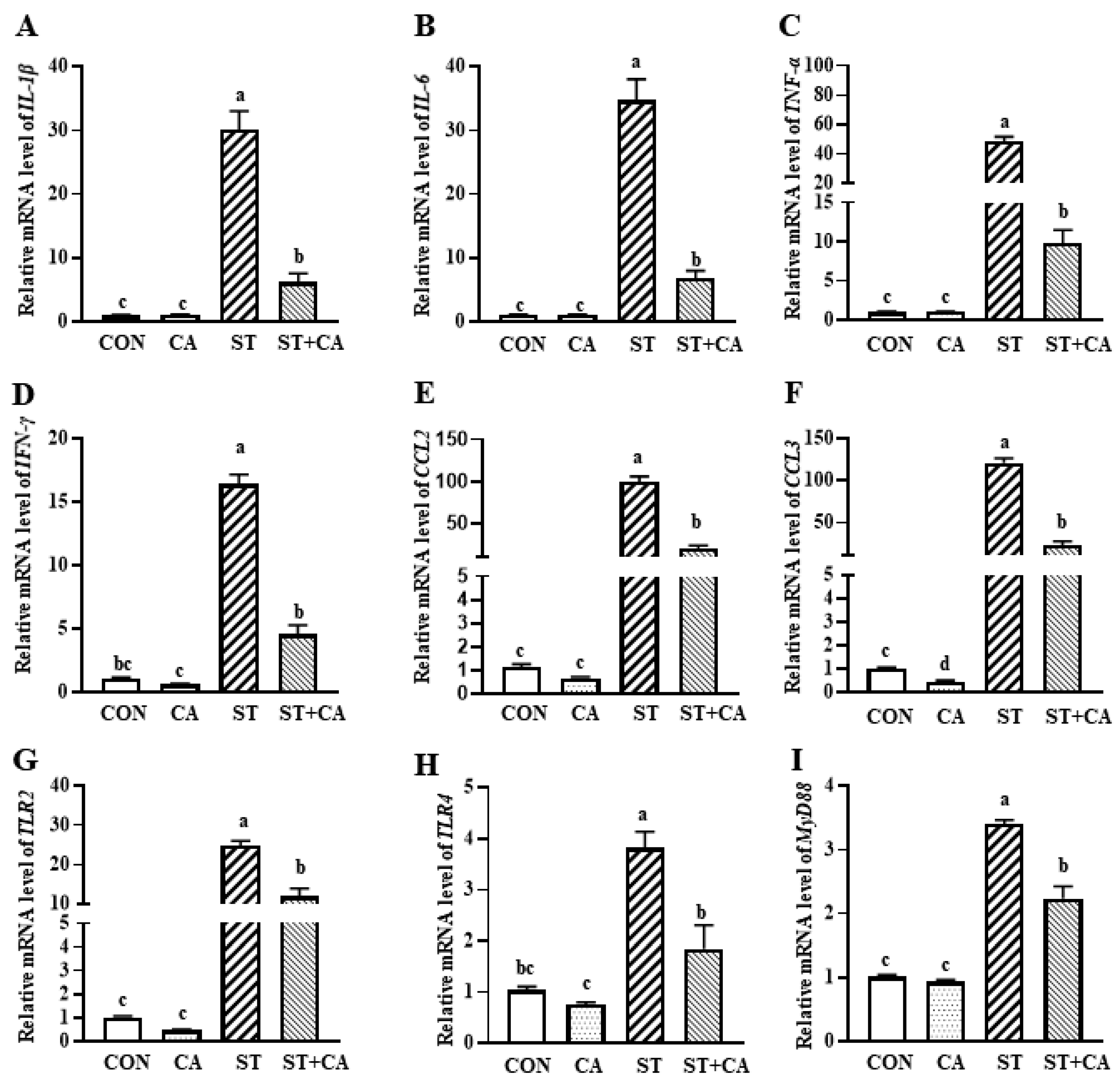
2.3. The Effect of Cinnamaldehyde and Salmonella typhimurium on the Expression of Inflammation-Related Genes in the Liver
2.4. Cinnamaldehyde Attenuated Salmonella typhimurium-Induced Hepatic Apoptosis in Mice
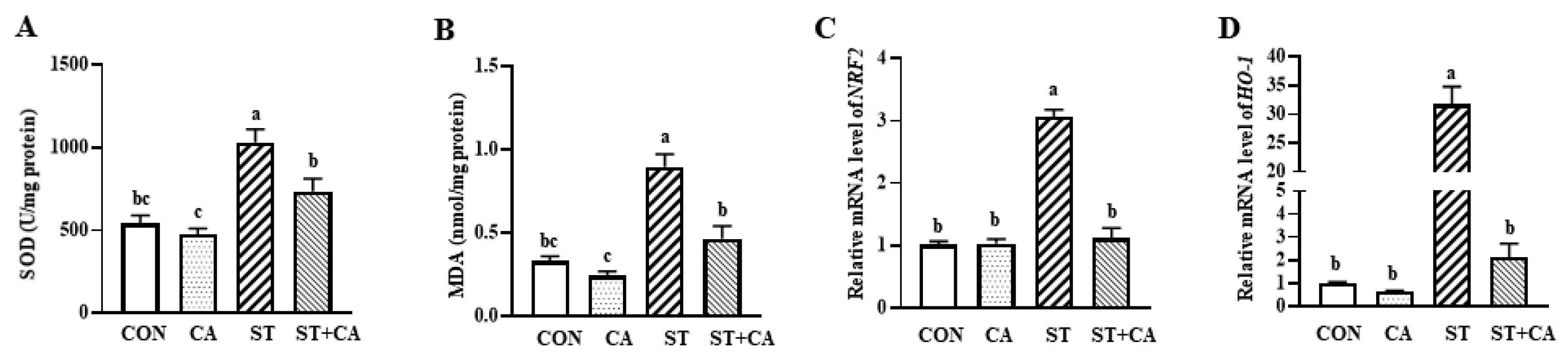
2.5. Cinnamaldehyde Attenuated Hepatic Oxidative Stress in Salmonella typhimurium-Infected Mice
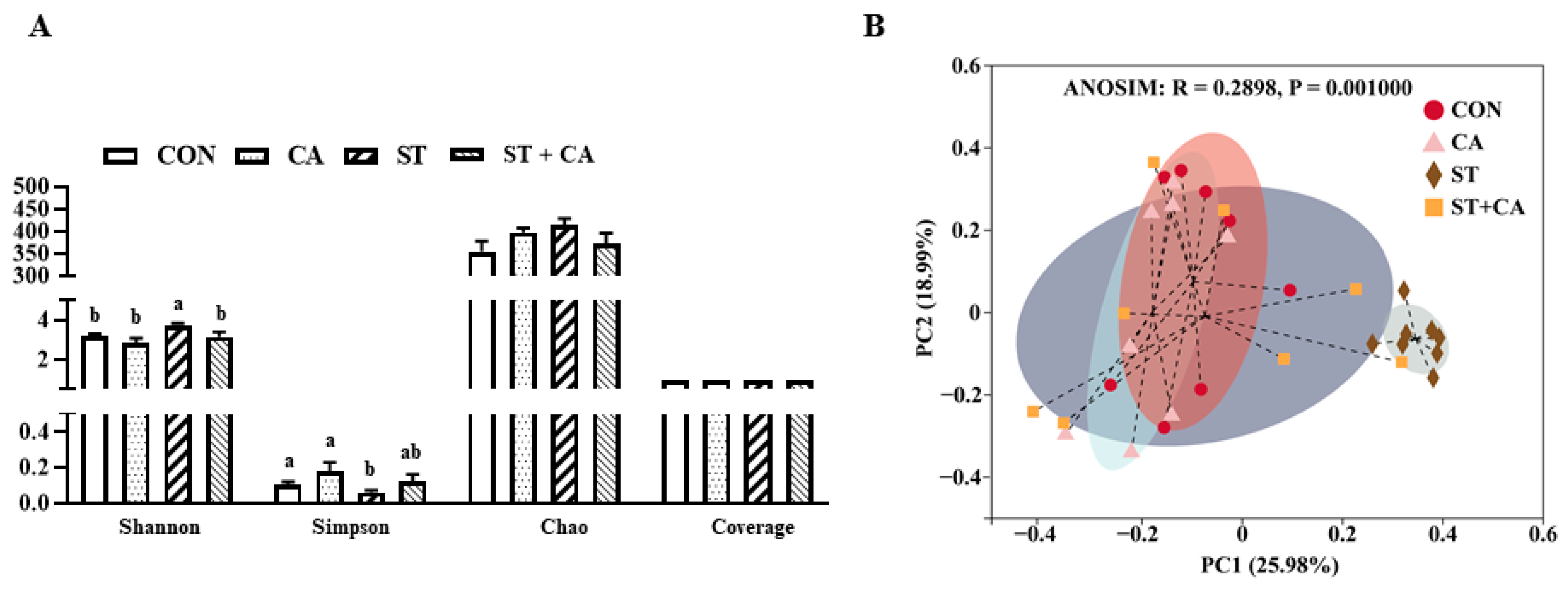
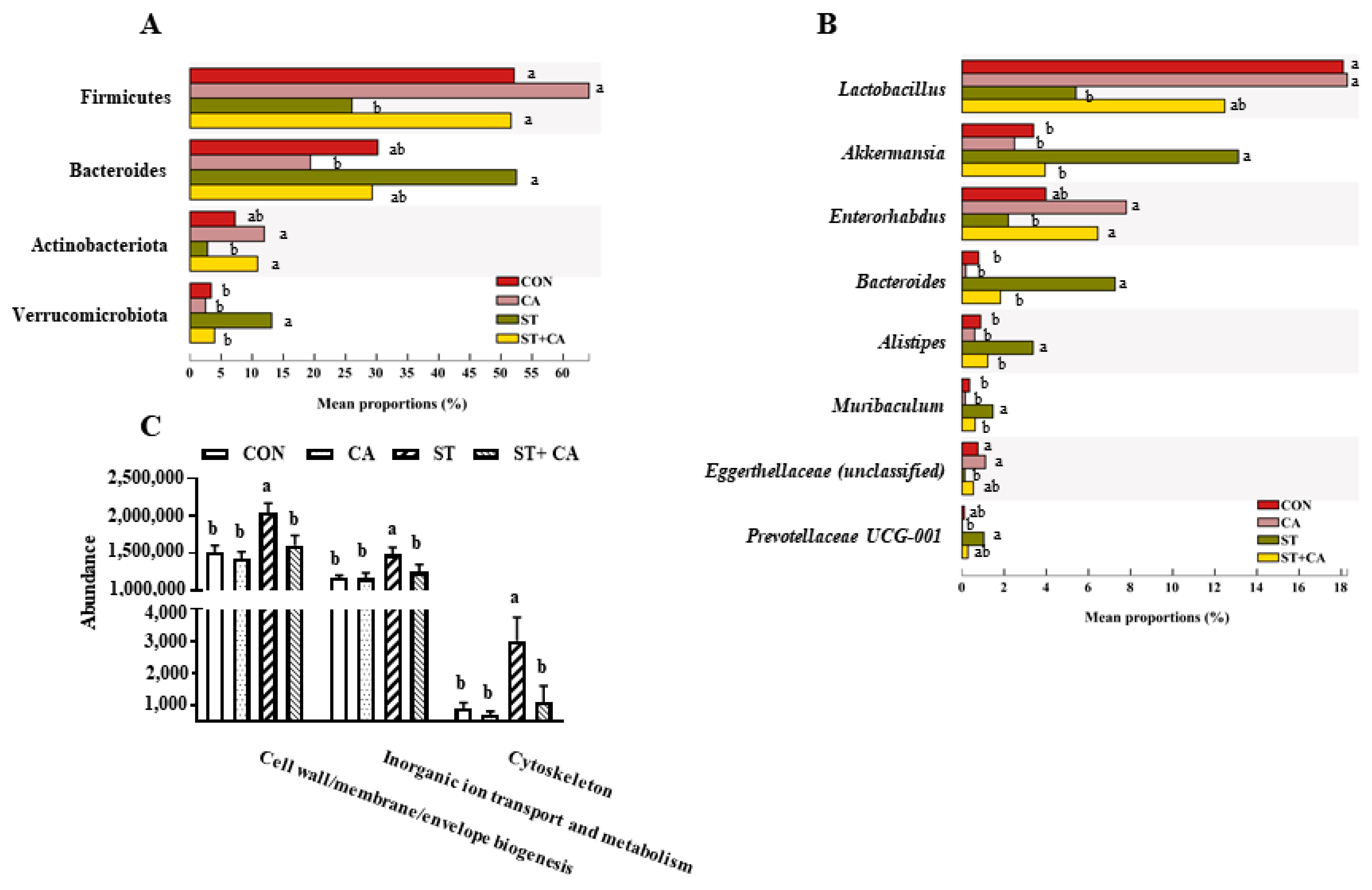
2.6. Effect of Cinnamaldehyde and Salmonella typhimurium on the Microbiota Community in Colonic Content
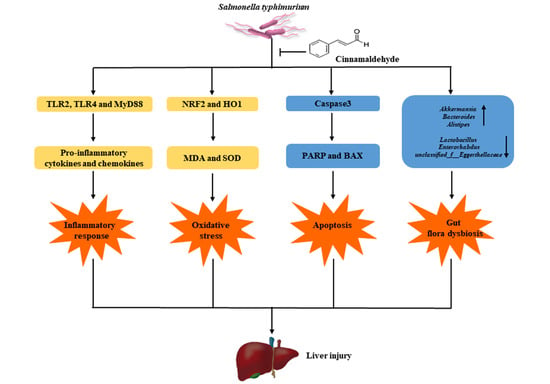
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Culture of Bacteria
4.3. Animal Experiments
4.4. Biochemical Index Estimation
4.5. Histological Analysis
4.6. TUNEL Assay
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. Western Blot Analysis
4.9. 16S rDNA Sequencing
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Nguyen, T.T.; Almon, R.R.; Dubois, D.C.; Sukumaran, S.; Jusko, W.J.; Androulakis, I.P. Tissue-specific gene expression and regulation in liver and muscle following chronic corticosteroid administration. Gene Regul. Syst. Biol. 2014, 8, 75–87. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, H.; Jin, Y.; Liu, N.; Chen, J.; Yang, Y.; Dai, Z.; Wang, C.; Wu, G.; Wu, Z. Glycine Attenuates LPS-Induced Apoptosis and inflammatory cell infiltration in mouse liver. J. Nutr. 2020, 150, 1116–1125. [Google Scholar] [CrossRef]
- Herrero-Fresno, A.; Olsen, J.E. Salmonella Typhimurium metabolism affects virulence in the host—A mini-review. Food Microbiol. 2018, 71, 98–110. [Google Scholar] [CrossRef]
- Dong, N.; Li, X.; Xue, C.; Wang, C.; Xu, X.; Bi, C.; Shan, A.; Li, D. Astragalus polysaccharides attenuated inflammation and balanced the gut microflora in mice challenged with Salmonella typhimurium. Int. Immunopharmacol. 2019, 74, 105681. [Google Scholar] [CrossRef]
