Synthesis, Quantification, and Characterization of Fatty Acid Amides from In Vitro and In Vivo Sources
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis of the Fatty Acid Amides
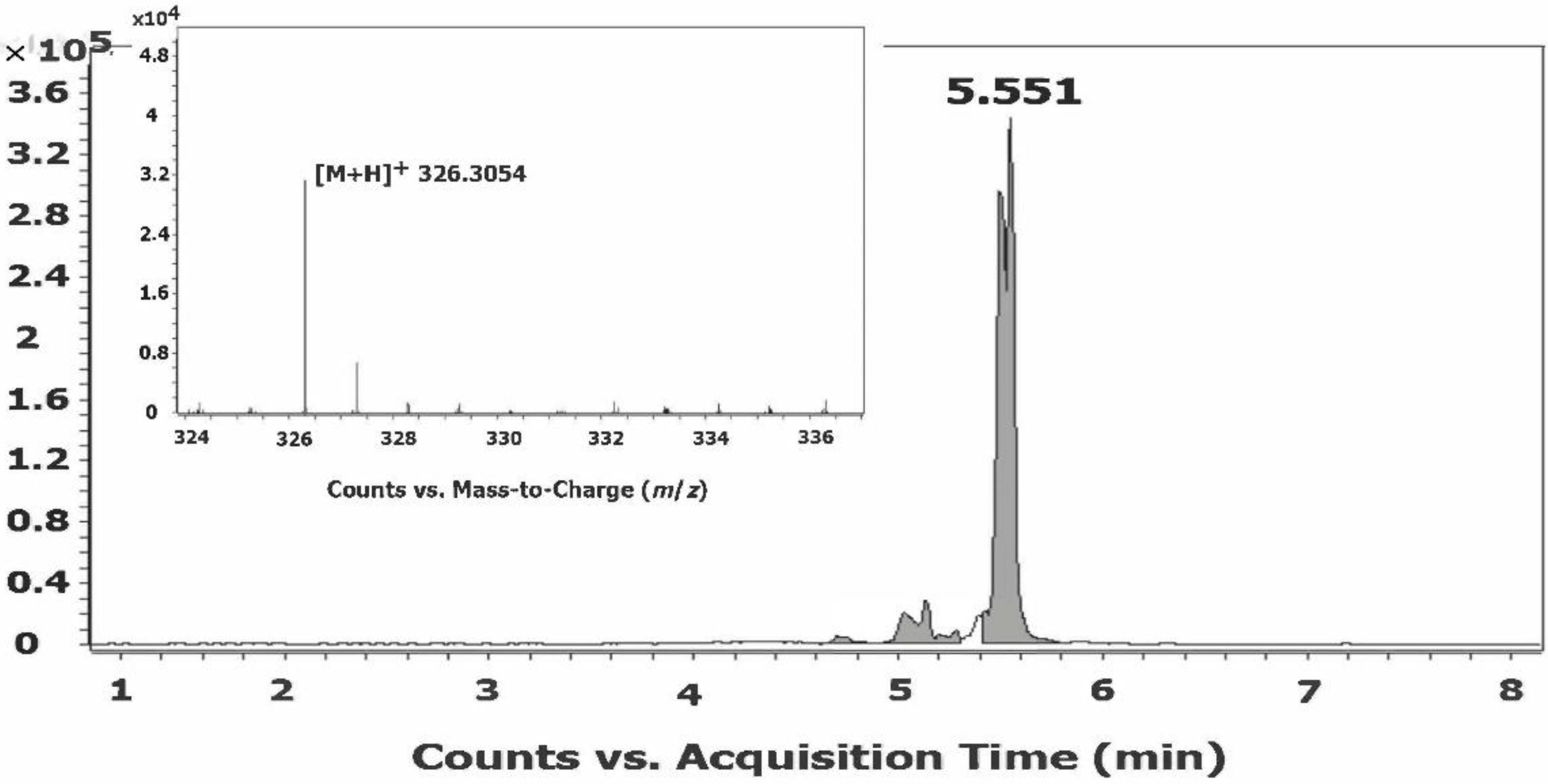
2.2. Structural Analysis of the Synthetically Prepared Fatty Acid Amides
2.3. The Fatty Acid Amidome from Mouse N18TG2 Cells and Sheep Choroid Plexus (SCP) Cells
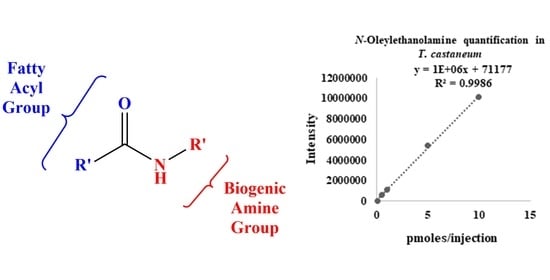
2.4. The Fatty Acid Amidome from Insects: Drosophila Melanogaster, Bombyx Mori, Apis Mellifera, and Tribolium Castaneum
2.5. Future Directions
3. Materials and Methods
3.1. General Information
3.2. N-(2-Hydroxyethyl)Oleamide (FA-1, N-Oleoylethanolamine)
3.3. N-Oleoylglycine (FA-2)
3.4. Palmitamide (FA-3)
3.5. N-Palmitoylglycine (FA-4)
3.6. Cells and Cell Culture
3.7. Insects
3.8. Characterization of the Fatty Acid Amidome
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bradshaw, H.B.; Leishman, E. Lipidomics: A Corrective Lens of Enzyme Mopia. Methods Enzymol. 2017, 593, 123–141. [Google Scholar]
- Bradshaw, H.B.; Walker, J.M. The Expanding Field of Cannabimimetic and Related Mediators. Br. J. Pharmacol. 2005, 144, 459–465. [Google Scholar] [CrossRef]
- Waluk, D.P.; Battistini, M.R.; Dempsey, D.R.; Farrell, E.K.; Jeffries, K.A.; Mitchell, P.; Hernandez, L.W.; McBride, J.C.; Merkler, D.J.; Hunt, M.C. Mammalian Fatty Acid Amides of the Brain and CNS. In Omega-3 Fatty Acids in Brain and Neurological Health; Watson, R.R., DeMeester, F., Eds.; Academic Press: London, UK, 2014; pp. 87–107. [Google Scholar]
- Maccarrone, M.; Bab, I.; Bíro, T.; Cabral, G.A.; Dey, S.K.; Di Marzo, V.; Konje, J.C.; Kunos, G.; Mechoulam, R.; Pacher, P.; et al. Endocannabinoid Signaling at the Periphery: 50 Years after THC. Trends Pharmacol. Sci. 2015, 36, 277–296. [Google Scholar] [CrossRef]
- Iannotti, F.A.; Di Marzo, V.; Petrosino, S. Endocannabinoids and Endocannabinoid-Related Mediators: Targets, Metabolism and Role in Neurological Disorders. Prog. Lipid Res. 2016, 62, 107–128. [Google Scholar] [CrossRef]
- Battista, N.; Bari, M.; Bisogno, T. N-Acyl Amino Acids: Metabolism, Molecular Targets, and Role in Biological Processes. Biomolecules 2019, 9, 822. [Google Scholar] [CrossRef]
- Llop, C.; Manrique, A.; Navarro, R.; Mijangos, C.; Reinecke, H. Control of the Migration Behavior of Slip Agents in Polyolefin-Based Films. Polym. Eng. Sci. 2011, 52, 1763–1769. [Google Scholar] [CrossRef]
- Hawthorne, J.N. A Note on the Life of J.L.W. Thudichum (1829–1901). Biochem. Soc. Trans. 1975, 3, 591. [Google Scholar] [CrossRef]
