A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157:H7 in an Aqueous Matrix
Abstract
1. Introduction
2. Results and Discussion
2.1. Selection of the TIR Protein Interaction Sequence Using Bioinformatics Tools
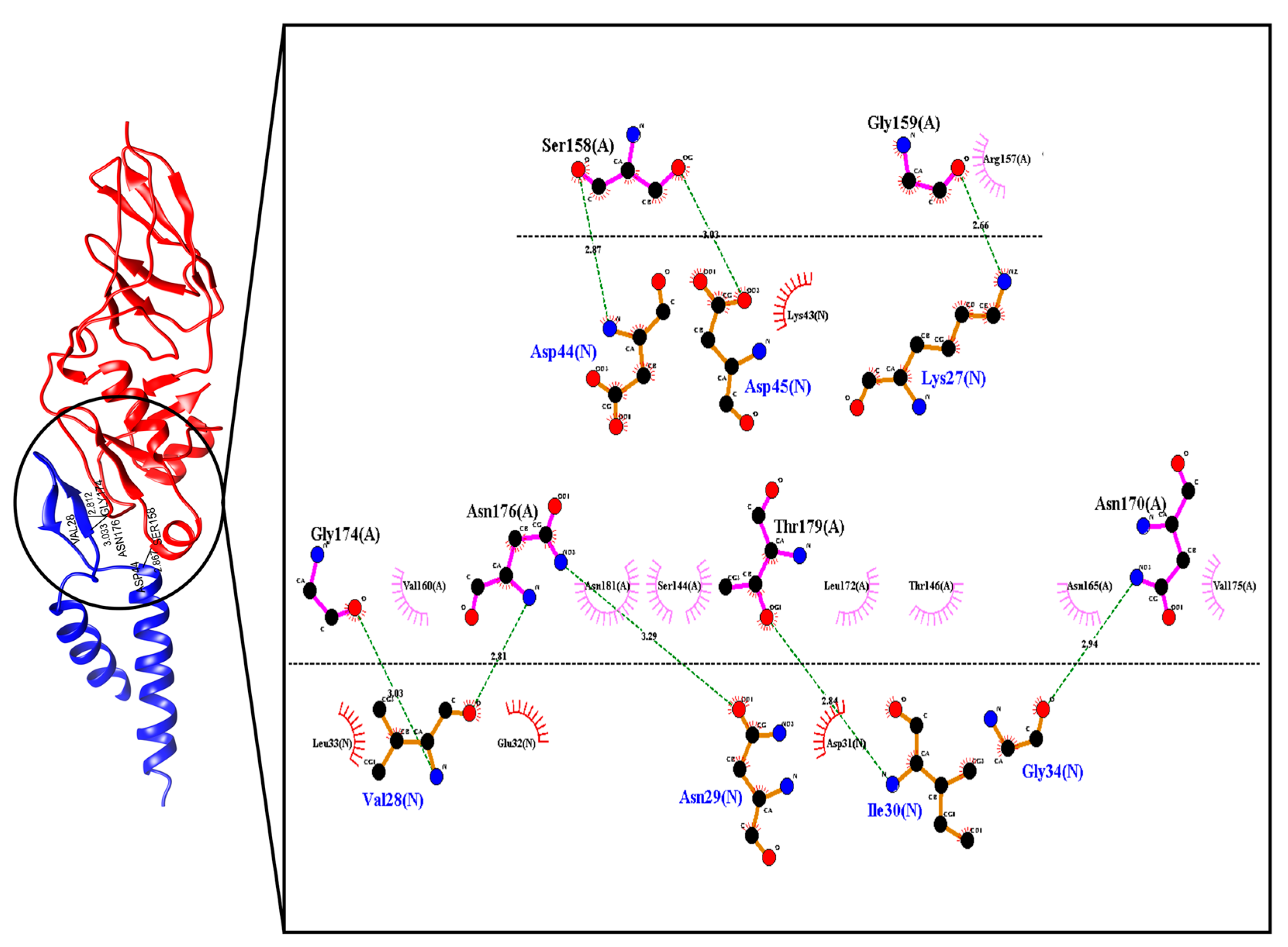
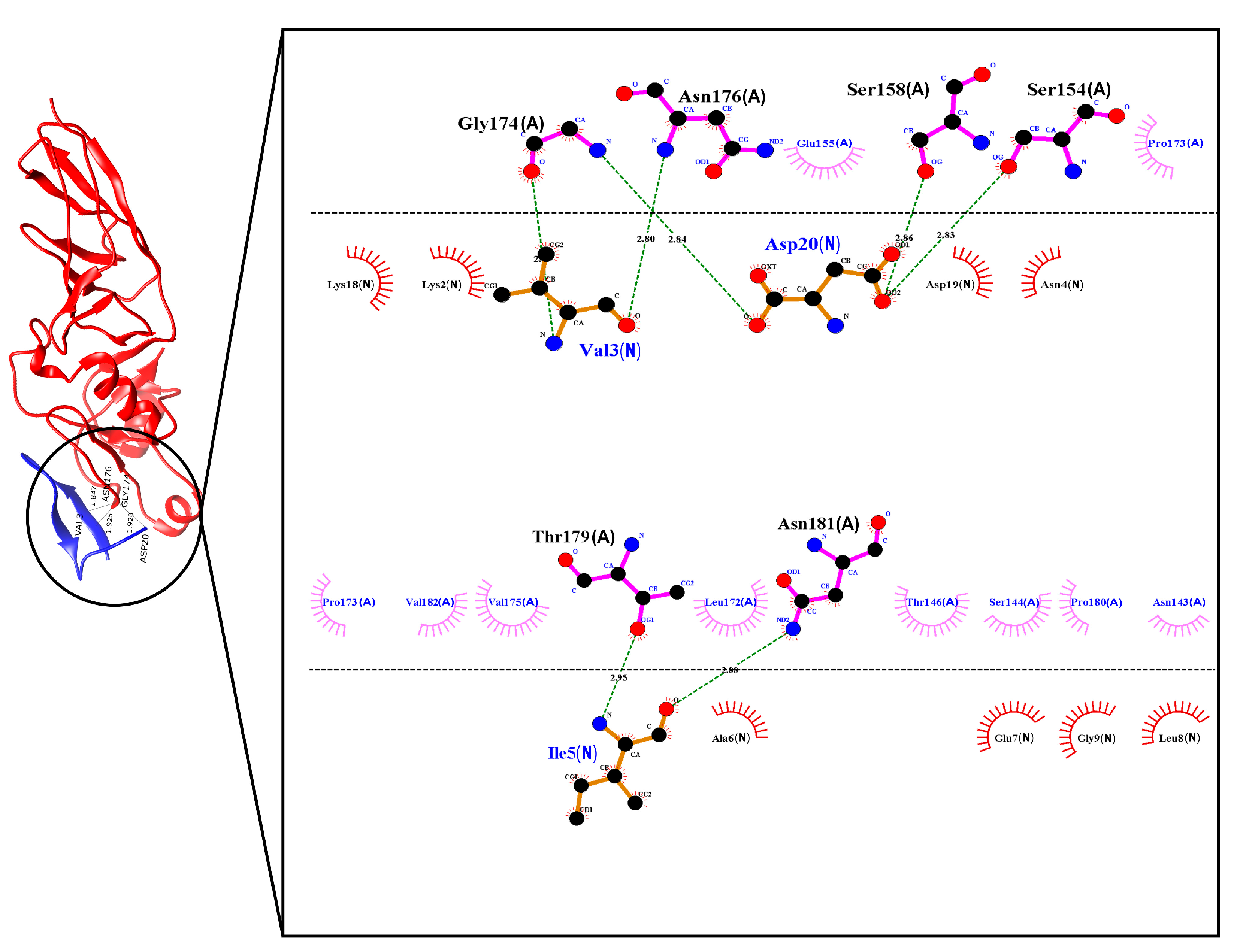
2.2. Selection of Ligands and Molecular Docking between the Peptide and the Intimin Protein
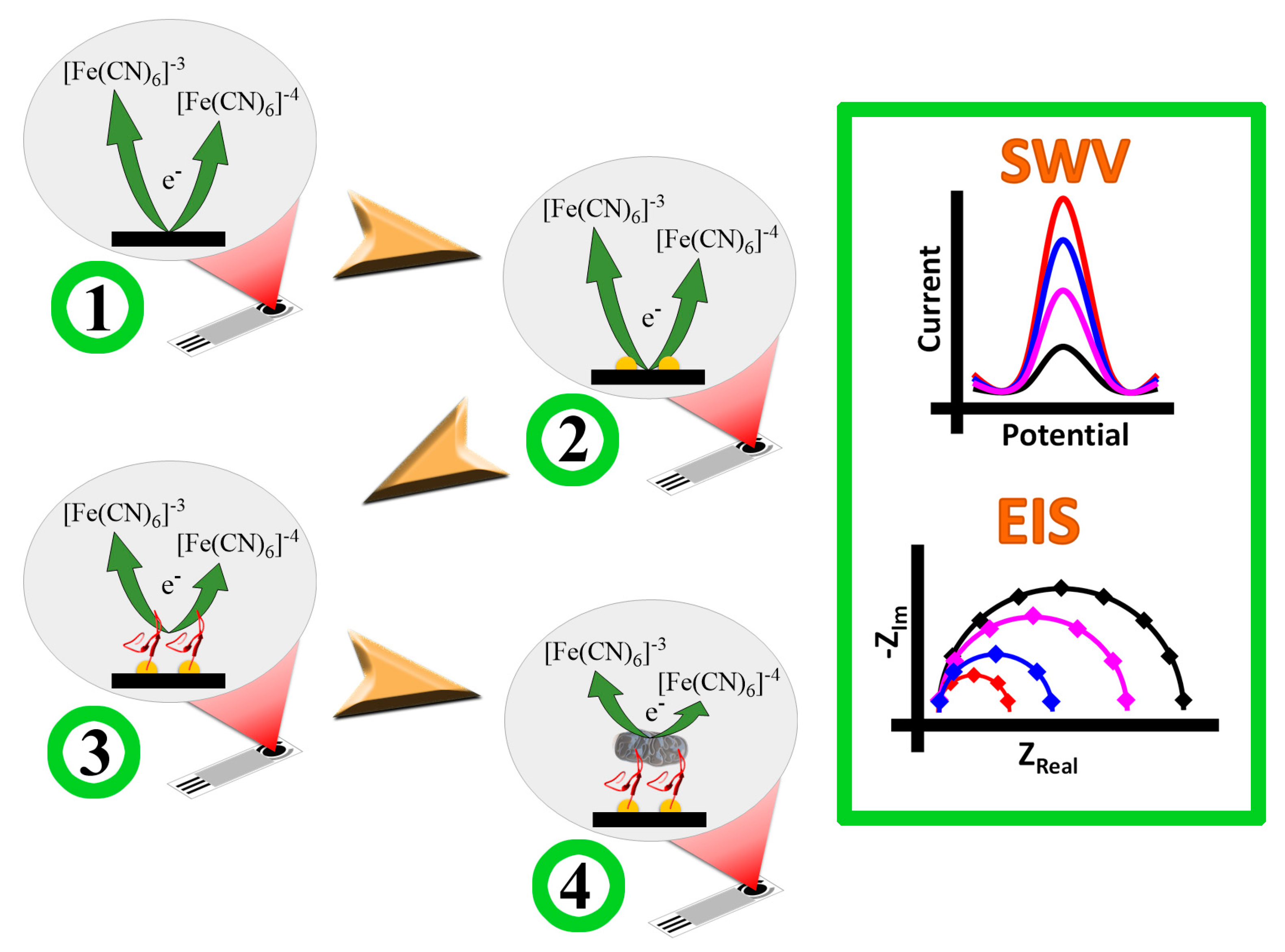
2.3. Preparation and Characterization of the Biosensor
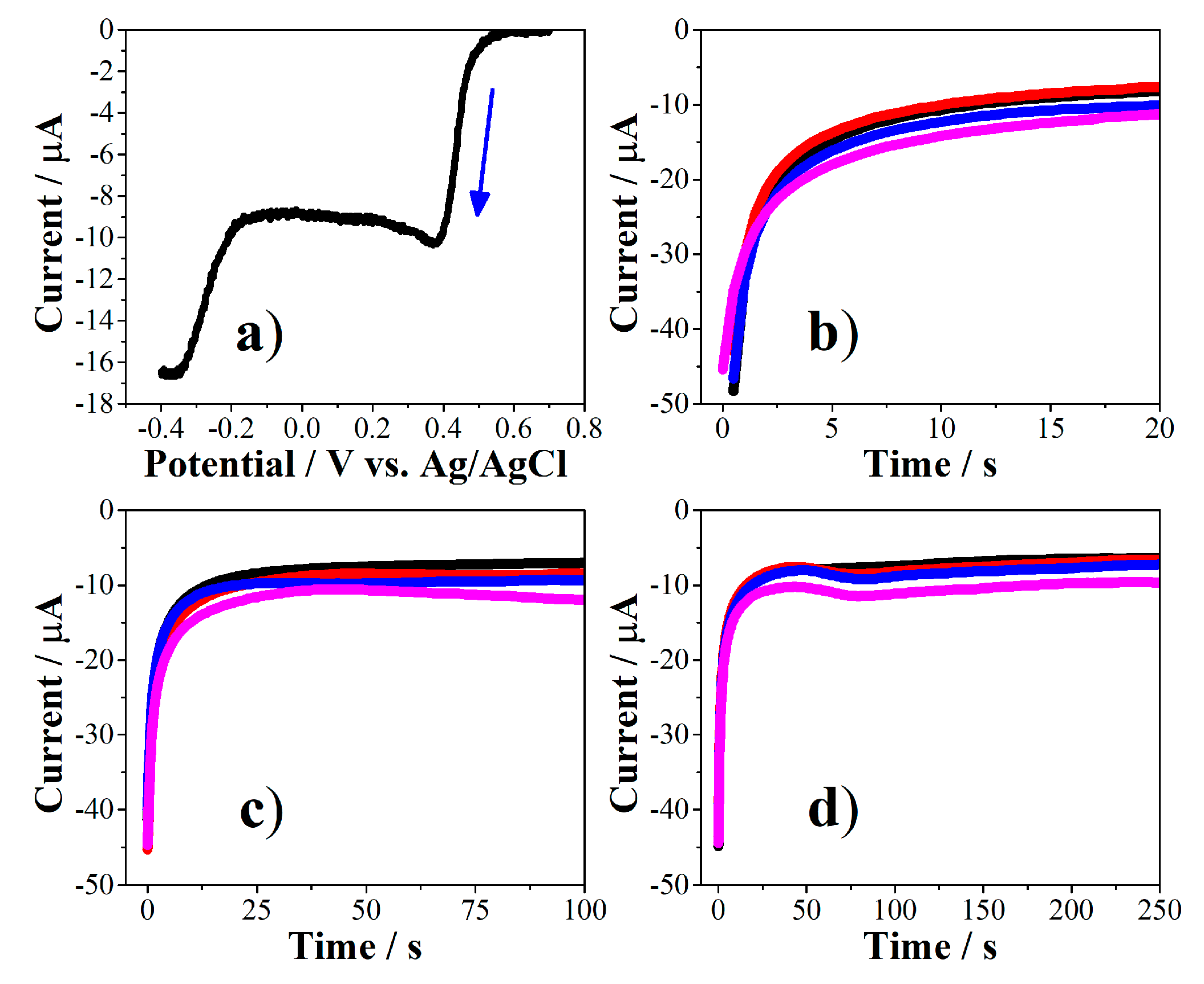
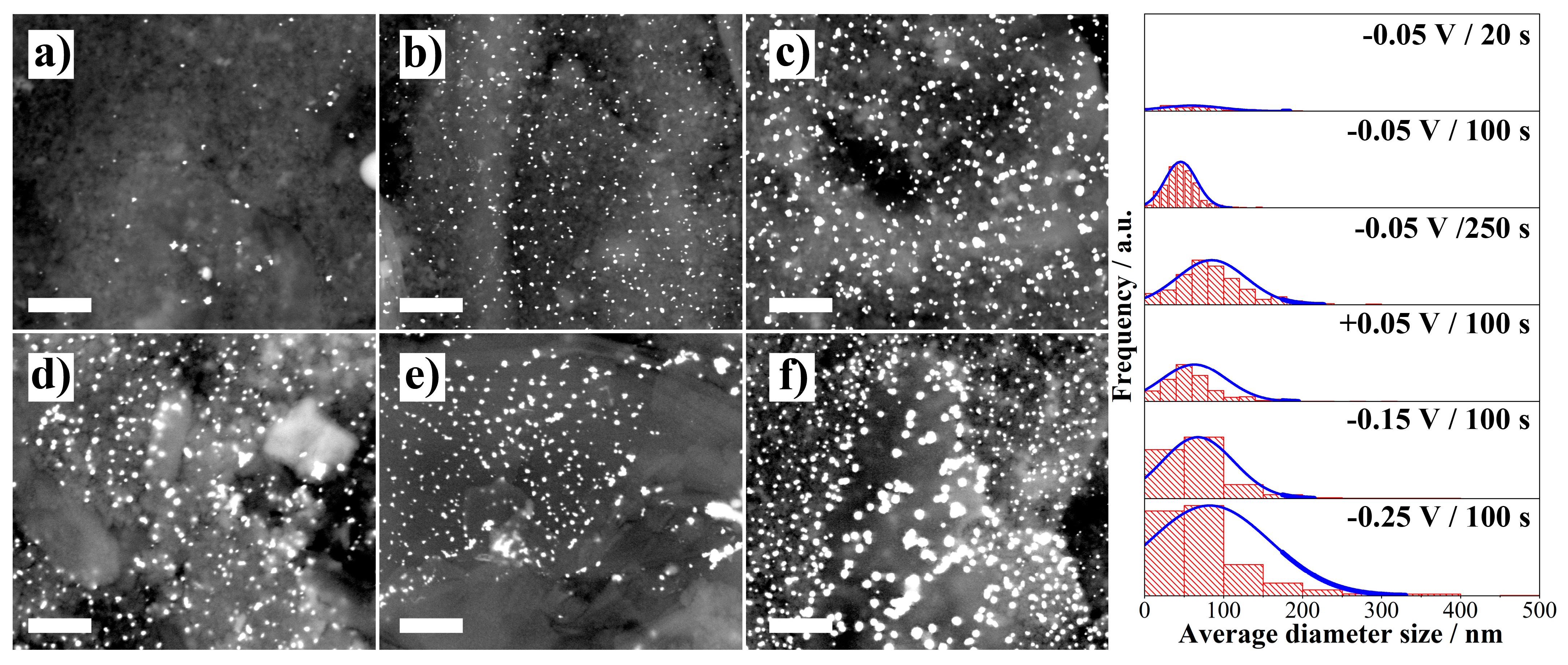
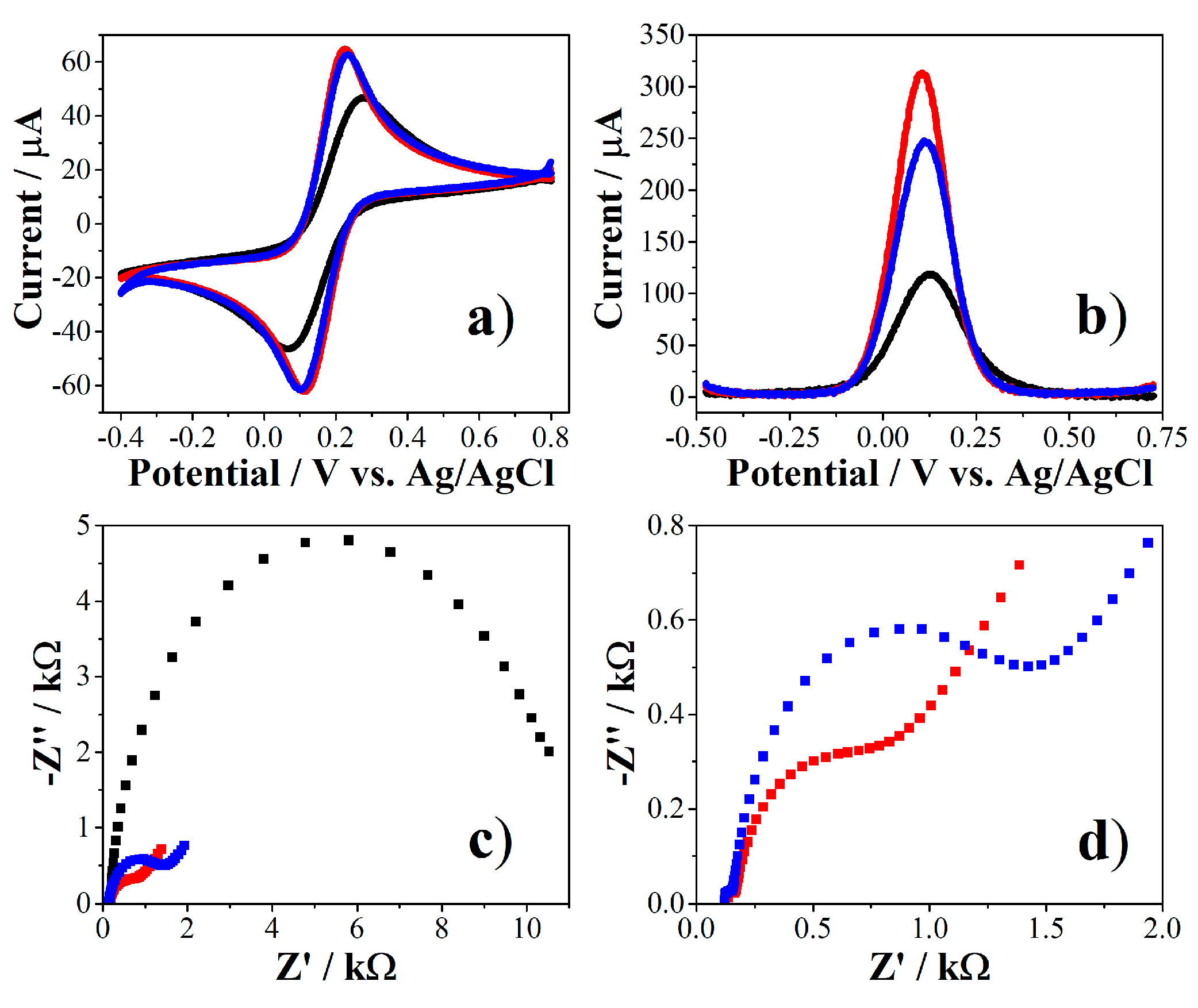
2.3.1. Electrodeposition of Gold Nanoparticles (AuNPs)
2.3.2. Immobilization of PEPTIR‑1.0 on AuNPs‑Modified Screen‑Printed Electrodes
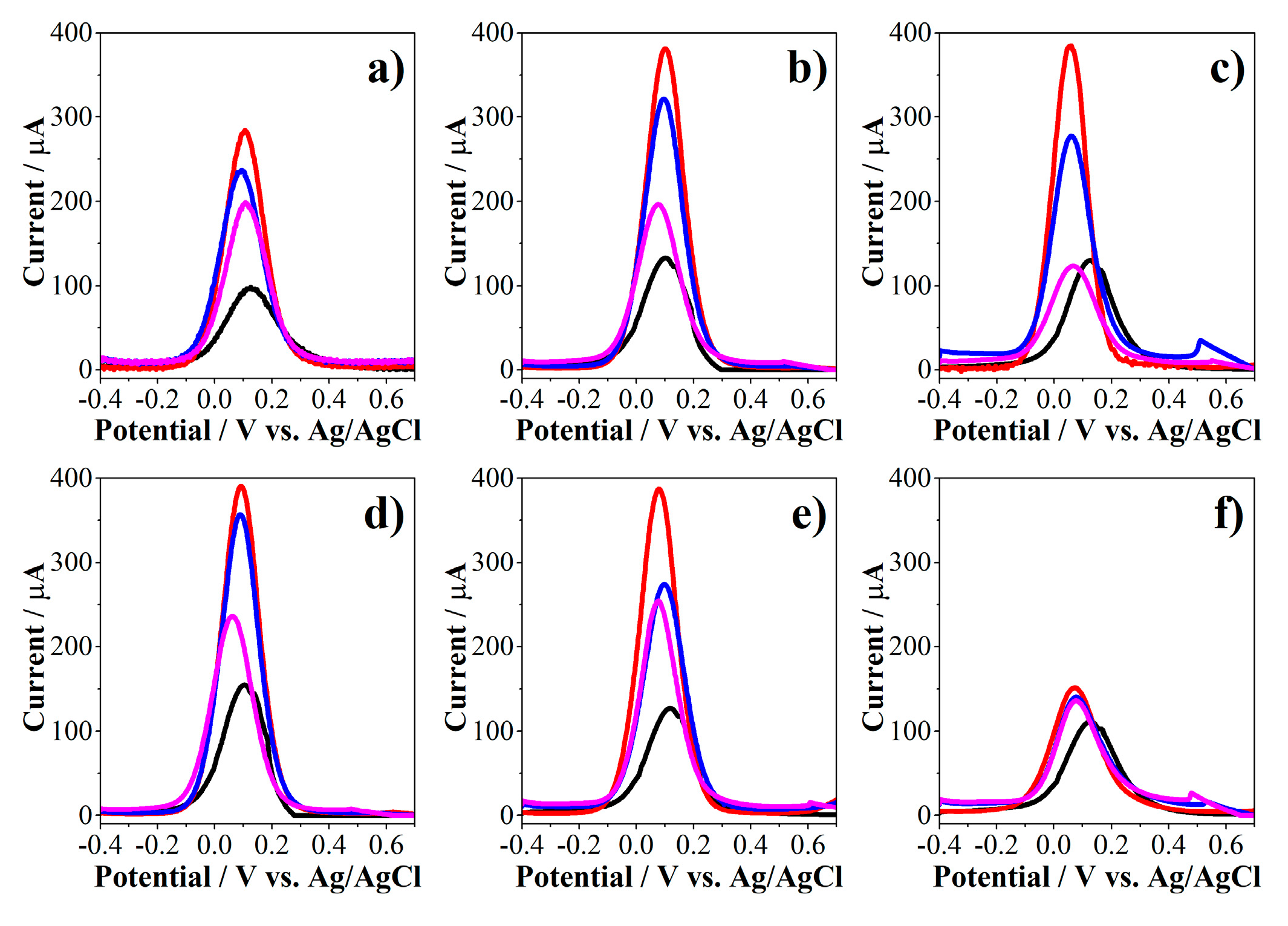
2.4. Evaluation of the Biosensor towards Detection of E. coli
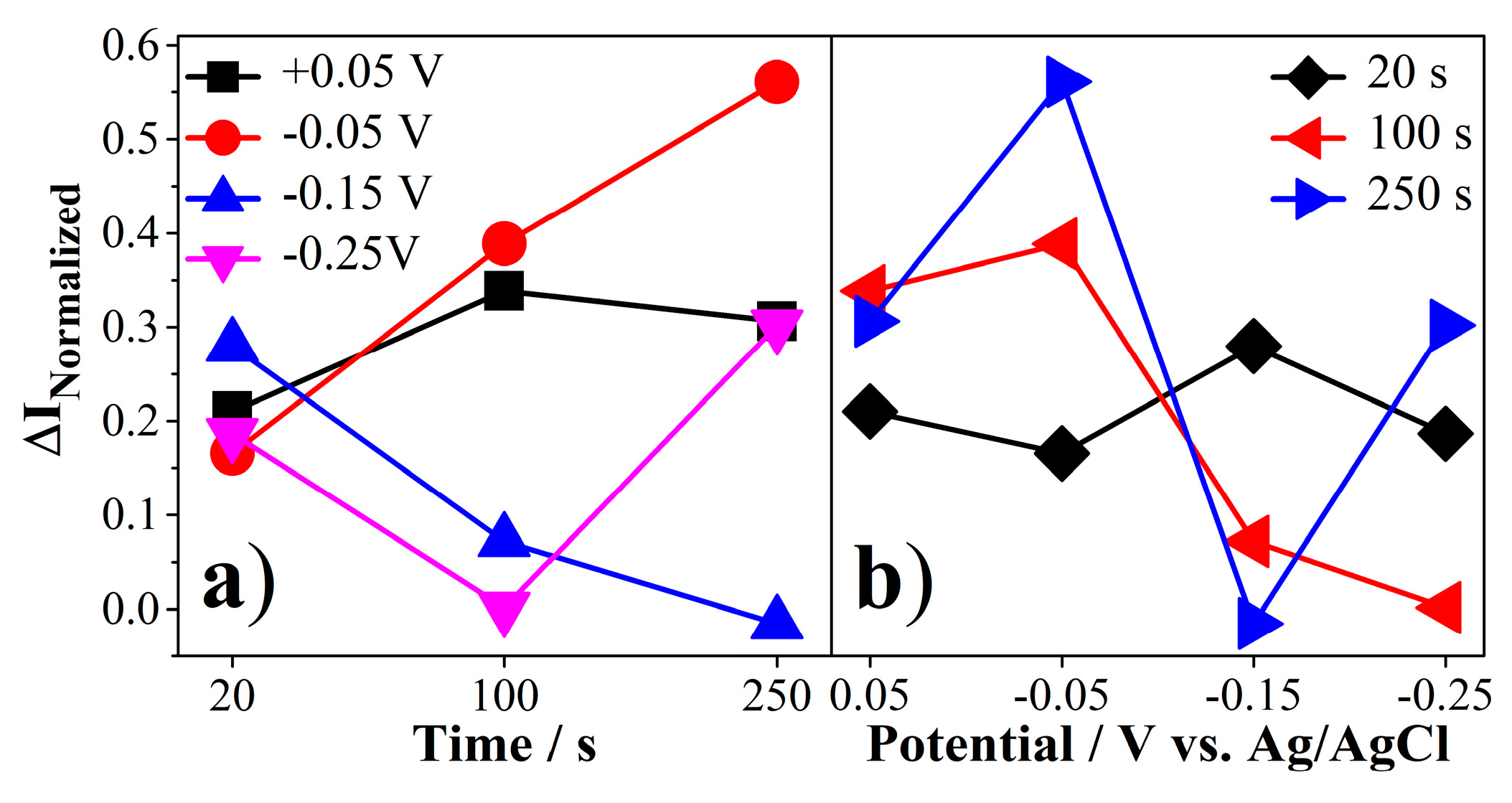
2.4.1. Evaluation of the Linear Response Range of the Biosensor
2.4.2. Evaluation of the Selectivity of the Biosensor