- Pradhan, B.; Guha, D.; Naik, A.K.; Banerjee, A.; Tambat, S.; Chawla, S.; Senapati, S.; Aich, P. Probiotics L. acidophilus and B. clausii modulate gut microbiota in Th1- and Th2-biased mice to ameliorate salmonella typhimurium-induced diarrhea. Probiotics Antimicrob. Proteins 2019, 11, 887–904. [Google Scholar] [CrossRef]
- Song, F.; Liu, J.; Zhao, W.; Huang, H.; Hu, D.; Chen, H.; Zhang, H.; Chen, W.; Gu, Z. Synergistic effect of eugenol and probiotic lactobacillus plantarum Zs2058 against salmonella infection in C57bl/6 mice. Nutrients 2020, 12, 1611. [Google Scholar] [CrossRef]
- Fàbrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Abatemarco Júnior, M.; Sandes, S.H.C.; Ricci, M.F.; Arantes, R.M.E.; Nunes, Á.C.; Nicoli, J.R.; Neumann, E. Protective effect of lactobacillus diolivorans 1Z, isolated from brazilian kefir, against salmonella enterica serovar typhimurium in experimental murine models. Front. Microbiol. 2018, 9, 2856. [Google Scholar] [CrossRef]
- Wei, S.; Huang, J.; Liu, Z.; Wang, M.; Zhang, B.; Lian, Z.; Guo, Y.; Han, H. Differential immune responses of C57BL/6 mice to infection by salmonella enterica serovar typhimurium strain SL1344, CVCC541 and CMCC50115. Virulence 2019, 10, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Rishi, P.; Kaur, P.; Virdi, J.S.; Shukla, G.; Koul, A. Amelioratory effects of zinc supplementation on Salmonella-induced hepatic damage in the murine model. Dig. Dis. Sci. 2008, 53, 1063–1070. [Google Scholar] [CrossRef]
- Borish, L.C.; Steinke, J.W. 2. Cytokines and chemokines. J. Allergy Clin. Immunol. 2003, 111 (Suppl. 2), S460–S475. [Google Scholar] [CrossRef]
- Lalmanach, A.C.; Lantier, F. Host cytokine response and resistance to Salmonella infection. Microb. Infect. 1999, 1, 719–726. [Google Scholar] [CrossRef]
- Eckmann, L.; Kagnoff, M.F. Cytokines in host defense against Salmonella. Microb. Infect. 2001, 3, 1191–1200. [Google Scholar] [CrossRef]
- Stoycheva, M.V.; Murdjeva, M.A. Cytokines in salmonella infections. Folia Med. 2004, 46, 5–10. [Google Scholar]
