Abstract
Interest in the use of non-Saccharomyces yeast in mixed cultures is increasing due to the perceived improvement in the quality and complexity of the resulting wines. The aim of the study was to determine the ability of monocultures and mixed yeast cultures for deacidification and improvement of the composition of cold climate grape wines. Fermentation of grape musts with increased total acidity was carried out with the use of monocultures of Saccharomyces cerevisiae MH020215 (Sc), Zygosaccharomyces bailii 749 (Zb) and Metschnikowia pulcherrima MG970690 (Mp), and their mixed cultures, inoculated simultaneously and sequentially. Oenological parameters, organic acids and volatile compounds profiles of obtained wines were characterized. The fermentation kinetics and analytical profiles of the obtained wines showed that the use of mixed yeast cultures contributed to the reduction of volatile acidity and acetic acid content in the wines, as well as obtaining a favorable aromatic profile of the wines. The dominant higher alcohols in all wines were 2-methyl-1-propanol, 3-methyl-1-butanol and 2-methyl-1-butanol. Significantly higher amounts of the first two compounds were found in wines obtained with M. pulcherrima MG070690, both in monoculture and in mixed cultures. The monocultures of M. pulcherrima MG070690 (Mp) compared with Z. bailli 749 (Zb) synthesized higher levels of esters in wines, including ethyl acetate, ethyl propionate, isobutyl acetate, ethyl pyroracemate and isoamyl acetate.
1. Introduction
Organic acids present in grapes are the main source of wine acidity. The chemical composition of the grapes affects the composition of juice, and finally-the quality of the product. One of the key reasons for excessive wine acidity is l-malic acid, the concentration of which in grapes varies from 1 to 16 g/L, depending on the climate, region, season, and grapevine variety [1,2]. l-malic acid is accumulated mainly in the peel of the grapes and its amount is much lower in the flesh and the grape juice. This tendency may change during the ripening of the berries and due to technical treatments [3,4]. l-Malic and l-tartaric acids usually constitute more than 90% of the total acidity of the grapes [5]. Content of l-malic acid in Vitis vinifera berries from cold climate wine regions are usually higher (15–16 g/L), as the low temperature favours the acid respiration process, unlike warm climate regions. l-Malic acid not only contributes to the increase in wine acidity but also serves as a substrate for the growth of bacteria and yeast, the presence of which leads to undesirable changes in the product after its bottling [6]. Therefore, removing excess l-malic acid is important to ensure physical, biochemical and microbiological stability, which in turn contributes to the quality improvement of the produced wine. A natural method for obtaining less sour wine is biological deacidification, which can be achieved through malolactic fermentation (MLF) or malo-ethanolic fermentation (MEF). During MLF, lactic acid bacteria, inter alia Oenococcus oeni, convert malic acid to lactate and CO2, while MEF is mainly performed by yeasts, such as Schizosaccharomyces pombe [1,7,8,9]. Malolactic fermentation is a traditional wine deacidification method. However, due to problems associated (production of biogenic amines, ethyl carbamate, changes in the organoleptic characteristics of wine), alternative techniques are sought to reduce the acidity of wines. Deacidification using yeast strains has many benefits. It contributes to the degradation of l-malic acid but also reduces the risk of the formation of undesired compounds. Moreover, simultaneous alcoholic fermentation and deacidification prevent wine spoilage due to oxidation and inhibit the growth of harmful microorganisms. Therefore, the deacidification of wines with yeast offers the possibility of introducing better strategies to optimize the vinification process.
A few decades ago, the term “non-Saccharomyces” was usually associated with wine spoilage, mainly due to the increase in volatile acidity or formation of undesirable compounds such as volatile phenols (for which representatives of Brettanomyces genus were responsible) [10,11]. However, over the past years, many studies have demonstrated the metabolic/enzymatic potential of certain non-Saccharomyces yeasts, and their role in improving some technological and sensory aspects of wine [12,13,14]. The application of non-Saccharomyces yeasts has become a trend in the modern wine industry. Currently, several species of non-Saccharomyces yeast are used on an industrial scale, including Torulaspora delbrueckii, Lachancea thermotolerans, Metschnikowia pulcherrima and Pichia kluyveri. The advantage of their application is, among others, modification of the aroma [15,16], control of acidity [17], improvement of color and mouthfeel properties [18], as well as decreasing the ethanol content in wines [19,20]. In recent years, the number of studies using non-Saccharomyces yeast has increased significantly. However, still many species have not yet been well investigated, including yeasts of Candida, Hanseniaspora/Kloeckera, Metschnikowia, Pichia, Torulaspora and Zygosaccharomyces genera.
The aim of the study was to determine the ability of yeast monocultures and mixed cultures to reduce the total acidity and improve the chemical composition of cold climate grape wines. Fermentation of grape musts with increased total acidity was carried out with the use of monocultures of Saccharomyces cerevisiae MH020215 (Sc), Zygosaccharomyces bailii 749 (Zb) and Metschnikowia pulcherrima MG970690 (Mp), and their mixed cultures, inoculated simultaneously and sequentially. Oenological parameters, organic acids and volatile compounds profiles of obtained wines were characterized. The non-Saccharomyces strains used in this research, although periodically mentioned in scientific literature as wine-related yeasts, have not been thoroughly characterized in terms of their deacidification properties. There are studies concerning the use of Z. bailli yeast to deacidify wines [21,22]. However, little information is available on deacidification of cold climate wines using M. pulcherrima and Z. bailli. Hence the idea to conduct new research to enrich this area of knowledge.