3. Materials and Methods
3.1. Reagents, Materials and Instruments
3.2. Selection of the TIR Protein Interaction Sequence and Its Modeling using Bioinformatics Tools
3.3. Selection of Ligands and Docking Molecular between the Peptide and the Intimin E Protein
3.4. Preparation and Characterization of the Biosensor
3.5. Detection of E. coli Cells Using the Prepared Electrochemical Biosensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef]
- Hald, T.; Aspinall, W.; Devleesschauwer, B.; Cooke, R.; Corrigan, T.; Havelaar, A.H.; Gibb, H.J.; Torgerson, P.R.; Kirk, M.D.; Angulo, F.J.; et al. World Health Organization Estimates of the Relative Contributions of Food to the Burden of Disease Due to Selected Foodborne Hazards: A Structured Expert Elicitation. PLoS ONE 2016, 11, e0145839. [Google Scholar] [CrossRef]
- Gracias, K.S.; McKillip, J.L. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can. J. Microbiol. 2004, 50, 883–890. [Google Scholar] [CrossRef]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.-M.; Chang, C.-L.; Hsu, M.-Y.; Lin, J.-Y.; Kuan, C.-M.; Wang, H.-K.; Huang, C.-T.; Chung, M.-C.; Huang, K.-C.; Hsu, C.-E.; et al. Paper-based ELISA to rapidly detect Escherichia coli. Talanta 2015, 145, 2–5. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Singh, P.; Mustapha, A. High-resolution melt curve PCR assay for specific detection of E. coli O157:H7 in beef. Food Control 2018, 86, 275–282. [Google Scholar] [CrossRef]
- Sharma, L.; Watts, E.; Singh, P. High resolution real-time PCR melting curve assay for identification of top five Penaeidae shrimp species. LWT 2020, 133, 109983. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; McDermott, P.F.; Schroeder, C.M.; White, D.G.; Meng, J. A DNA microarray for identification of virulence and antimicrobial resistance genes in Salmonella serovars and Escherichia coli. Mol. Cell. Probes 2005, 19, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Kosack, C.S.; Page, A.-L.; Klatser, P.R. A guide to aid the selection of diagnostic tests. Bull. World Health Organ. 2017, 95, 639–645. [Google Scholar] [CrossRef]