- Liu, Y.; Zhang, Y.; Zhou, Y.; Wang, T.; Deng, X.; Chu, X.; Zhou, T. Cinnamaldehyde inhibits type three secretion system in Salmonella enterica serovar Typhimurium by affecting the expression of key effector proteins. Vet. Microbiol. 2019, 239, 108463. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.P.; Lee, S.J.; Nam, S.H.; Friedman, M. The composition of a bioprocessed shiitake (Lentinus edodes) mushroom mycelia and rice bran formulation and its antimicrobial effects against Salmonella enterica subsp. enterica serovar Typhimurium strain SL1344 in macrophage cells and in mice. BMC Complement Altern. Med. 2018, 18, 322. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Liu, H.; Liu, C.; Wang, L.; Ma, R.; Chen, B.; Li, L.; Niu, J.; Fu, M.; Zhang, D.; et al. Cinnamaldehyde in diabetes: A review of pharmacology, pharmacokinetics and safety. Pharmacol. Res. 2017, 122, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.E.; Na, J.Y.; Lee, J.S. Anti-inflammatory effects of trans-cinnamaldehyde on lipopolysaccharide-stimulated macrophage activation via MAPKs pathway regulation. Immunopharmacol. Immunotoxicol. 2018, 40, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-Y.; Ahn, S.-G.; Kim, S.-A. Cinnamaldehyde protects human dental pulp cells against oxidative stress through the Nrf2/HO-1-dependent antioxidant response. Eur. J. Pharmacol. 2017, 815, 73–79. [Google Scholar] [CrossRef]
- Yang, G.; Jin, T.; Yin, S.; Guo, D.; Zhang, C.; Xia, X.; Shi, C. trans-Cinnamaldehyde mitigated intestinal inflammation induced by Cronobacter sakazakii in newborn mice. Food Funct. 2019, 10, 2986–2996. [Google Scholar] [CrossRef]
- McGill, M.R. The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI J. 2016, 15, 817–828. [Google Scholar]
- Kengni, F.; Fodouop, S.P.C.; Tala, D.S.; Djimeli, M.N.; Fokunang, C.; Gatsing, D. Antityphoid properties and toxicity evaluation of Harungana madagascariensis Lam (Hypericaceae) aqueous leaf extract. J. Ethnopharmacol. 2016, 179, 137–145. [Google Scholar] [CrossRef]