2. Results and Discussion
2.1. The Dynamics of the Fermentation Process
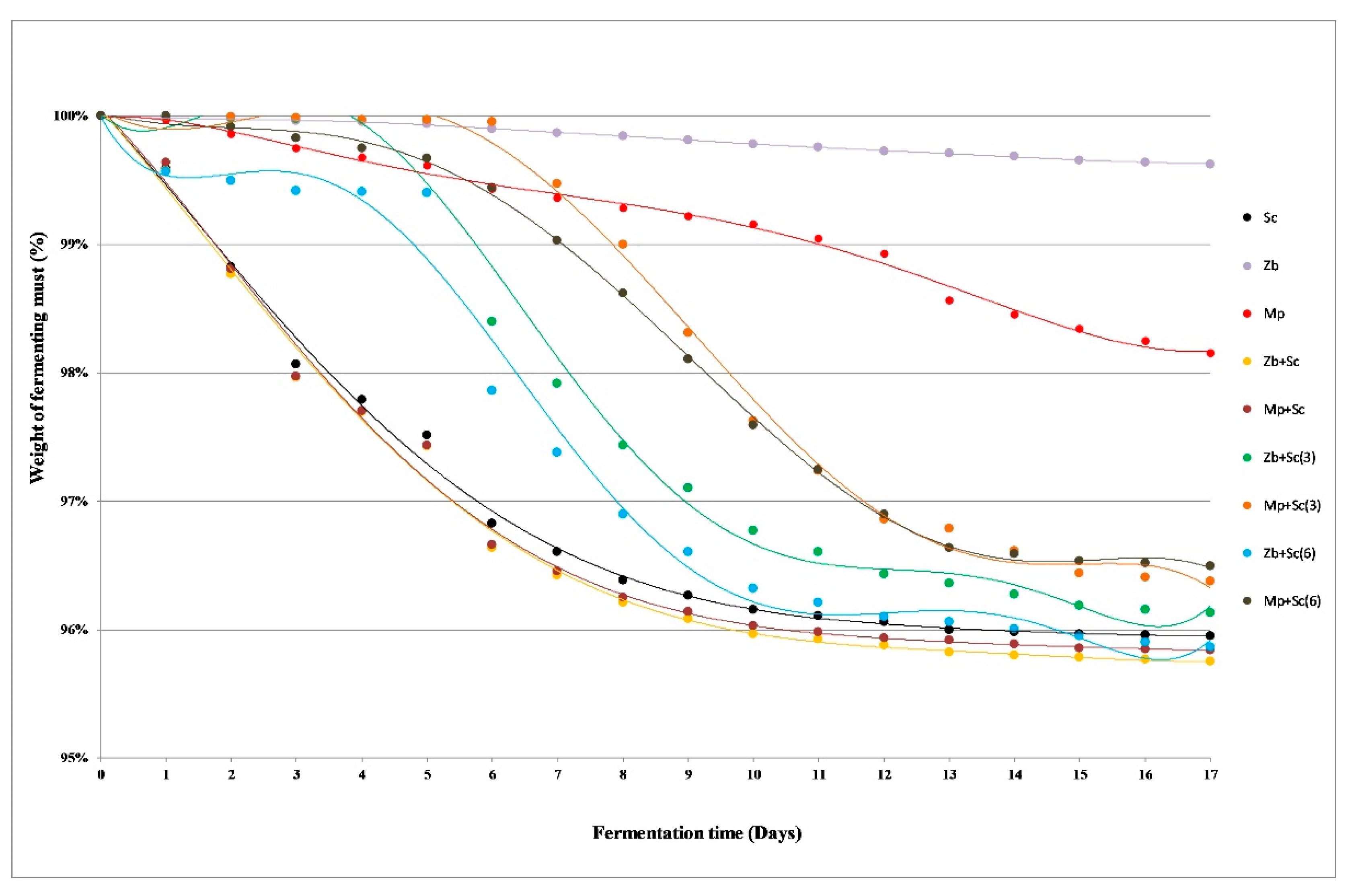
Ethyl alcohol, carbon dioxide and a wide range of by-products are produced as a result of carbohydrate metabolism during the must fermentation. The weight losses of fermenting batches are the effect of carbon dioxide release [23]. The samples inoculated with S. cerevisiae MH020215 (Sc) showed clearly the best fermentation dynamics among analysed monocultures (Figure 1). The weight losses of the batches were noticeable from the first day, although most of the carbon dioxide was released between the third and seventh day of the fermentation. A reduction in the fermentation rate was observed from around the 10th day. The small mass loss of the batches inoculated with Z. bailii 749 monoculture confirmed that this strain exhibits low ethanol fermentation abilities (Figure 1). The fermentation activity of Z. bailli yeasts is much lower than that of the S. cerevisiae, which is related to the low growth rate [24]. Z. bailii however, has a much higher tolerance for ethanol and some acids, therefore fermentation proceeds longer, and the cells do not degrade [25]. M. pulcherrima yeast are the natural microbiota of grapes. It participates in the initial stages of spontaneous fermentation, accounting up to 20% of microorganisms involved [26]. In the case of grape musts inoculated with M. pulcherrima MG970690 (Mp), the significant weight loss of the samples was observed (Figure 1). The fermentation dynamics of simultaneous fermentation of Z. bailii 749 and S. cerevisiae MH020215 (Zb+Sc), as well as M. pulcherrima MG970690 and S. cerevisiae MH020215 (Mp+Sc) proceeded similarly. Weight losses of the fermented batches were analogous as in S. cerevisiae MH020215 (Sc) monoculture (Figure 1).
Figure 1.
The fermentation dynamics of grape musts (abbreviations are described in Section 3.1.2. Grape Musts Fermentation).
Non-Saccharomyces yeast gain dominance during the initial stages of fermentation, which causes an increase in their amount. They are unable to produce high concentrations of alcohol, but they impart the desired sensory characteristics to the wines [27]. A rapid weight loss of the batches in sequential fermentation samples occurred after adding S. cerevisiae MH020215 yeast (Figure 1). For all trials inoculated on the third and sixth day, this phenomenon was observed on the next day of the process. The mixed cultures with Z. bailli 749 were characterized by a slightly better degree of attenuation (Figure 1).
2.2. Oenological Parameters
The term Free Amino Nitrogen (FAN) describes the content of amino acids and short-chain proteins in the must, which can be used by yeast as a building material. This parameter is a measure of the free nitrogen that is assimilable by the yeast cells. FAN content in wines depends on many factors, such as grapevine variety, soil quality, winegrowing practices and the usage of plant protection products [28]. The main sources of nitrogen needed for the growth of microorganisms during the fermentation process are free amino acids and ammonium ions. Literature data indicates that some juices may contain insufficient amounts of these components, which are required to ensure optimal growth of microorganisms and proper fermentation. Additionally, nitrogen requirement increases in must with the sugar concentration [7,29]. The content of free amino nitrogen in the grape must was 106 mg/L (Table 1). As expected, the lowest FAN values were found in wines subjected to simultaneous and sequential fermentation, as well as inoculated with yeast S. cerevisiae MH020215. The Z. bailli 749 (Zb) and M. pulcherrima MG970690 (Mp) monocultures utilized much lower amounts of FAN during the fermentation process (Table 2). This suggests that these species require less nitrogen compounds.

Table 1.
Characterization of fresh grape must (mean of 3 series ± standard deviation).

Table 2.
Characterization of analysed wines (mean of 3 series ± standard deviation).
All produced wines were subjected to chemical analysis to determine the content of alcohol, real extract and free amino nitrogen. The wine obtained by fermentation with the monoculture of S. cerevisiae MH020215 (Sc) (Table 2) was characterized by the highest ethyl alcohol concentration. Comparable values were acquired for samples inoculated simultaneously (Zb+Sc, Mp+Sc) (Table 2). As is known, S. cerevisiae dominates in the later stages of fermentation. The high initial biomass of non-Saccharomyces yeast gradually decreases, and often even completely disappears. Such a high ethanol content in the samples inoculated simultaneously was caused by the presence of S. cerevisiae in the fermenting must. Relatively high ethanol concentrations were also found in the case of sequential fermentation. Wines obtained with Z. bailli 749 (Zb) monoculture were characterized by low alcohol content, which confirmed also fermentation dynamics (Table 2, Figure 1). According to the literature, Z. bailli produces small amounts of ethanol. Its content in obtained wine was only 4 g/L. This result was over 60% lower than that of S. cerevisiae [30]. M. pulcherrima MG970690 (Mp) monoculture produced approximately 7% alcohol in wine (Table 2). These yeasts demonstrated superior fermentation capacity as compared to Z. bailii. During spontaneous fermentation they persist in the initial stages of the process, afterwards, the dominance of S. cerevisiae strains occurs. However, it has been shown, that the concentration of alcohol produced by M. pulcherrima yeast can reach even 10% [31].