- Kuehl, F.A., Jr.; Jacob, T.A.; Ganley, O.H.; Ormond, R.E.; Meisinger, M.A.P. The Identification of N-(2-Hydroxyethyl)-Palmitamide as a Naturally Occurring Anti-Inflammatory Agent. J. Am. Chem. Soc. 1957, 79, 5577–5578. [Google Scholar] [CrossRef]
- Arafat, E.S.; Trimble, J.W.; Andersen, R.N.; Dass, C.; Desiderio, D.M. Identification of Fatty Acid Amides in Human Plasma. Life Sci. 1989, 45, 1679–1687. [Google Scholar] [CrossRef]
- Devane, W.A.; Hanuš, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and Structure of a Brain Constituent That Binds to the Cannabinoid Receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- Alborn, H.T.; Turlings, T.C.J.; Jones, T.H.; Stenhagen, J.H.; Loughrin, J.H.; Tumlinson, J.H. An Elicitor of Plant Volatiles from Beet Armyworm Oral Secretion. Science 1997, 276, 945–949. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Prospero-Garcia, O.; Siuzdak, G.; Gilula, N.B.; Henriksen, S.J.; Boger, D.L.; Lerner, R.A. Chemical Characterization of a Family of Brain Lipids that Induce Sleep. Science 1995, 268, 1506–1509. [Google Scholar] [CrossRef] [PubMed]
- Merkler, D.J.; Merkler, K.A.; Stern, W.; Fleming, F.F. Fatty Acid Amide Biosynthesis: A Possible New Role for Peptidylglycine α-Amidating Enzyme and Acyl-CoA:Glycine N-Acyltransferase. Arch. Biochem. Biophys. 1996, 330, 430–434. [Google Scholar] [CrossRef]
- Farrell, E.K.; Chen, Y.; Barazanji, M.; Jeffries, K.A.; Cameroamortegui, F.; Merkler, D.J. Primary Fatty Acid Amide Metabolism: Conversion of Fatty Acids and an Ethanolamine in N18TG2 and SCP Cells. J. Lipid Res. 2012, 53, 247–256. [Google Scholar] [CrossRef]
- Jeffries, K.A.; Dempsey, D.R.; Farrell, E.K.; Anderson, R.L.; Garbade, G.J.; Gurina, T.S.; Gruhonjic, I.; Gunderson, C.A.; Merkler, D.J. Glycine N-Acyltransferase-like 3 is Responsible for Long-chain N-Acylglycine Formation in N18TG2 Cells. J. Lipid Res. 2016, 57, 781–790. [Google Scholar] [CrossRef]
- Dempsey, D.R.; Jeffries, K.A.; Anderson, R.L.; Carpenter, A.-M.; Rodriquez Ospina, S.; Merkler, D.J. Identification of an Arylalkylamine N-Acyltransferase from Drosophila melanogaster that Catalyzes the Formation of Long-chain N-Acylserotonins. FEBS Lett. 2014, 588, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, K.A.; Dempsey, D.R.; Behari, A.L.; Anderson, R.L.; Merkler, D.J. Drosophila melanogaster as a Model System to Study Long-chain Fatty Acid Amide Metabolism. FEBS Lett. 2014, 588, 1596–1602. [Google Scholar] [CrossRef]