- Wang, L.; Li, L.; Lv, Y.; Chen, Q.; Feng, J.; Zhao, X. Lactobacillus plantarum restores intestinal permeability disrupted by salmonella infection in newly-hatched chicks. Sci. Rep. 2018, 8, 2229. [Google Scholar] [CrossRef] [PubMed]
- Czaja, A.J. Review article: Chemokines as orchestrators of autoimmune hepatitis and potential therapeutic targets. Aliment. Pharmacol. Ther. 2014, 40, 261–279. [Google Scholar] [CrossRef]
- Dai, C.; Xiao, X.; Li, D.; Tun, S.; Wang, Y.; Velkov, T.; Tang, S. Chloroquine ameliorates carbon tetrachloride-induced acute liver injury in mice via the concomitant inhibition of inflammation and induction of apoptosis. Cell Death Dis. 2018, 9, 1164. [Google Scholar] [CrossRef] [PubMed]
- Marra, F.; Tacke, F. Roles for chemokines in liver disease. Gastroenterology 2014, 147, 577–594.e1. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K.; Kaisho, T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2001, 2, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Godec, J.; Lau, L.; Sivick, K.E.; McLaughlin, L.M.; Jones, M.B.; Dracheva, T.; Peterson, S.N.; Monack, D.M.; Barton, G.M. TLR signaling is required for salmonella typhimurium virulence. Cell 2011, 144, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Silk, E.; Zhao, H.; Weng, H.; Ma, D. The role of extracellular histone in organ injury. Cell Death Dis. 2017, 8, e2812. [Google Scholar] [CrossRef]
- Seibert, S.A.; Mex, P.; Köhler, A.; Kaufmann, S.H.; Mittrücker, H.W. TLR2-, TLR4- and Myd88-independent acquired humoral and cellular immunity against Salmonella enterica serovar Typhimurium. Immunol. Lett. 2010, 127, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.K.; Mittal, S.; Tiwari, N.; Maurya, A.K.; Singh, D.; Pandey, A.K.; Pal, A. Plasmodium-salmonella coinfection induces intense inflammatory response, oxidative stress, and liver damage: A mice model study for therapeutic strategy. Shock 2018, 50, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Roche, L.; Mesta, F. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) Infection. Arch. Med. Res. 2020, 51, 384–387. [Google Scholar] [CrossRef]