The real extract consists of compounds remaining after simple distillation and it is a good indicator of sugars content and the degree of their utilization. The content of the real extract is related to the concentration of ethyl alcohol and residual sugars. The higher concentration of ethanol is produced, the less sugars remain after fermentation, and therefore the real extract is lower [32]. The value of this parameter in the analysed wines ranged from 20.6–25.8 g/L, except wines obtained with Z. bailii 749 (Zb) and M. pulcherrima MG970690 (Mp) monocultures (Table 2). Despite different methods of inoculation, S. cerevisiae MH020215 utilizes similar amounts of sugar. The addition of other cultures, such as Z. bailii 749 or M. pulcherrima MG970690 for deacidification does not reduce fermentation efficiency. In some cases, it is even slightly improved [14]. The yeast Z. bailli 749 showed very favorable deacidifying properties. Sequential fermentation resulted in a greater reduction of total acidity in the obtained wines, as compared to the simultaneous fermentation. On the other hand, M. pulcherrima MG970690 yeast did not deacidify the wines. However, the monoculture showed good fermentation properties (Table 2).
2.3. Total Acidity and Organic Acids
Total acidity is one of the basic parameters determining the winemaking process and affecting wine quality, as well as balanced taste. In the finished product, it should not exceed 10 g/L [33]. The initial acidity of the must before the fermentation was adjusted for every batch to 10 g/L, expressed as a malic acid concentration (g malic acid/L), (Table 1). Z. bailii 749 showed very favourable deacidification properties (Table 2). In contrast to S. cerevisiae, Z. bailii cells are able to assimilate l-malic acid in a relatively short time, but they require a carbon source in the form of sugars for the process [34]. Sequential fermentation resulted in a greater reduction in the acidity of obtained wines, compared to the simultaneous inoculation (Table 2). Based on the obtained results, it can be supposed that intensively growing S. cerevisiae MH020215 cells use the sugar from the must for the alcoholic fermentation. It causes depletion in carbon source for Z. bailii 749 and stops the acids decomposition. Delayed inoculation with S. cerevisiae MH020215 (Zb+Sc(3) and Zb+Sc(6)) seems to have a positive effect on the deacidification capacity of the mixed culture. However, according to the literature, the extended period of Z. bailii activity may cause the formation of acetic acid, which negatively affects the sensory characteristics of the wine [14]. S. cerevisiae yeasts individually are able to break down the excess of this compound in amounts from 3 to even 45%. However, this ability primarily depends on the varied levels of expression of the gene encoding the malic enzyme, which in turn is related to several environmental factors [3]. The analysis of the acidity of wines produced using mixed cultures revealed a clear relationship that later inoculation of S. cerevisiae MH020215 decreases the value of this parameter in the finished product (Table 2).
Volatile acidity is another factor determining the quality of the wine. It is usually expressed as acetic acid concentration in the beverage. This compound is formed as a result of the conversion of ethyl alcohol to acetaldehyde, followed by its transformation with the participation of the aldehyde dehydrogenase. Acetic acid is responsible for 90% of the volatile acidity of wines while the remaining fatty acids (propanoic and butanoic acid) are present in small amounts [7]. Acetic acid is usually formed in amounts of 0.2–0.8 g/L [33]. The levels of volatile acidity in analysed wines were low (Table 2). The highest volatile acidity was found in the samples fermented by M. pulcherrima MG970690 monoculture (Mp)-0.3 g/L. A slightly lower value was found in wines obtained with Z. bailii 749 (Zb) monoculture (Table 2). Some non-Saccharomyces yeast (such as Hanseniaspora, Zygosaccharomyces, Schizosaccharomyces) are producers of a considerable amount of acetic acid [35,36,37]. However, most non-Saccharomyces yeast are not capable of producing high amounts of volatile acids. Their content rarely exceeds 0.2 g/L after fermentation with M. pulcherrima or Z. bailli [38]. Similar levels of the analysed parameter were achieved both in simultaneous and sequential fermentations, ranging from 0.16 to 0.19 g/L (Table 2). Also, relatively low volatile acidity values were obtained by Comitini et al. [39] who used mixed cultures of M. pulcherrima and S. cerevisiae in the fermentation (0.30–0.34 g/L).
The main organic acids found in grapes are l-tartaric and l-malic acids. Although these compounds have a similar chemical structure, they are synthesized from glucose in fruits using different metabolic pathways. l-malic acid is formed in the reaction of glycolysis and the tricarboxylic acid (TCA) cycle, while ascorbate is an intermediate product of l-tartaric acid biosynthesis. Tartaric acid is usually present in grapes in concentrations from 5 to 10 g/L, while the content of l-malic acid in ripe grapes ranges most often between 2 and 6.5 g/L [8]. In fresh grape must used for fermentation, 2.54 g/L tartaric acid was detected (Table 1). Tartaric acid is not metabolized in the fermentation process, so its amounts are usually constant [40]. This has been confirmed in our research (Table 3). Tartaric acid is very stable, but in rare cases, it may decrease as a result of the precipitation of potassium tartrate [41]. Vonach et al. [42] indicate the presence of this component in wine in the amount of 1.06–1.68 g/L. Other literature sources report that its content can be as high as 4.55 g/L [43]. On the other hand, in Slovak wines, the concentration of l-tartaric acid ranged from 0.95 to 2.58 g/L. Dobrowolska-Iwanek et al. [44] noted over 10 times lower tartaric acid content in wine obtained from the Leon Millot variety (0.224 g/L) compared to our research. Similarly, low values of the analyzed component were found in the Rondo and Regent wines. In turn, white wines were characterized by higher tartaric acid content.

Table 3.
Characterization of organic acids profiles in obtained wines (mean of 3 series ± standard deviation).
Cold climate wines are characterized by a higher content of l-malic acid compared to tartaric acid. In turn, the reduction of malic acid content takes place during the malolactic fermentation process, as a result of which malic acid is converted into lactic acid [40]. The greatest decrease in malic acid was found in the wines inoculated sequentially (Zb+Sc(3), Mp+Sc(3), Zb+Sc(6), Mp+Sc(6)) (Table 3). In the experiments carried out by Soyer et al. [2], the content of l-malic acid in grapes ranged from 1.43 to 3.40 g/L. Much lower amounts of acid were found in grapevines from Chile (0.39–1.8 g/L) [45]. On the other hand, the l-malic acid content in cold climate wines reaches even 6.07 g/L [46]. Research by Dobrowolska-Iwanek et al. [44] showed a different ratio of tartaric and malic acid in cold climate wines. In contrast to white wines, in the case of wines obtained from the Leon Millot, Merechal Foch and Regent varieties, a greater amount of tartaric acid was observed compared to l-malic acid. It is worth mentioning that by the decision of the Council of Europe of December 20, 2005, the territory of Poland was included in zone A of viticulture in Europe. Lower temperatures result in lower sugar content in the fruit and increased acidity. Nevertheless, the chemical composition of the grapes grown in the cold climate zone allows for the achievement of high-quality wines that can be perceived as more harmonized and fresh, thanks to the different proportion of acids and sugars contained in grapes.
Despite the fact that citric acid is present in trace amounts in grapes (0.5–1 g/L), it plays an important role in biochemical and metabolic processes (Krebs cycle). l-Tartaric acid is resistant to degradation by microorganisms during the fermentation process, while malic and citric acids can be partially metabolized by yeast and bacteria, which results in the reduction of wine acidity [47,48]. In all analysed wines, citric acid was found at a similar level (Table 3).