- Anderson, R.L.; Battistini, M.R.; Wallis, D.J.; Shoji, C.; O’Flynn, B.G.; Dillashaw, J.E.; Merkler, D.J. Bm-iAANAT and Its Potential Role in Fatty Acid Amide Biosynthesis in Bombyx mori. Prostaglandins Leukot. Essent. Fatty Acids 2018, 135, 10–17. [Google Scholar] [CrossRef]
- Jeffries, K.A.; Farrell, E.K.; Anderson, R.L.; Suarez, G.; Osborne, A.J.G.; Heide, M.K.; Merkler, D.J. Characterization and Quantification of the Fatty Acid Amidome. In Metabolomics; Wood, P.L., Ed.; Humana Press: New York, NY, USA, 2021; pp. 143–153. [Google Scholar]
- Mitchell, P.R., Jr. The Detection and Quantitative Analysis of Endocannabinoids and Endogenous Fatty Acid Amides in Apis mellifera and Tribolium castneum. Master’s Thesis, University of South Florida, Tampa, FL, USA, 2015. [Google Scholar]
- Morales-Sanfrutos, J.; Megia-Fernandez, A.; Hernandez-Mateo, F.; Giron-Gonzalez, D.; Salto-Gonzalez, R.; Santoyo-Gonzalez, F. Alkyl Sulfonyl Derivatized PAMAM-G2 Dendrimers as Nonviral Gene Delivery Vectors with Improved Transfection Efficiencies. Org. Biomol. Chem. 2011, 9, 851–864. [Google Scholar] [CrossRef]
- Ohsedo, Y.; Oono, M.; Saruhashi, K.; Watanabe, H.; Miyamoto, N. Thixotropic Stiff Hydrogels from a New Class of Oleoyl-D-Glutamine-Based Low-Molecular-Weight Gelators. RSC Adv. 2017, 7, 41686. [Google Scholar] [CrossRef]
- Ji, Y.-F.; Yan, H.; Jiang, Q.-B. Effective Nitration of Anilides and Acrylamides by tert-Butyl Nitrite. Eur. J. Org. Chem. 2015, 2051–2060. [Google Scholar] [CrossRef]
- Bisogno, T.; Sepe, N.; De Petrocellis, L.; Mechoulam, R.; Di Marzo, V. The Sleep Inducing Factor Oleamide is Produced by Mouse Neuroblastoma Cells. Biochem. Biophys. Res. Commun. 1997, 239, 473–479. [Google Scholar] [CrossRef]
- Di Marzo, V.; De Petrocellis, L.; Sepe, N.; Buono, A. Biosynthesis of Anandamide and Related Acylethanolamides in Mouse J774 Macrophages and N18 Neuroblastoma Cells. Biochem. J. 1996, 316, 977–984. [Google Scholar] [CrossRef]
- Ritenour-Rodgers, K.J.; Driscoll, W.J.; Merkler, K.A.; Merkler, D.J.; Mueller, G.P. Induction of Peptidylglycine α-Amidating Monooxygenase in N18TG2 Cells: A Model for Studying Oleamide Biosynthesis. Biochem. Biophys. Res. Commun. 2000, 267, 521–526. [Google Scholar] [CrossRef]
- Merkler, D.J.; Chew, G.H.; Gee, A.J.; Merkler, K.A.; Sorondo, J.-P.O.; Johnson, M.E. Oleic Acid Derived Metabolites on Mouse Neuroblastoma N18TG2 Cells. Biochemistry 2004, 43, 12667–12674. [Google Scholar] [CrossRef]
- Lun, M.P.; Monuki, E.S.; Lehtinen, M.K. Development and Functions of the Choroid Plexus-Cerebrospinal Fluid System. Nat. Rev. Neurosci. 2015, 16, 445–457. [Google Scholar] [CrossRef]