- Wu, C.-T.; Deng, J.-S.; Huang, W.-C.; Shieh, P.-C.; Chung, M.-I.; Huang, G.-J. Salvianolic acid C against acetaminophen-induced acute liver injury by attenuating inflammation, oxidative stress, and apoptosis through inhibition of the Keap1/Nrf2/HO-1 signaling. Oxid. Med. Cell. Longev. 2019, 2019, 9056845. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of oxidative stress in inflammatory bowel disease and potential antioxidant therapies. Oxid. Med. Cell. Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef]
- Labat-Moleur, F.; Guillermet, C.; Lorimier, P.; Robert, C.; Lantuejoul, S.; Brambilla, E.; Negoescu, A. TUNEL apoptotic cell detection in tissue sections: Critical evaluation and improvement. J. Histochem. Cytochem. 1998, 46, 327–334. [Google Scholar] [CrossRef]
- Sairanen, T.; Szepesi, R.; Karjalainen-Lindsberg, M.L.; Saksi, J.; Paetau, A.; Lindsberg, P.J. Neuronal caspase-3 and PARP-1 correlate differentially with apoptosis and necrosis in ischemic human stroke. Acta Neuropathol. 2009, 118, 541–552. [Google Scholar] [CrossRef]
- Omonijo, F.A.; Ni, L.; Gong, J.; Wang, Q.; Lahaye, L.; Yang, C. Essential oils as alternatives to antibiotics in swine production. Anim. Nutr. 2018, 4, 126–136. [Google Scholar] [CrossRef]
- Zhang, L.; Gui, S.; Liang, Z.; Liu, A.; Chen, Z.; Tang, Y.; Xiao, M.; Chu, F.; Liu, W.; Jin, X.; et al. Musca domestica cecropin (Mdc) alleviates salmonella typhimurium-induced colonic mucosal barrier impairment: Associating with inflammatory and oxidative stress response, tight junction as well as intestinal flora. Front. Microbiol. 2019, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.A.; Wellington, M.O.; González-Vega, J.C.; Htoo, J.K.; Van Kessel, A.G.; Columbus, D.A. Functional amino acid supplementation, regardless of dietary protein content, improves growth performance and immune status of weaned pigs challenged with Salmonella Typhimurium. J. Anim. Sci. 2021, 99, skaa365. [Google Scholar] [CrossRef]