The concentration of lactic acid in wine is usually between 0.2–0.4 g/L, although some sources mention as much as 6 g/L [49]. This component was not noted in most of the analysed wines. In wines obtained with the Z. bailli 749 (Zb) and M. pulcherrima MG970690 (Mp) monocultures, the highest concentration of this component was found (2.30 and 1.61 g/L). Some yeasts are able to produce malic and lactic acid affecting the pH of wines. Some non-Saccharomyces yeasts can produce it in high concentration, even in oenological conditions at a variable range of pH [8,50,51].
Succinic acid was found in all analysed wines (Table 3). The succinic acid concentration in wine is at the level of 0.5–1.5 g/L. Values above 3.0 g/L may be present in some red wines and may not necessarily negatively affect the taste of the wine [52]. The Z. bailli strain is known for the high production of succinic and acetic acid in wine [53]. On the other hand, the use of mixed cultures of M. pulcherrima MG970690 and S. cerevisiae MH020215 (Mp+Sc, Mp+Sc (3), Mp+Sc (6)), both as a result of simultaneous and sequential fermentation, resulted in obtaining a lower level of acetic acid in wines, in comparison with the monoculture of M. pulcherrima MG970690 (Table 3). Low acetic acid production in wines with the participation of non-Saccharomyces yeast in mixed cultures with S. cerevisiae was also demonstrated in the studies of other authors [16,31,39,54,55].
2.4. Volatile Compounds
Wine is one of the products with the most complex flavour profile. The aroma is influenced by the variety of grapevine used, the conditions of cultivation and harvest, while the characteristic bouquet is the result of the fermentation process, maturation and aging process of the wine. Most of the aroma compounds that are released into wine from grapes are present in the form of odourless glycosides [56].
Table 4 characterizes the components present in fresh grape must. It is known that higher alcohols are produced in the wine fermentation process, affecting the sensory profile of the drink [57]. However, literature sources also indicate trace amounts of these components in grapes, mainly in skins [58]. In fresh grape must, small amounts were noted, including 1-hexanol, 3-hexen-1-ol, 1-undecanol and 1-nonanol, as well as other compounds, including acetals (Table 4). The concentration of these compounds varies depending on the grapevine variety, climatic and agrotechnical factors [48].

Table 4.
Characterization of volatile compounds of grape must (mean of three series ± standard deviation).
Table 5 present the volatile components of obtained wines. One of the main group of compounds synthesized by yeast are higher alcohols, also known as fusels. Their molecules contain more than two carbon atoms and are characterized by higher molecular mass, as well as boiling point in comparison to ethanol. They are produced during the fermentation process, and their concentration reaches approx. 150–550 mg/L. In chemical terms, they can be divided into aliphatic and aromatic alcohols. The first group consists of inter alia propanol, isobutanol and amyl alcohols. The second includes: 2-phenylethanol, tyrosol or tryptophol. Fusel alcohols have an intense aroma that plays an important role in forming a wine bouquet. At low concentrations (below 300 mg/L) they positively affect its aroma whereas higher contents mask the proper aroma of the beverage [33]. 2-methyl-1-propanol, 3-methyl-1-butanol and 2-methyl-1-butanol were prevailing higher alcohols in all of the samples (Table 5). In the case of the two first listed components, significantly higher concentrations were found in wines inoculated sequentially (additional inoculation on the sixth day of fermentation). M. pulcherrima MG970690 (Mp) monoculture produced higher amounts of these compounds in the analysed wines, as compared to S. cerevisiae MH020215 (Sc). Obtained data do not match the results of other authors, who claim that non-Saccharomyces yeasts are responsible for the formation of lower concentrations of higher alcohols (n-propanol, isobutanol, amyl alcohols) in relation to S. cerevisiae strains [59,60]. However, the formation of these components is assessed based on a number of factors, including oxygen content, sugar content in grapes, temperature and maceration time [4,48,61]. The beneficial effect of non-Saccharomyces has been repeatedly demonstrated in mixed culture with wine strains. Wines obtained with the participation of S. cerevisiae, T. delbrueckii and M. pulcherrima were characterized by a high content of higher alcohols [39]. A combination of microorganisms can be used for obtaining unique wine aromas that could not be obtained with monocultures. Non-Saccharomyces yeast produced higher levels of 1-decanol, 1-undecanol, 2-dodecanol, 1-dodecanol and 1-hexanol in wines (Table 5). 3-methylpentanol and isohexanol were found only in wines inoculated with S. cerevisiae MH020215 yeast. Phenylethanol was detected in significant amounts in wines that were at least partially inoculated with S. cerevisiae MH020215. Significantly lower concentrations were found in samples fermented with M. pulcherrima MG970690 (Mp) and Z. bailii 749 (Zb) monocultures (Table 5). Escribano et al. [62] determined the fermentation capacity and aroma formation for several non-Saccharomyces yeasts. They suggested that M. pulcherrima owns good properties for wine fermentation and creates high concentrations of 2-phenylethyl alcohol and 2-phenylethyl acetate [63].

Table 5.
A heat map analysis of 49 volatiles [μg L−1] in wines produced using mono- and mixed cultures of yeasts (mean of three series). The highest content is in the darkest green and the lowest content is in the darkest red.
Esters are a broad group of by-products found in fermented beverages. They can be formed as a result of chemical condensation of carboxylic acids and alcohols, however mainly are the products of yeast metabolism. The enzymatic synthesis of esters is catalysed by many microbial enzymes (esterases, lipases), including acetyltransferases. Alcohol and acetyl coenzyme A are the substrates for this reaction. Currently, approximately 160 specific esters have been recognized. Depending on the substrates from which they are formed, volatile and non-volatile esters can be distinguished. Compounds belonging to the first group positively affect wine bouquet, while those from the second group act as a component of taste [64]. Among all identified esters in the tested samples, ethyl acetate was found in the highest quantities. Substantial amounts of this compound were identified in wines inoculated with M. pulcherrima MG970690 (Mp) and S. cerevisiae MH020215 (Sc) monocultures. Lower concentrations were found in samples inoculated with Z. bailii 749 (Zb) monoculture (Table 5). Both simultaneous and sequential fermentation increased quantities of ethyl acetate in the analysed wines compared to monocultures (Table 4). Similar results were obtained by Comitini et al. [39], using a mixed culture of S. cerevisiae and M. pulcherrima for the fermentation of the grape must. Yeast belonging to the Saccharomyces genus synthesizes mainly ethyl and acetate esters. Studies show that small quantities of ethyl acetate (50–80 mg/L) have a positive effect on the quality of the drink, whereas too high concentrations can lead to abnormal aftertaste [4]. Content of isoamyl acetate, 2-phenylethyl acetate and isobutyl acetate was several dozen, or even several hundred times lower than that of ethyl acetate (Table 5). According to the literature data, the use of mixed strains of M. pulcherrima and S. cerevisiae yeast allows for obtaining higher amounts of isoamyl acetate compared to Saccharomyces strains [39]. Our studies showed the opposite tendency (Table 5). Our previous research [65] concerned the physicochemical characterization of wines produced using indigenous yeasts from cold climate grapes. S. cerevisiae strain MH020215 has been shown to produce comparable amounts of isoamyl acetate and slightly lower ethyl acetate in cold climate wines, compared to the results in this article [65]. Other esters such as ethyl hexanoate, ethyl butanoate, ethyl pyroracemate, ethyl octanoate and diethyl succinate were identified in slightly lower concentrations in the analysed samples (Table 5). The wines obtained as a result of simultaneous and sequential fermentation were characterized by higher levels of these components (Table 4), similarly as in the case of the previously mentioned compounds. The remaining esters present in the wines occurred at a relatively low level (Table 5). According to the literature data, M. pulcherrima produces significant amounts of esters [16], especially ethyl octanoate [57,66]. The conducted research did not confirm this fact. In the wine inoculated with M. pulcherrima MG970690 (Mp) monoculture, this compound was not found (Table 5).