- Hajihosseini, S.; Nasirizadeh, N.; Hejazi, M.S.; Yaghmaei, P. A sensitive DNA biosensor fabricated from gold nanoparticles and graphene oxide on a glassy carbon electrode. Mater. Sci. Eng. C 2016, 61, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Bachan Upadhyay, L.S.; Verma, N. Enzyme Inhibition Based Biosensors: A Review. Anal. Lett. 2012, 46, 225–241. [Google Scholar] [CrossRef]
- Zhao, S.; Wen, H.; Ou, Y.; Li, M.; Wang, L.; Zhou, H.; Di, B.; Yu, Z.; Hu, C. A new design for living cell-based biosensors: Microgels with a selectively permeable shell that can harbor bacterial species. Sens. Actuators B Chem. 2021, 334, 129648. [Google Scholar] [CrossRef]
- Vanova, V.; Mitrevska, K.; Milosavljevic, V.; Hynek, D.; Richtera, L.; Adam, V. Peptide-based electrochemical biosensors utilized for protein detection. Biosens. Bioelectron. 2021, 180, 113087. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef] [PubMed]
- Castro-Ortiz, L.P.; Luna Pabello, V.M.; Villalobos Pietrini, R. Estado del arte y perspectivas del uso de biosensores ambientales en México. Rev. Int. Contam. Ambient. 2007, 23, 35–45. [Google Scholar]
- Wei, Z.; Cai, X.; Zhang, J.; Fan, J.; Xu, J.; Xu, L. High Sensitive Immunoelectrochemical Measurement of Lung Cancer Tumor Marker ProGRP Based on TiO2-Au Nanocomposite. Molecules 2019, 24, 656. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zhao, H.; Xu, M.; Ma, Q.; Ai, S. A label-free electrochemical impedance immunosensor based on AuNPs/PAMAM-MWCNT-Chi nanocomposite modified glassy carbon electrode for detection of Salmonella typhimurium in milk. Food Chem. 2013, 141, 1980–1986. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H.-B. Impedance based detection of pathogenic E. coli O157:H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Yi, H.; Xue, S.; Yuan, R.; Xu, W. A ‘signal on-off’ electrochemical peptide biosensor for matrix metalloproteinase 2 based on target induced cleavage of a peptide. RSC Adv. 2015, 5, 65725–65730. [Google Scholar] [CrossRef]