- Gee, P.; Rhodes, C.H.; Fricker, L.D.; Angeletti, R.H. Expression of Neuropeptide Processing Enzymes and Neurosecretory Proteins in Ependyma and Choroid Plexus Epithelium. Brain Res. 1993, 617, 238–248. [Google Scholar] [CrossRef]
- Tsukamoto, T.; Noguchi, M.; Kayama, H.; Watanabe, T.; Asohi, T.; Yamamoto, T. Increased Peptidylglycine α-Amidating Monooxygease Activity in Cerebrospinal Fluid of Patients with Multiple Schlerosis. Intern. Med. 1995, 34, 229–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gulaya, N.M.; Volkov, G.L.; Klimashevsky, V.M.; Glovseeva, N.N.; Melnik, A.A. Changes in Lipid Composition of Neuroblastoma C1300 N18 Cell During Differentiation. Neuroscience 1989, 30, 153–164. [Google Scholar] [CrossRef]
- Marchioni, C.; de Souza, I.D.; Junior, V.R.A.; de Souza Crippa, J.A.; Tumas, V.; Queiroz, M.E.C. Recent Advances in LC-MS/MS Methods to Determine Endocannabinoids in Biological Samples: Application in Neurodegenerative Diseases. Anal. Chim. Acta 2018, 1044, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, H.B.; Rimmerman, N.; Hu, S.S.-J.; Burstein, S.; Walker, J.M. Novel Endogenous N-Acyl Glycines: Identification and Characterization. Vitam. Horm. 2009, 81, 191–205. [Google Scholar]
- Bradshaw, H.B.; Lee, S.H.; McHugh, D. Orphan Endogenous Lipids and Orphan GPCRs: A Good Match. Prostaglandins Other Lipid Mediat. 2009, 89, 131–134. [Google Scholar] [CrossRef]
- O’Flynn, B.G.; Suarez, G.; Hawley, A.J.; Merkler, D.J. Insect Arylalkylamine N-Acyltransferases: Mechanism and Role in Fatty Acid Amide Biosynthesis. Front. Mol. Biosci. 2018, 5, 66. [Google Scholar] [CrossRef]
- McParland, J.; Di Marzo, V.; De Petrocellis, L.; Mercer, A.; Glass, M. Cannabinoid Receptors are Absent in Insects. J. Comp. Neurol. 2001, 436, 423–429. [Google Scholar] [CrossRef]
- Suarez, G.; Merkler, D.J. University of South Florida, Tampa, FL, USA. Unpublished work. 2021. [Google Scholar]
- Tortoriello, G.; Rhodes, B.P.; Takacs, S.M.; Stuart, J.M.; Basnet, A.; Raboune, S.; Widlanski, T.S.; Doherty, P.; Harkany, T.; Bradshaw, H.B. Target Lipidomic in Drosophila melanogaster Identified Novel 2-Monoacylglycerols and N-Acyl Amides. PLoS ONE 2013, 8, e67865. [Google Scholar] [CrossRef]
- Meng, X.; Zhu, F.; Chen, K. Silkworm: A Promising Model Organism in Life Science. J. Insect Sci. 2017, 17, 1–6. [Google Scholar]
- Dissanayaka, D.M.S.K.; Sammani, A.M.P.; Wijayaratne, L.K.W. Response of Different Population Sizes to Traps and Effect of Spinosad on the Trap Catch and Progeny Adult Emergence in Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). J. Stored Prod. Res. 2020, 86, 101576. [Google Scholar] [CrossRef]