The main carbonyl compounds that occur in wine are aldehydes, and in smaller quantities, ketones. They emerge as by-products of fermentation, but also in the process of wine aging. Their concentration can vary considerably, depending on the conditions of the wine production and storage. The content of carbonyl compounds is higher in sweet wines than in dry wines, which is probably related to the oxidation of sugars. According to the literature the most important are acetaldehyde (3–494 mg/L), acetoin (0.7–350 mg/L) and diacetyl (0.1–7.5 mg/L) [67]. The aldehydes present in grapes are involved in the creation of varietal aromas, and their amount in young wines usually does not exceed 75 mg/L. The most important compound of this group is acetaldehyde, followed by propionic, isobutyric and isovaleric aldehyde. Higher amounts of acetaldehyde were detected in beverages fermented with S. cerevisiae MH020215 (Sc) monoculture, as well as those obtained with the use of mixed yeast cultures (Table 5). Much higher amounts of the analysed component were found in wines fermented sequentially with M. pulcherrima MG970690 (Mp+Sc(3), Mp+Sc(6)), in relation to Z. bailli 749 (Table 5). The profiles of volatile compounds in cold climate wines were analysed in our previous studies [65]. In wines produced with the use of the S. cerevisiae strain MH020215, higher acetaldehyde values (27.4–33.7 mg/L) were noted compared to the results obtained in this study. Also, Li and de Orduña [68] analyzed the production of acetaldehyde in fermenting grape must by yeast strains of oenological importance. It was found that both the S. cerevisiae and non-Saccharomyces strains showed similar metabolic kinetics with the highest acetaldehyde content at the beginning of fermentation followed by its reutilization. In wines obtained with the participation of M. pulcherrima yeast, the amount of acetaldehyde was low (<2 mg/L), while the amount of acetaldehyde was higher for Z. bailli (<25 mg/L) [68]. In our research, the content of acetaldehyde in wines fermented with the monocultures of M. pulcherrima MG970690 and Z. bailli 749 did not exceed 1 mg/L (Table 5). Clemente-Jimenez et al. [66] demonstrated the synthesis of significant amounts of acetaldehyde in wine by strains of C. stellata, H. uvarum, I. orientalis, I. terricola and M. pulcherrima in relation to S. cerevisiae. Similar results were achieved by Mateos et al. [69], indicating higher levels of acetaldehyde in wines produced with native microbiota compared to Saccharomyces yeast. Nonenal and decanal amounts were considerably lower in wines investigated. In the aspect of analysed compounds no substantial differences were found between the wines inoculated with non-Saccharomyces yeast and the strains of S. cerevisiae MH020215 (Table 5).
The presence of carboxylic acids was also noted in the analysed wines (Table 5). Octanoic acid was present in wines fermented with the M. pulcherrima MG970690 monoculture. In greater amounts, this component was present in wines fermented simultaneously and sequentially (Mp+Sc, Mp+Sc (3)). The exception was the wine that was inoculated with S. cerevisiae MH020215 yeast on the sixth day of fermentation (Table 5). M. pulcherrima yeast is capable of producing acetic acid in wines, which results in a rancid and pungent odour. These yeasts also produce hexanoic acid, which gives the wines the smell of fat and cheese [51]. In our research, no presence of hexanoic acid was found in wines fermented with non-Saccharomyces monocultures (Table 5).
The presence of terpenes was reported in the tested wines. Larger amounts of α-terpineol were found in wine fermented with the monoculture M. pulcherrima MG970690 (Table 5). Significant amounts of this compound were detected in simultaneously and sequentially fermented wines (Mp+Sc, Mp+Sc (3)). A similar relationship was noted for damascenone. Interestingly, lower amounts of these components were detected during sequential fermentation in which the yeast S. cerevisiae MH020215 was inoculated on the sixth day of the process (Mp+Sc(6)), (Table 5). Non-Saccharomyces yeasts, including M. pulcherrima, are capable of releasing distinct aromas from precursors such as glycosylated terpenes or bonded thiols via ß-glucosidase or C-S-lyase. Non-Saccharomyces yeast allows obtaining freshness of wine by synthesizing and subsequently free enzymes capable of releasing volatile thiols or terpenes (geraniol, linalool) [51,70].
3. Materials and Methods
3.1. Materials
Strains of Saccharomyces cerevisiae MH020215, Zygosaccharomyces bailii 749 and Metschnikowia pulcherrima MG970690 from the collection of Department of Fermentation Technology and Microbiology, Faculty of Food Technology, University of Agriculture in Krakow (Poland) were used in this research. Metschnikowia pulcherrima MG970690 and Saccharomyces cerevisiae MH020215 strains used for the research were isolated from spontaneous grape fermentation [71,72,73].
3.1.1. Inoculum Preparation
The three-step propagation was used to multiply cultures. In the first stage, the strains were propagated on Sabouraud agar slants (Biocorp, Warsaw, Poland) for 24 h, then were introduced to 10 mL of liquid Sabouraud broth (Biocorp). After another 24 h, dynamic propagation was conducted in 200 mL of liquid Sabouraud broth (Biocorp) for 48 h on a water bath shaker (120 rpm, 28 °C). After cultivation was completed, dry weight of yeast was determined with a moisture analyser (Radwag WPS 210S, Zakład Mechaniki Produkcyjnej, Radom, Poland), and then the adequate amount of yeast slurry was centrifuged (10 min, 4989× g/min). The precipitate was washed with sterile water, centrifuged again under the same conditions and introduced into the grape must.
3.1.2. Grape Musts Fermentation
The raw material used for fermentation was a Leon Millot grape juice, obtained by pressing the grapes using a basket press. The juice was poured into 4-L bottles and pasteurized. The total acidity of the must was set to 10 g/L, using l-malic acid. Each of 500 mL flasks was filled with 300 mL of juice. The yeast slurry was introduced into the batch in the amount of 0.5 g d.w/L. After closing the fermentation flasks and fixing fermentation tubes filled with glycerin, the setup was additionally sealed with a parafilm. The fermentation process was carried out for 28 days at 20 °C.
The samples were inoculated in three repetitions, according to the following scheme:
Monocultures: S. cerevisiae MH020215 (0.5 g d.w./L; Sc), Z. bailii 749 (0.5 g d.w./L; Zb), M. pulcherrima MG970690 (0.5 g d.w./L; Mp),
Simultaneous fermentation: S. cerevisiae MH020215 (day zero; 0.4 g d.w./L) + Z. bailii 749 (day zero; 0.1 g d.w./L; Zb+Sc); S. cerevisiae MH020215 (day zero; 0.4 g d.w./L) + M. pulcherrima MG970690 (day zero; 0.1 g d.w./L; Mp+Sc),
Sequential fermentation: Z. bailii 749 (day zero; 0.1 g d.w./L) + S.cerevisiae MH020215 (day three; 0.4 g d.w/L; Zb+Sc(3)), M. pulcherrima MG970690 (day zero; 0.1 g d.w./L) + S. cerevisiae MH020215 (day three; 0.4 g d.w/L; Mp+Sc(3)), Z. bailii 749 (day zero; 0.1 g d.w./L) + S. cerevisiae MH020215 (day six; 0.4 g d.w/L; Zb+Sc(6)), M. pulcherrima MG970690 (day zero; 0.1 g d.w./L) + S. cerevisiae MH020215 (day six; 0.4 g d.w/L; Mp+Sc(6)).