- El Kamouni, S.; El Kebbaj, R.; Andreoletti, P.; El Ktaibi, A.; Rharrassi, I.; Essamadi, A.; El Kebbaj, M.S.; Mandard, S.; Latruffe, N.; Vamecq, J.; et al. Protective effect of argan and olive oils against LPS-induced oxidative stress and inflammation in mice livers. Int. J. Mol. Sci. 2017, 18, 2181. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ren, Y.; Lu, J.; Bartlett, M.; Chen, L.; Zhang, Y.; Guo, X.; Liu, C. A novel prebiotic blend product prevents irritable bowel syndrome in mice by improving gut microbiota and modulating immune response. Nutrients 2017, 9, 1341. [Google Scholar] [CrossRef]
- Xu, X.; Gong, L.; Wang, B.; Wu, Y.; Wang, Y.; Mei, X.; Xu, H.; Tang, L.; Liu, R.; Zeng, Z.; et al. Glycyrrhizin attenuates salmonella enterica serovar typhimurium infection: New insights into its protective mechanism. Front. Immunol. 2018, 9, 2321. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, F.; Xu, Y.; Zhang, J.; Qi, J.; Liu, X.; Wang, R. Thymol alleviates lipopolysaccharide-stimulated inflammatory response via downregulation of RhoA-mediated NF-κB signalling pathway in human peritoneal mesothelial cells. Eur. J. Pharmacol. 2018, 833, 210–220. [Google Scholar] [CrossRef]
- Arguello, H.; Estelle, J.; Zaldivar-Lopez, S.; Jimenez-Marin, A.; Carvajal, A.; Lopez-Bascon, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early salmonella typhimurium infection in pigs disrupts microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018, 8, 7788. [Google Scholar] [CrossRef]
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007, 5, 2177–2189. [Google Scholar] [CrossRef]
- Bescucci, D.M.; Moote, P.E.; Ortega Polo, R.; Uwiera, R.R.E.; Inglis, G.D. Serovar typhimurium temporally modulates the enteric microbiota and host responses to overcome colonization resistance in swine. Appl. Environ. Microbiol. 2020, 86, e01569-20. [Google Scholar] [CrossRef]