- Lu, H.-C.; Mackie, K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatry 2016, 79, 516–525. [Google Scholar] [CrossRef]
- Prospéro-García, O.; Amancio-Belmont, O.; Meléndez, A.L.B.; Ruiz-Contreras, A.E.; Méndez-Díaz, M. Endocannabinoids and Sleep. Neurosci. Biobehav. Rev. 2016, 71, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Alhouayek, M.; Muccioli, G.G. Harnessing the Anti-Inflammatory Potential of Palmitoylethanolamide. Drug Discov. Today 2014, 19, 1632–1639. [Google Scholar] [CrossRef]
- Niphakis, M.J.; Lum, K.M.; Cognetta, A.B., III; Correia, B.E.; Ichu, T.-A.; Olucha, J.; Brown, S.J.; Kundu, S.; Piscitelli, F.; Rosen, H.; et al. A Global Map of Lipid-Binding Proteins ant Their Ligandability in Cells. Cell 2015, 161, 1668–1680. [Google Scholar] [CrossRef] [PubMed]
- Merkler, D.J.; Leahy, J.W. Binding-Based Proteomic Profiling and the Fatty Acid Amides. Trends Res. 2018, 1. [Google Scholar] [CrossRef]
- Sun, Y.X.; Tsuboi, K.; Okamoto, Y.; Tonai, T.; Murakami, M.; Kudo, I.; Ueda, N. Biosynthesis of Anandamide and N-Palmitoylethanolamine by Sequential Actions of Phospholipase A2 and Lysophospholipase D. Biochem. J. 2004, 380, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Ueda, N.; Tsuboi, K.; Uyama, T. Enzymological Studies on the Biosynthesis of N-Acylethanolamines. Biochim. Biophys. Acta 2010, 1801, 1274–1285. [Google Scholar] [CrossRef]
- Maccarrone, M. Metabolism of the Endocannabinoid Anandamide: Open Questions after 25 Years. Front. Mol. Neurosci. 2017, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Waluk, D.P.; Schultz, N.; Hunt, M.C. Identification of Glycine N-Acyltransferase-like 2 (GLYATL2) as a Transferase that Produces N-Acyl Glycines in Humans. FASEB J. 2010, 24, 2795–2803. [Google Scholar] [CrossRef]
- Aneetha, H.; O’Dell, D.K.; Tan, B.; Walker, J.M.; Hurley, T.D. Alcohol Dehydrogenase-Catalyzed In Vitro Oxidation of Anandamide to N-Arachidonoyl Glycine, a Lipid Mediator: Synthesis of N-Acyl Glycinals. Bioorg. Med. Chem. Lett. 2009, 19, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.R.; Bond, J.D.; Carpenter, A.-M.; Ospina, S.R.; Merkler, D.J. Expression, Purification, and Characterization of Mouse Glycine N-Acyltransferase in Esherichia coli. Protein Expr. Purif. 2014, 97, 23–28. [Google Scholar] [CrossRef]