3.2. Analytical Methods
3.2.1. Determination of Fermentation Dynamics
Fermentation dynamics were determined based on the weight losses of the batches during the process. Weight losses were measured daily. The process was completed at the moment of achieving daily weight losses below 0.01 g/L.
3.2.2. Determination of Biomass Growth Yield
Biomass after centrifugation was washed with distilled water, and then dried in a moisture analyzer to a constant mass. Yeast dry mass was expressed in g/L.
3.2.3. Determination of Ethyl Alcohol Content, Real Extract, Total Acidity and Volatile Acidity
The analysis was performed in accordance with the official International Organisation of Vine and Wine methodology (OIV 2012) [32]. The alcohol concentration in finished wine was determined using the pycnometric method. For this purpose, the distillation of samples after fermentation was performed. The obtained distillate was filled up to 100 mL with distilled water, its density was determined, and the concentration of ethanol was obtained from the adequate tables.
In order to determine real extract content, distillation residues were quantitatively transferred to a 100 mL volumetric flask, filled up to 100 mL using distilled water and the procedure was analogous.
The potentiometric method was applied to determine total acidity, titrating a sample with 0.1 M NaOH solution to obtain pH = 7. Volatile acids were separated from the wine by steam distillation and titrated using standard sodium hydroxide solution.
3.2.4. Determination of Nitrogen Compounds
To determine free amino nitrogen (FAN) content, the samples were diluted in distilled water and 2 mL of diluted samples were transferred to glass tubes using a pipette. 1 mL of colour ninhydrin reagent was added, followed by boiling for 16 min in a boiling water bath. After cooling, 5 mL of the diluting agent was added into the tubes and the absorbance was measured at a wavelength of λ = 575 nm. A glycine solution containing 2 mg/L of nitrogen was used as a standard solution. The result was calculated using formulas to determine the amount of nitrogen in the sample:
3.2.5. Determination of Volatile Compounds (SPME-GC-MS)
In order to determine the headspace volatile compounds, 1 g of NaCl and a 2 mL sample of must/wine were placed into a 10 mL vial. Next, an internal standard solution was added (0.57 mg/L 4-methyl-2-pentanol, 0.2 mg/L anethol and 1.48 mg/L ethyl nonanoate, Sigma-Aldrich, St. Louis, MO, USA). The SPME device (Supelco Inc., Bellefonte, PA, USA) coated with PDMS (100 μm) fiber was first conditioned by inserting it into the gas chromatograph injector port at 250 °C for 1 h. For sampling, the fiber was inserted into the headspace under stirring (300 rpm) for 30 min at 60 °C. Subsequently, the SPME device was introduced into the injector port of a 7890B chromatography system (Agilent Technologies) equipped with a Pegasus High Throughput TOFMS (LECO,) and kept in the inlet for 3 min. The SPME process was automated using the MultiPurpose Sampler (MPS, GERSTEL, Linthicum, WA, USA). The tested components were separated on a Rtx-1ms capillary column (Crossbond 100% dimethyl polysiloxane, 30 m × 0.53 mm × 0.5 μm). The detector was 250 °C, and the column was heated using the following temperature program: 40 °C for 3 min at an increment of 8 °C/min to 230 °C, then maintaining a constant temperature for 9 min. Carrier: Helium at 1.0 mL/min constant flow. EIMS electron energy 70 eV; ion source temperature and connection parts: 250 °C. The analyte transfer was performed in splitless mode; the MSD was set to scan mode from m/z = 40 to m/z = 400. Compounds were identified using mass spectral libraries and linear retention indices, calculated based on a series of n-alkanes from C6 to C30. The qualitative and quantitative identification of volatile substances (ethyl acetate, ethyl butanoate, isoamyl acetate, ethyl hexanoate, ethyl octanoate, 2-phenylethyl acetate, ethyl decanoate, ethyl dodecanoate, ethyl tetradecanoate, ethyl hexadecanoate, ethyl octadecanoate, 2-methyl-1-propanol, 2-methyl-1-butanol, 1-hexanol, 3-methyl-1-butanol, 2-phenylethanol, 1- nonanol, 1-decanol, hexanoic acid, octanoic acid, n-decanoic acid, α-terpineol, ß-damascenone, decanal; Sigma-Aldrich, St. Louis, MO, USA) were based on a comparison of retention times and peak surface area reads from samples and standard chromatograms. Other detected components were determined semiquantitatively (μg/L) by measuring the relative peak area of each identified compound according to the National Institute of Standards and Technology (NIST) database in relation to that of the internal standard. Each of the tests were performed three times.
3.3. Statistical Analysis
The results are presented as the means of three independent repetitions with the determination of the standard deviation. Additionally, one-way analysis of variance (ANOVA) and a test of the difference between means (Duncan’s test) were also performed (StatSoft Poland, Cracow, Poland). A heat map was prepared using the statistical package SPSS 18.0 (SPSS Inc., Chicago, IL, USA).
4. Conclusions
The fermentation dynamics and analytical profiles of the obtained wines showed that the use of mixed yeast cultures contributed to the reduction of volatile acidity and acetic acid content in the wines, as well as obtaining a favorable aroma profile of the wines. The yeast Z. bailli 749 showed very favorable deacidifying properties. Sequential fermentation resulted in a greater reduction of total acidity in the obtained wines, as compared to the simultaneous fermentation. The dominant higher alcohols in all wines were 2-methyl-1-propanol, 3-methyl-1-butanol and 2-methyl-1-butanol. Significantly higher amounts of the first two compounds were found in wines obtained with M. pulcherrima MG070690, both in monoculture and in mixed cultures. The monocultures of M. pulcherrima MG070690 (Mp) compared with Z. bailli 749 (Zb) synthesized higher levels of esters in wines, including ethyl acetate, ethyl propionate, isobutyl acetate, ethyl pyroracemate and isoamyl acetate. Octanoic acid was present in wines fermented with the M. pulcherrima MG970690 monoculture. In greater amounts, this component was present in wines fermented simultaneously and sequentially (Mp+Sc, Mp+Sc (3)). A similar relationship were noted for α-terpineol and damascenone A. Mixed fermentations of non-Saccharomyces yeasts in combination with S. cerevisiae can therefore be used as a tool to modulate flavour profiles and improve oenological parameters of wines.