- Saulnier, D.M.; Riehle, K.; Mistretta, T.A.; Diaz, M.A.; Mandal, D.; Raza, S.; Weidler, E.M.; Qin, X.; Coarfa, C.; Milosavljevic, A.; et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.J.; Liu, J.; Wang, W.X.; Wang, A.T.; Yang, X.Y.; Guan, H.S.; Wang, C.Y.; Yan, D. Berberine treatment-emergent mild diarrhea associated with gut microbiota dysbiosis. Biomed Pharm. 2019, 116, 109002. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Z.; Lu, W.; Hu, D.; Zhao, X.; Huang, H.; Zhang, H.; Zhao, J.; Chen, W. Multiple mechanisms applied by Lactobacillus pentosus AT6 to mute the lethal effects of Salmonella in a mouse model. Food Funct. 2018, 9, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Di Pasqua, R.; Betts, G.; Hoskins, N.; Edwards, M.; Ercolini, D.; Mauriello, G. Membrane toxicity of antimicrobial compounds from essential oils. J. Agric. Food Chem. 2007, 55, 4863–4870. [Google Scholar] [CrossRef] [PubMed]
- Keelara, S.; Thakur, S.; Patel, J. Biofilm formation by environmental isolates of salmonella and their sensitivity to natural antimicrobials. Foodborne Pathog. Dis. 2016, 13, 509–516. [Google Scholar] [CrossRef]
- Sun, K.; Lei, Y.; Wang, R.; Wu, Z.; Wu, G. Cinnamicaldehyde regulates the expression of tight junction proteins and amino acid transporters in intestinal porcine epithelial cells. J. Anim. Sci. Biotechnol. 2017, 8, 66. [Google Scholar] [CrossRef]
- Baranova, I.N.; Souza, A.C.P.; Bocharov, A.V.; Vishnyakova, T.G.; Hu, X.; Vaisman, B.L.; Amar, M.J.; Chen, Z.; Kost, Y.; Remaley, A.T.; et al. Human SR-BI and SR-BII potentiate lipopolysaccharide-induced inflammation and acute liver and kidney injury in mice. J. Immunol. 2016, 196, 3135–3147. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Dai, Z.; Sun, S.; Ma, X.; Yang, Y.; Tso, P.; Wu, G.; Wu, Z. Hydroxyproline attenuates dextran sulfate sodium-induced colitis in mice: Involvment of the NF-κB signaling and oxidative stress. Mol. Nutr. Food Res. 2018, 62, 1800494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward Primer (5′to 3′) | Reverse Primer (5′to 3′) |