- Rouzer, C.A.; Marnett, L.J. Endocannabinoid Oxygenation by Cyclooxygenases, Lipoxygenases, and Cytochromes P450: Cross-Talk between the Eicosanoid and Endocannabinoid Signaling Pathways. Chem. Rev. 2011, 111, 5899–5921. [Google Scholar] [CrossRef] [PubMed]
- Plastina, P.; Meijerink, J.; Vincken, J.-P.; Gruppen, H.; Witkamp, R.; Gabriele, B. Selective Synthesis of Unsaturated N-Acylethanolamines by Lipase-Catalyzed N-Acylation of Ethanolamine with Unsaturated Fatty Acids. Lett. Org. Chem. 2009, 6, 444. [Google Scholar] [CrossRef]
- Goujard, L.; Figueroa, M.C.; Villeneuve, P. Chemo-Enzymatic Synthesis of N-Arachidonoyl Glycine. Biotechnol. Lett. 2004, 26, 1211–1216. [Google Scholar] [CrossRef]
- Vandevoorde, S.; Jonsson, K.-O.; Fowler, C.J.; Lambert, D.M. Modifications of the Ethanolamine Head in N-Palmitoylethanolamine Synthesis and Evaluation of New Agents Interfering with the Metabolism of Anandamide. J. Med. Chem. 2003, 46, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.T.; Kang, G.J.; Yoo, E.S.; Hong, J.; Choi, J.S.; Kim, H.S.; Chung, H.Y.; Jung, J.H. Evaluation of Endogenous Fatty Acid Amides with Their Synthetic Analogues as Potential Anti-inflammatory Leads. Bioorg. Med. Chem. 2011, 19, 1520–1527. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; Johnson, M.E. Sample Preparation and Gas Chromatography of Primary Fatty Acid Amides. J. Chromatogr. A 2006, 1101, 278–285. [Google Scholar] [CrossRef] [PubMed]

| Fatty Acid Amide | N18TG2 Cells | SCP Cells 5 |
|---|---|---|
| ➤ N-Acylglycines | ||
| N-Palmitoylglycine 2 | + | - |
| N-Oleoylglycine 2 | ++ | - |
| ➤ N-Acylethanolamines | ||
| N-Palmitoylethanolamine 3 | + | - |
| N-Oleoylethanolamine 2,3 | + | - |
| N-Stearoylethanolamine 3 | + | - |
| N-Linoleoylethanolamine 3 | + | - |
| Anandamide 3 | + | - |
| ➤ Primary Fatty Acid Amides | ||
| Palmitoleamide 2 | + | ++ |
| Palmitamde 2 | +++ | +++ |
| Oleamide 2,4 | +++ | +++ |
| Linoleamide 2 | ++ | ++ |
| ➤ N-Acyldopamines | ||
| N-Palmitoyldopamine 2 | ++ | - |
| N-Oleoyldopamine 2 | + | - |
| N-Archidonoyldopamine 2 | + | - |
| Fatty Acid Amide | Larvae 2,3 | Head 4 | Thorax-Abdomen 4 |
|---|---|---|---|
| ➤ N-Acylalanine | |||
| N-Palmitoylalanine | detected | - | - |
| N-Stearoylalanine | detected | - | - |
| N-Oleoylalanine | detected | - | - |
| N-Linoleoylalanine | detected | - | - |
| ➤ N-Acyl-γ-aminobutyrates | |||
| N-Oleoyl-γ-aminobutyrate | detected | - | - |
| N-Linoleoyl-γ-aminobutyrate | detected | - | - |
| ➤ N-Acylglycines | |||
| N-Palmitoylglycine | + | + | + |
| N-Stearoylglycine | detected | not detected | not detected |
| N-Oleoylglycine | + | ++ | + |