Author Contributions
Conceptualization, M.C.-S.; methodology, M.C.-S.; software, M.C.-S., S.S.; validation, M.C.-S., M.G. and S.S..; formal analysis, M.C.-S., M.G.; investigation, M.C.-S.; resources, M.C.-S.; data curation, M.C.-S.; writing—original draft preparation, M.C.-S., M.G.; writing—review and editing, M.C.-S., P.S., K.K., S.S.; visualization, M.C.-S., K.K., S.S.; supervision, M.C.-S., P.S.; project administration, M.C.-S.; funding acquisition, M.C.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
References
- Redzepovic, S.; Orlic, S.; Majdak, A.; Kozina, B.; Volschenk, H.; Viljoen-Bloom, M. Differential malic acid degradation by selected strains of Saccharomyces during alcoholic fermentation. Int. J. Food Microbiol. 2003, 83, 49–61. [Google Scholar] [CrossRef]
- Soyer, Y.; Koca, N.; Karadeniz, F. Organic acid profile of Turkish white grapes and grape juices. J. Food Compos. Anal. 2003, 16, 629–636. [Google Scholar] [CrossRef]
- Coloretti, F.; Zambonelli, C.; Castellari, L.; Tini, V.; Rainieri, S. The effect of DL-malic acid on the metabolism of L-malic acid during wine alcoholic fermentation. Food Technol. Biotechnol. 2002, 40, 317–320. [Google Scholar]
- Jackson, R.S. Wine Science: Principles and Applications; Academic Press: Cambridge, MA, USA, 2008; ISBN 0080568742. [Google Scholar]
- Kunicka-Styczynska, A. Glucose, L-Malic acid and pH effect on fermentation products in biological deacidification. Czech. J. Food Sci. 2009, 27, 319–322. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine Microbiology and Biotechnology; CRC Press: Boca Raton, FL, USA, 1993; ISBN 0415278503. [Google Scholar]
- Volschenk, H.; Van Vuuren, H.J.J.; Viljoen-Bloom, M. Malic acid in wine: Origin, function and metabolism during vinification. S. Afr. J. Enol. Vitic. 2006, 27, 123–136. [Google Scholar] [CrossRef]
- Zoecklein, B.; Fugelsang, K.C.; Gump, B.H.; Nury, F.S. Wine Analysis and Production; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; ISBN 1475769679. [Google Scholar]
- le Roux, S.I. The microbiology of South African wine making. Part VIII. The microflora of healthy and Botrytis cinerea infected grapes. Phytophylactica 1973, 5, 51–54. [Google Scholar]
- Rankine, B.C. Influence of yeast strain and malo-lactic fermentation on composition and quality of table wines. Am. J. Enol. Vitic. 1972, 23, 152–158. [Google Scholar]
- Balmaseda, A.; Bordons, A.; Reguant, C.; Bautista-Gallego, J. Non-Saccharomyces in wine: Effect upon Oenococcus oeni and malolactic fermentation. Front. Microbiol. 2018, 9, 534. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M. Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Ruiz, J.; Belda, I.; Beisert, B.; Navascués, E.; Marquina, D.; Calderón, F.; Rauhut, D.; Santos, A.; Benito, S. Analytical impact of Metschnikowia pulcherrima in the volatile profile of Verdejo white wines. Appl. Microbiol. Biotechnol. 2018, 102, 8501–8509. [Google Scholar] [CrossRef] [PubMed]
- Sadoudi, M.; Tourdot-Maréchal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacón, J.J.; Ballester, J.; Vichi, S.; Guérin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Conchillo, L.B.; Ruiz, J.; Navascués, E.; Marquina, D.; Santos, A. Selection and use of pectinolytic yeasts for improving clarification and phenolic extraction in winemaking. Int. J. Food Microbiol. 2016, 223, 1–8. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of Non-Saccharomyces Yeasts for the Reduction of Alcohol Content in Wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Ortega, N.; Martín-Santamaría, M.; Acedo, A.; Marquina, D.; Pascual, O.; Rozès, N.; Zamora, F.; Santos, A.; Belda, I. Occurrence and enological properties of two new non-conventional yeasts (Nakazawaea ishiwadae and Lodderomyces elongisporus) in wine fermentations. Int. J. Food Microbiol. 2019, 305, 1–9. [Google Scholar] [CrossRef]
- Vilela-Moura, A.; Schuller, D.; Mendes-Faia, A.; Côrte-Real, M. Reduction of volatile acidity of wines by selected yeast strains. Appl. Microbiol. Biotechnol. 2008, 80, 881. [Google Scholar] [CrossRef]
- Thornton, R.J.; Rodriguez, S.B. Deacidification of red and white wines by a mutant of Schizosaccharomyces malidevorans under commercial winemaking conditions. Food Microbiol. 1996, 13, 475–482. [Google Scholar] [CrossRef]
- Tang, K.; Li, Q. Biochemistry of wine and beer fermentation. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2017; pp. 281–304. [Google Scholar]
- Fernandes, L.; Côrte-Real, M.; Loureiro, V.; Loureiro-Dias, M.C.; Leao, C. Glucose respiration and fermentation in Zygosaccharomyces bailii and Saccharomyces cerevisiae express different sensitivity patterns to ethanol and acetic acid. Lett. Appl. Microbiol. 1997, 25, 249–253. [Google Scholar] [CrossRef]
- Thomas, D.S.; Davenport, R.R. Zygosaccharomyces bailii—A profile of characteristics and spoilage activities. Food Microbiol. 1985, 2, 157–169. [Google Scholar] [CrossRef]
- Pawlikowska, E.; Gregiel, D. Niekonwencjonalne drożdże Metschnikowia pulcherrima i ich zastosowanie w biotechnologii. Postępy Mikrobiol. 2017, 56, 4. [Google Scholar]
- Pitt, J.I.; Miller, M.W. The parasexual cycle in yeasts of the genus Metschnikowia. Mycologia 1970, 62, 462–473. [Google Scholar] [CrossRef]
- Fugelsang, K.; Edwards, C. Wine Microbiology; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Spayd, S.E.; Andersen-Bagge, J. Free amino acid composition of grape juice from 12 Vitis vinifera cultivars in Washington. Am. J. Enol. Vitic. 1996, 47, 389–402. [Google Scholar]
- Xu, Y.; Zhi, Y.; Wu, Q.; Du, R. Zygosaccharomyces bailii is a potential producer of various flavor compounds in Chinese Maotai-flavor liquor fermentation. Front. Microbiol. 2017, 8, 2609. [Google Scholar] [CrossRef]
- Sadoudi, M.; Rousseaux, S.; David, V.; Alexandre, H.; Tourdot-Maréchal, R. Metschnikowia pulcherrima influences the expression of genes involved in PDH bypass and glyceropyruvic fermentation in Saccharomyces cerevisiae. Front. Microbiol. 2017, 8, 1137. [Google Scholar] [CrossRef] [PubMed]
- OIV. Compendium of International Methods of Wine and Must Analysis. In Organisation Internationale de la Vigne et du Vin; OIV: Paris, France, 2012. [Google Scholar]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Handbook of Enology, Volume 1: The Microbiology of Wine and Vinifications; John Wiley & Sons: New York, NY, USA, 2006; Volume 1, ISBN 0470010355. [Google Scholar]
- Volschenk, H.; Van Vuuren, H.J.J.; Viljoen–Bloom, M. Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr. Genet. 2003, 43, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Du Toit, M.; Pretorius, I.S. Microbial spoilage and preservation of wine: Using weapons for nature’s own arsenal. S. Afr. J. Enol. Vitic. 2000, 21, 74–96. [Google Scholar] [CrossRef]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Mendoza, L.M.; de Nadra, M.C.M.; Farías, M.E. Kinetics and metabolic behavior of a composite culture of Kloeckera apiculata and Saccharomyces cerevisiae wine related strains. Biotechnol. Lett. 2007, 29, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Zott, K.; Thibon, C.; Bely, M.; Lonvaud-Funel, A.; Dubourdieu, D.; Masneuf-Pomarede, I. The grape must non-Saccharomyces microbial community: Impact on volatile thiol release. Int. J. Food Microbiol. 2011, 151, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Harding, J. The Oxford Companion to Wine; American Chemical Society: Oxford, UK, 2015; ISBN 0198705387. [Google Scholar]