- Rodovalho, V.R.; Araujo, G.R.; Vaz, E.R.; Ueira-Vieira, C.; Goulart, L.R.; Madurro, J.M.; Brito-Madurro, A.G. Peptide-based electrochemical biosensor for juvenile idiopathic arthritis detection. Biosens. Bioelectron. 2018, 100, 577–582. [Google Scholar] [CrossRef]
- Hoyos-Nogués, M.; Brosel-Oliu, S.; Abramova, N.; Bratov, A.; Mas-Moruno, C.; Gil, F.J. Sensor impedimétrico para la detección de bacterias patogénicas mediante péptidos antimicrobianos. Biomecánica 2018, 24, 32–38. [Google Scholar] [CrossRef]
- Malvano, F.; Pilloton, R.; Albanese, D. A novel impedimetric biosensor based on the antimicrobial activity of the peptide nisin for the detection of Salmonella spp. Food Chem. 2020, 325, 126868. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, Y. Peptide-Based Biosensors. In Nano-Inspired Biosensors for Protein Assay with Clinical Applications; Elsevier: Amsterdam, The Netherlands, 2019; pp. 167–185. ISBN 9780128150535. [Google Scholar]
- Karimzadeh, A.; Hasanzadeh, M.; Shadjou, N.; Guardia, M. De la Peptide based biosensors. TrAC Trends Anal. Chem. 2018, 107, 1–20. [Google Scholar] [CrossRef]
- Farfán-García, A.E.; Ariza-Rojas, S.C.; Vargas-Cárdenas, F.A.; Vargas-Remolina, L.V. Mecanismos de virulencia de Escherichia coli enteropatógena. Rev. Chil. Infectología 2016, 33, 438–450. [Google Scholar] [CrossRef] [PubMed]
- DeVinney, R.; Stein, M.; Reinscheid, D.; Abe, A.; Ruschkowski, S.; Finlay, B.B. Enterohemorrhagic Escherichia coliO157:H7 Produces Tir, Which Is Translocated to the Host Cell Membrane but Is Not Tyrosine Phosphorylated. Infect. Immun. 1999, 67, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, btw514. [Google Scholar] [CrossRef]
- Rosetta Interface Analyzer. Available online: https://new.rosettacommons.org/docs/latest/application_documentation/analysis/interface-analyzer (accessed on 10 February 2021).