| N-Linoleoylglycine | + | ++ | + |
| N-Arachidonoylglycine 5 | +, not detected | not detected | not detected |
| ➤ N-Acylleucines | |||
| N-Palmitoylleucine | detected | - | - |
| N-Stearoylleucine | detected | - | - |
| N-Oleoylleucine | detected | - | - |
| N-Linoleoylleucine | detected | - | - |
| ➤ N-Acylmethionines | |||
| N-Palmitoylmethionine | detected | - | - |
| N-Oleoylmethionine | detected | - | - |
| N-Linoleoylmethionine | detected | - | - |
| ➤ N-Acylphenylalanines | |||
| N-Palmitoylphenylalanine | detected | - | - |
| N-Stearoylphenylalanine | detected | - | - |
| N-Oleoylphenylalanine | detected | - | - |
| N-Linoleoylphenylalanine | detected | - | - |
| ➤ N-Acylprolines | |||
| N-Palmitoylproline | detected | - | - |
| N-Stearoylproline | detected | - | - |
| N-Oleoylproline | detected | - | - |
| N-Linoleoylproline | detected | - | - |
| ➤ N-Acylserines | |||
| N-Palmitoylserine | detected | - | - |
| N-Stearoylserine | detected | - | - |
| N-Oleoylserine | detected | - | - |
| N-Linoleoylserine | detected | - | - |
| ➤ N-Acyltryptophans | |||
| N-Palmitoyltryptophan | detected | - | - |
| N-Stearoyltryptophan | detected | - | - |
| N-Oleoyltryptophan | detected | - | - |
| N-Linoleoyltryptophan | detected | - | - |
| ➤ N-Acyltyrosines | |||
| N-Palmitoyltyrosine | detected | - | - |
| N-Stearoyltyrosine | detected | - | - |
| N-Oleoyltyrosine | detected | - | - |
| N-Linoleoyltyrosine | detected | - | - |
| ➤ N-Acylvalines | |||
| N-Palmitoylvaline | detected | - | - |
| N-Stearoylvaline | detected | - | - |
| N-Oleoylvaline | detected | - | - |
| N-Linoleoylvaline | detected | - | - |
| ➤ N-Acylethanolamines | |||
| N-Palmitoylethanolamine | detected | not detected | not detected |
| N-Stearoylethanolamine | detected | not detected | not detected |
| N-Oleoylethanolamine | + | + | + |
| N-Linoleoylethanolamine | detected | not detected | not detected |
| Anandamide 5 | +, not detected | + | + |
| ➤ N-Acyldopamines | |||
| N-Palmitoyldopamine | + | + | + |
| N-Oleoyldopamine | + | not detected | + |
| N-Arachidonoyldopamne | + | + | not detected |
| ➤ N-Acylserotonins | |||
| N-Palmitoylserotonin | + | - | - |
| N-Oleoylserotonin | + | - | - |
| N-Arachidonoylserotonin | + | - | - |
| ➤ Primary Fatty Acid Amides | |||
| Palmitamide | + | + | not detected |
| Palmitoleamide | + | + | not detected |
| Oleamide | + | ++ | not detected |
| Linoleamide | + | + | not detected |
| Fatty Acid Amide | Head | Thorax | Abdomen |
|---|---|---|---|
| ➤ N-Acylglycines | |||
| N-Palmitoylglycine | + | + | ++ |