- Dequin, S.; Baptista, E.; Barre, P. Acidification of grape musts by Saccharomyces cerevisiae wine yeast strains genetically engineered to produce lactic acid. Am. J. Enol. Vitic. 1999, 50, 45–50. [Google Scholar]
- Vonach, R.; Lendl, B.; Kellner, R. High-performance liquid chromatography with real-time Fourier-transform infrared detection for the determination of carbohydrates, alcohols and organic acids in wines. J. Chromatogr. A 1998, 824, 159–167. [Google Scholar] [CrossRef]
- Henick-Kling, T.; Edinger, W.; Daniel, P.; Monk, P. Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J. Appl. Microbiol. 1998, 84, 865–876. [Google Scholar] [CrossRef]
- Dobrowolska-Iwanek, J.; Gąstol, M.; Wanat, A.; Krośniak, M.; Jancik, M.; Zagrodzki, P. Wine of cool-climate areas in South Poland. S. Afr. J. Enol. Vitic. 2014, 35, 1–9. [Google Scholar] [CrossRef]
- Muñoz-Robredo, P.; Robledo, P.; Manríquez, D.; Molina, R.; Defilippi, B.G. Characterization of sugars and organic acids in commercial varieties of table grapes. Chil. J. Agric. Res. 2011, 71, 452. [Google Scholar] [CrossRef]
- Kordis-Krapez, M.; Abram, V.; Kac, M.; Ferjancic, S. Determination of organic acids in white wines by RP-HPLC. Food Technol. Biotechnol. 2001, 39, 93–100. [Google Scholar]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C. Wine Chemistry and Biochemistry, 1st ed.; Springer: New York, NY, USA, 2009; Volume 735, ISBN 978-0-387-74118-5. [Google Scholar]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Mendes Ferreira, A.; Mendes-Faia, A. The role of yeasts and lactic acid bacteria on the metabolism of organic acids during winemaking. Foods 2020, 9, 1231. [Google Scholar] [CrossRef] [PubMed]
- Morata, A.; Escott, C.; Bañuelos, M.A.; Loira, I.; Del Fresno, J.M.; González, C.; Suárez-Lepe, J.A. Contribution of non-Saccharomyces yeasts to wine freshness. A review. Biomolecules 2020, 10, 34. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Schmidt, S.; Henschke, P.A.; Curtin, C.; Varela, C. The application of non-Saccharomyces yeast in fermentations with limited aeration as a strategy for the production of wine with reduced alcohol content. Int. J. Food Microbiol. 2015, 205, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Hutkins, R.W. Microbiology and Technology of Fermented Foods; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 22, ISBN 047027624X. [Google Scholar]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii–Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Dolci, P.; Giacosa, S.; Torchio, F.; Tofalo, R.; Torriani, S.; Suzzi, G.; Rolle, L.; Cocolin, L. Candida zemplinina can reduce acetic acid produced by Saccharomyces cerevisiae in sweet wine fermentations. Appl. Environ. Microbiol. 2012, 78, 1987–1994. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y. The influence of yeast strains on the volatile flavour compounds of Chinese rice wine. J. Inst. Brew. 2010, 116, 190–196. [Google Scholar] [CrossRef]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and its importance to wine aroma-a review. S. Afr. J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef]
- Jackson, D.I.; Lombard, P.B. Environmental and management practices affecting grape composition and wine quality-a review. Am. J. Enol. Vitic. 1993, 44, 409–430. [Google Scholar]
- Romano, P.; Suzzi, G. Higher alcohol and acetoin production by Zygosaccharomyces wine yeasts. J. Appl. Bacteriol. 1993, 75, 541–545. [Google Scholar] [CrossRef]
- Zironi, R.; Romano, P.; Suzzi, G.; Battistutta, F.; Comi, G. Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermondii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol. Lett. 1993, 15, 235–238. [Google Scholar] [CrossRef]
- Tominac, V.P.; Ganić, K.K.; KomeS, D.; Gracin, L.; Banović, M.; Marić, V. Influence of media composition and temperature on volatile aroma production by various wine yeast strains. Czech J. Food Sci. 2008, 26, 376–382. [Google Scholar] [CrossRef]
- Escribano, R.; González-Arenzana, L.; Portu, J.; Garijo, P.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R. Wine aromatic compound production and fermentative behaviour within different non-Saccharomyces species and clones. J. Appl. Microbiol. 2018, 124, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Ivit, N.N.; Kemp, B. The impact of non-Saccharomyces yeast on traditional method sparkling Wine. Fermentation 2018, 4, 73. [Google Scholar] [CrossRef]
- Sumby, K.M.; Grbin, P.R.; Jiranek, V. Microbial modulation of aromatic esters in wine: Current knowledge and future prospects. Food Chem. 2010, 121, 1–16. [Google Scholar] [CrossRef]
- Cioch-Skoneczny, M.; Satora, P.; Skoneczny, S.; Klimczak, K. Physicochemical characterization of wines produced using indigenous yeasts from cold climate grapes. Eur. Food Res. Technol. 2021, 201–209. [Google Scholar] [CrossRef]
- Clemente-Jimenez, J.M.; Mingorance-Cazorla, L.; Martínez-Rodríguez, S.; Las Heras-Vázquez, F.J.; Rodríguez-Vico, F. Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol. 2004, 21, 149–155. [Google Scholar] [CrossRef]
- Elias, R.J.; Laurie, V.F.; Ebeler, S.E.; Wong, J.W.; Waterhouse, A.L. Analysis of selected carbonyl oxidation products in wine by liquid chromatography with diode array detection. Anal. Chim. Acta 2008, 626, 104–110. [Google Scholar] [CrossRef]
- Li, E.; Mira de Orduña, R. Acetaldehyde kinetics of enological yeast during alcoholic fermentation in grape must. J. Ind. Microbiol. Biotechnol. 2017, 44, 229–236. [Google Scholar] [CrossRef]
- Mateos, J.A.R.; Pérez-Nevado, F.; Fernández, M.R. Influence of Saccharomyces cerevisiae yeast strain on the major volatile compounds of wine. Enzyme Microb. Technol. 2006, 40, 151–157. [Google Scholar] [CrossRef]
- Morata, A.; Loira, I.; Escott, C.; del Fresno, J.M.; Bañuelos, M.A.; Suárez-Lepe, J.A. Applications of Metschnikowia pulcherrima in wine biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef]
- Cioch-Skoneczny, M.; Satora, P.; Skoneczny, S.; Skotniczny, M. Yeasts associated with the spontaneously fermented grape musts obtained from cool climate white grape varieties. J. Food Nutr. Res. 2019, 58, 295–306. [Google Scholar]
- Cioch-Skoneczny, M.; Satora, P.; Skotniczny, M.; Skoneczny, S. Quantitative and qualitative composition of yeast microbiota in spontaneously fermented grape musts obtained from cool climate grape varieties Rondo and Regent. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Cioch-Skoneczny, M.; Satora, P.; Skoneczny, S.; Skotniczny, M. Biodiversity of yeasts isolated during spontaneous fermentation of cool climate grape musts. Arch. Microbiol. 2021, 203, 153–162. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).