- Kanyong, P.; Rawlinson, S.; Davis, J. Gold nanoparticle modified screen-printed carbon arrays for the simultaneous electrochemical analysis of lead and copper in tap water. Microchim. Acta 2016, 183, 2361–2368. [Google Scholar] [CrossRef]
- López-Lorente, Á.I.; Valcárcel, M.; López-Lorente, Á.I. Gold Nanoparticles in Analytical Chemistry, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 9780444632852. [Google Scholar]
- Majzik, A.; Fülöp, L.; Csapó, E.; Bogár, F.; Martinek, T.; Penke, B.; Bíró, G.; Dékány, I. Functionalization of gold nanoparticles with amino acid, β-amyloid peptides and fragment. Colloids Surfaces B Biointerfaces 2010, 81, 235–241. [Google Scholar] [CrossRef]
- Labib, M.; Zamay, A.S.; Kolovskaya, O.S.; Reshetneva, I.T.; Zamay, G.S.; Kibbee, R.J.; Sattar, S.A.; Zamay, T.N.; Berezovski, M.V. Aptamer-Based Impedimetric Sensor for Bacterial Typing. Anal. Chem. 2012, 84, 8114–8117. [Google Scholar] [CrossRef] [PubMed]
- Castillo, G.; Pribransky, K.; Mező, G.; Kocsis, L.; Csámpai, A.; Németh, K.; Keresztes, Z.; Hianik, T. Electrochemical and Photometric Detection of Plasmin by Specific Peptide Substrate. Electroanalysis 2015, 27, 789–798. [Google Scholar] [CrossRef]
- Puiu, M.; Bala, C. Peptide-based electrochemical biosensors. In Electrochemical Biosensors; Elsevier: Amsterdam, The Netherlands, 2019; pp. 277–306. ISBN 9780128164914. [Google Scholar]
- Rezaei, B.; Irannejad, N. Electrochemical detection techniques in biosensor applications. In Electrochemical Biosensors; Elsevier: Amsterdam, The Netherlands, 2019; pp. 11–43. ISBN 9780128164914. [Google Scholar]
- Brosel-Oliu, S.; Ferreira, R.; Uria, N.; Abramova, N.; Gargallo, R.; Muñoz-Pascual, F.X.; Bratov, A. Novel impedimetric aptasensor for label-free detection of Escherichia coli O157:H7. Sens. Actuators B Chem. 2018, 255, 2988–2995. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, H.; Hao, H.; Gong, Q.; Nie, K. Detection of Escherichia coli with a label-free impedimetric biosensor based on lectin functionalized mixed self-assembled monolayer. Sens. Actuators B Chem. 2016, 229, 297–304. [Google Scholar] [CrossRef]
- Yi, Y.; Ma, Y.; Gao, F.; Mao, X.; Peng, H.; Feng, Y.; Fan, Z.; Wang, G.; Guo, G.; Yan, J.; et al. Crystal Structure of EHEC Intimin: Insights into the Complementarity between EPEC and EHEC. PLoS ONE 2010, 5, e15285. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Maupetit, J.; Derreumaux, P.; Tuffery, P. PEP-FOLD: An online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009, 37, W498–W503. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version Open-Source PyMOL 1.8.x., Schrödinger, LLC. Available online: https://github.com/schrodinger/pymol-open-source (accessed on 11 September 2020).
- Raveh, B.; London, N.; Schueler-Furman, O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins Struct. Funct. Bioinform. 2010, 78, 2029–2040. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Uria, N.; Abramova, N.; Bratov, A. Impedimetric Sensors for Bacteria Detection. In Biosensors—Micro and Nanoscale Applications; InTechOpen: London, UK, 2015. [Google Scholar]



| Model | RMSD |
|---|---|
| 1 | 4834 |
| 2 | 7722 |
| 3 | 4252 |
| 4 | 8359 |
| 5 | 6011 |
| Model | ΔG (Kcal/mol) 1 | Kd (mol/L) 2 | Interactions | dG_Sep (Rosetta Energy) | dSASA_int (Å2) | Per_res_Energy (Rosetta Energy) |
|---|---|---|---|---|---|---|
| TIR (Original) | −11.50 | 3.7 × 10−9 | 62 | −11.36 | 1174.01 | +2.148 |
| 3.1 | −10.9 | 9.6 × 10−9 | 52 | −25.08 | 984.05 | −1.873 |
| 3.2 | −10.6 | 1.6 × 10−8 | 53 | −25.19 | 981.98 | −1.751 |
| 3.3 | −10.8 | 1.3 × 10−8 | 47 | −24.99 | 890.17 | −1.845 |
| 3.4 | −10.8 | 1.2 × 10−8 | 49 | −22.52 | 998.71 | −1.568 |
| 3.5 | −10.5 | 2.1 × 10−8 | 54 | −21.99 | 963.32 | −1.745 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ropero-Vega, J.L.; Redondo-Ortega, J.F.; Galvis-Curubo, Y.J.; Rondón-Villarreal, P.; Flórez-Castillo, J.M. A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157:H7 in an Aqueous Matrix. Molecules 2021, 26, 2559. https://doi.org/10.3390/molecules26092559
Ropero-Vega JL, Redondo-Ortega JF, Galvis-Curubo YJ, Rondón-Villarreal P, Flórez-Castillo JM. A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157:H7 in an Aqueous Matrix. Molecules. 2021; 26(9):2559. https://doi.org/10.3390/molecules26092559
Chicago/Turabian StyleRopero-Vega, Jose Luis, Joshua Felipe Redondo-Ortega, Yuli Juliana Galvis-Curubo, Paola Rondón-Villarreal, and Johanna Marcela Flórez-Castillo. 2021. "A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157:H7 in an Aqueous Matrix" Molecules 26, no. 9: 2559. https://doi.org/10.3390/molecules26092559
APA StyleRopero-Vega, J. L., Redondo-Ortega, J. F., Galvis-Curubo, Y. J., Rondón-Villarreal, P., & Flórez-Castillo, J. M. (2021). A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157:H7 in an Aqueous Matrix. Molecules, 26(9), 2559. https://doi.org/10.3390/molecules26092559