|---|---|---|
| IL-1β | TGCACTACAGGCTCCG | TGCCGTCTTTCATTACAC |
| IL-6 | TTGTGCAATGGCAATTCTG | CGGACTCTGGCTTTGTCTTTC |
| TNF-α | TTCTCATTCCTGCTTGTGG | CACTTGGTGGTTTGCTACG |
| IFN-γ | TCAAGTGCGATAGATGTGGAAGAA | TGGCTCTGCAGGATTTTCATG |
| CCL-2 | CAGCTCTCTCTTCCTCCACC | TGGGATCATCTTGCTGGTGA |
| CCL-3 | CCAGCCAGGTGTCATTTTCC | AGGCATTCAGTTCCAGGTCA |
| TLR2 | CTGAGAATGATGTGGGCGTG | TTAAAGGGCGGGTCAGAGTT |
| TLR4 | TCTAACTTCCCTCCTGCGAC | ACGATCTGTAACTGGTGGCA |
| MyD88 | CCATTGCCAGCGAGCTAATT | TCTGTTGGACACCTGGAGAC |
| NRF2 | TCCATTTACGGAGACCCACC | GGCCGTTCTGTTTGACACTT |
| HO-1 | CAGGTGTCCAGAGAAGGCTT | GCTTGTTGCGCTCTATCTCC |
| GAPDH | AAGCCCATCACCATCTTCCA | CACCAGTAGACTCCACGACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Li, S.; Jia, H.; Si, X.; Lei, Y.; Lyu, J.; Dai, Z.; Wu, Z. Protective Effects of Cinnamaldehyde on the Inflammatory Response, Oxidative Stress, and Apoptosis in Liver of Salmonella typhimurium-Challenged Mice. Molecules 2021, 26, 2309. https://doi.org/10.3390/molecules26082309
Wang R, Li S, Jia H, Si X, Lei Y, Lyu J, Dai Z, Wu Z. Protective Effects of Cinnamaldehyde on the Inflammatory Response, Oxidative Stress, and Apoptosis in Liver of Salmonella typhimurium-Challenged Mice. Molecules. 2021; 26(8):2309. https://doi.org/10.3390/molecules26082309
Chicago/Turabian StyleWang, Renjie, Senlin Li, Hai Jia, Xuemeng Si, Yan Lei, Jirong Lyu, Zhaolai Dai, and Zhenlong Wu. 2021. "Protective Effects of Cinnamaldehyde on the Inflammatory Response, Oxidative Stress, and Apoptosis in Liver of Salmonella typhimurium-Challenged Mice" Molecules 26, no. 8: 2309. https://doi.org/10.3390/molecules26082309
APA StyleWang, R., Li, S., Jia, H., Si, X., Lei, Y., Lyu, J., Dai, Z., & Wu, Z. (2021). Protective Effects of Cinnamaldehyde on the Inflammatory Response, Oxidative Stress, and Apoptosis in Liver of Salmonella typhimurium-Challenged Mice. Molecules, 26(8), 2309. https://doi.org/10.3390/molecules26082309