| N-Oleoylglycine | + | + | + |
| N-Arachidonoylglycine | + | detected 3 | |
| ➤ N-Acylethanolamines | |||
| N-Oleoylethanolamine | + | + | +++ |
| Anandamide | + | + | detected 3 |
| ➤ N-Acyldopamines | |||
| N-Palmitoyldopamine | + | + | ++ |
| N-Oleoyldopamine | + | detected 3 | detected 3 |
| N-Arachidonoyldopamine | detected 3 | detected 3 | detected 3 |
| ➤ N-Acylserotonin | |||
| N-Oleoylserotonin | + | + | ++ |
| ➤ Primary Fatty Acid Amides | |||
| Palmitamide | ++ | + | + |
| Palmitoleamide | ++ | +++ | +++ |
| Oleamide | ++ | +++ | ++ |
| Linoleamide | + | + | ++ |
| Fatty Acid Amide | T. Castaneum | B. Mori |
|---|---|---|
| ➤ N-Acylglycines | ||
| N-Palmitoylglycine | + | not detected |
| N-Oleoylglycine | + | + |
| N-Arachidonoylglycine | + | not detected |
| ➤ N-Acylethanolamines | ||
| N-Oleoylethanolamine | + | + |
| Anandamide | + | not detected |
| ➤ N-Acyldopamines | ||
| N-Palmitoyldopamine | + | not detected |
| N-Oleoyldopamine | + | + |
| N-Arachidonoyldopamine | + | not detected |
| ➤ N-Acylserotonins | ||
| N-Palmitoylserotonin | not detected | + |
| N-Stearoylserotonin | not detected | + |
| N-Oleoylserotonin | detected 4 | + |
| ➤ Primary Fatty Acid Amides | ||
| Palmitamide | + | + |
| Palmitoleamide | ++ | + |
| Oleamide | + | + |
| Linoleamide | detected 4 | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, R.; Bhandari, S.; Mitchell, P.R., Jr.; Suarez, G.; Patel, N.B.; Lamb, K.; Bisht, K.S.; Merkler, D.J. Synthesis, Quantification, and Characterization of Fatty Acid Amides from In Vitro and In Vivo Sources. Molecules 2021, 26, 2543. https://doi.org/10.3390/molecules26092543
Ni R, Bhandari S, Mitchell PR Jr., Suarez G, Patel NB, Lamb K, Bisht KS, Merkler DJ. Synthesis, Quantification, and Characterization of Fatty Acid Amides from In Vitro and In Vivo Sources. Molecules. 2021; 26(9):2543. https://doi.org/10.3390/molecules26092543
Chicago/Turabian StyleNi, Ruidong, Suzeeta Bhandari, Perry R. Mitchell, Jr., Gabriela Suarez, Neel B. Patel, Kara Lamb, Kirpal S. Bisht, and David J. Merkler. 2021. "Synthesis, Quantification, and Characterization of Fatty Acid Amides from In Vitro and In Vivo Sources" Molecules 26, no. 9: 2543. https://doi.org/10.3390/molecules26092543
APA StyleNi, R., Bhandari, S., Mitchell, P. R., Jr., Suarez, G., Patel, N. B., Lamb, K., Bisht, K. S., & Merkler, D. J. (2021). Synthesis, Quantification, and Characterization of Fatty Acid Amides from In Vitro and In Vivo Sources. Molecules, 26(9), 2543. https://doi.org/10.3390/molecules26092543