Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics
Abstract
1. Introduction
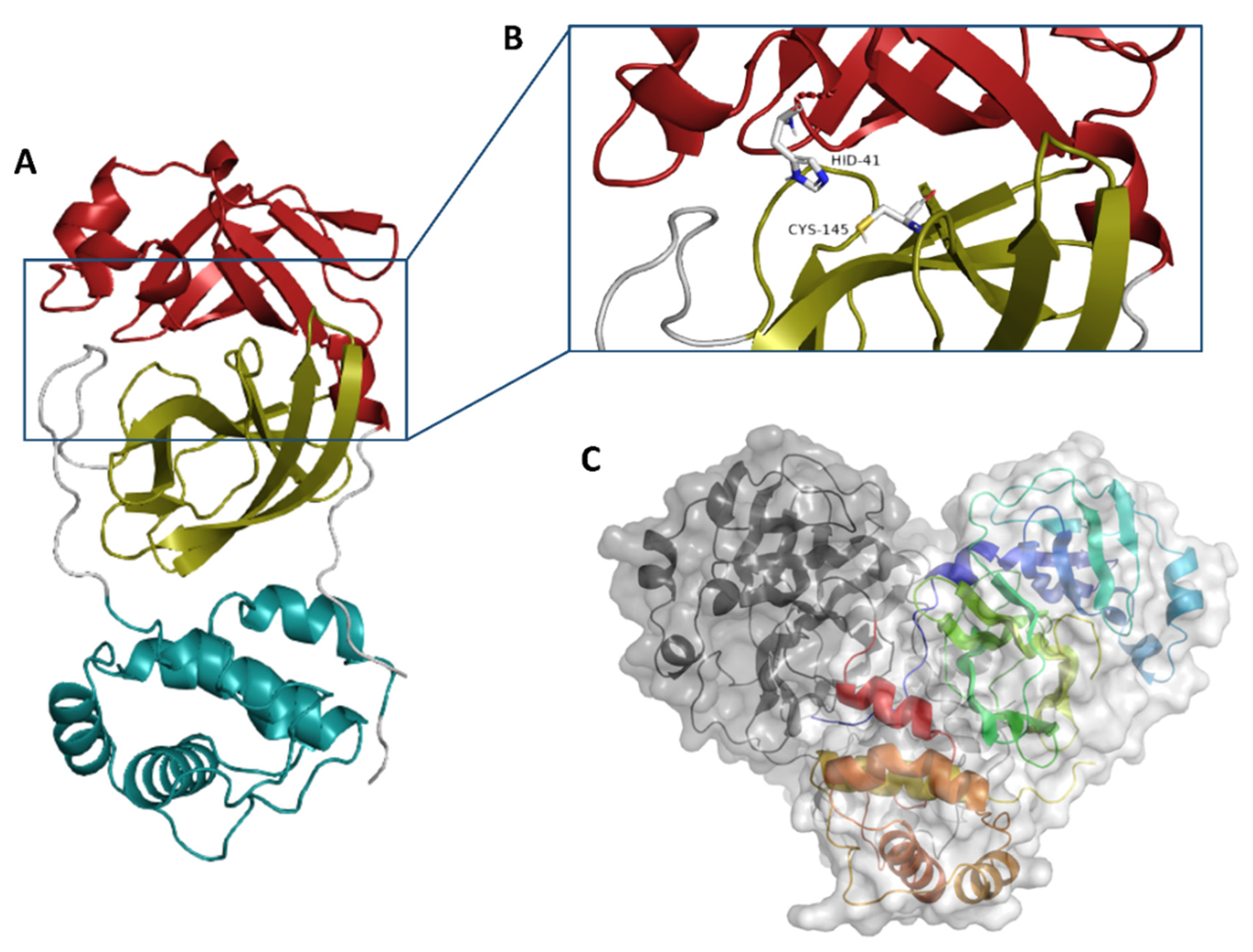
2. Results and Discussion
3. Materials and Methods
3.1. Isolation of Compounds
3.2. Ensemble Docking
3.3. Molecular Dynamic Simulation
3.4. In Vitro Enzyme Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Sayed, A.M.; Khattab, A.R.; AboulMagd, A.M.; Hassan, H.M.; Rateb, M.E.; Zaid, H.; Abdelmohsen, U.R. Nature as a treasure trove of potential anti-SARS-CoV drug leads: A structural/mechanistic rationale. RSC Adv. 2020, 10, 19790–19802. [Google Scholar] [CrossRef]
- Adamson, C.S.; Chibale, K.; Goss, R.J.; Jaspars, M.; Newman, D.J.; Dorrington, R.A. Antiviral drug discovery: Preparing for the next pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, B.; Jiang, X.M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A. Protein structural heterogeneity: A hypothesis for the basis of proteolytic recognition by the main protease of SARS-CoV and SARS-CoV-2. Biochimie 2021, 182, 177–184. [Google Scholar] [CrossRef]
- Pavlova, A.; Lynch, D.L.; Daidone, I.; Zanetti-Polzi, L.; Smith, M.D.; Chipot, C.; Gumbart, J.C. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021, 12, 1513–1527. [Google Scholar] [CrossRef]
- Sayed, A.M.; Alhadrami, H.A.; El-Gendy, A.O.; Shamikh, Y.I.; Belbahri, L.; Hassan, H.M.; Rateb, M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro). Microorganisms 2020, 8, 970. [Google Scholar] [CrossRef]
- Chang, H.P.; Chou, C.Y.; Chang, G.G. Reversible unfolding of the severe acute respiratory syndrome coronavirus main protease in guanidinium chloride. Biophys. J. 2007, 92, 1374–1383. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Zhou, Q.A.; Kato-Weinstein, J.; Li, Y.; Deng, Y.; Granet, R.; Garner, L.; Liu, C.; Polshakov, D.; Gessner, C.; Watkins, S. Potential therapeutic agents and associated bioassay data for COVID-19 and related human coronavirus infections. ACS Pharmacol. Transl. Sci. 2020, 3, 813–834. [Google Scholar] [CrossRef]
- Wen, C.C.; Kuo, Y.H.; Jan, J.T.; Liang, P.H.; Wang, S.Y.; Liu, H.G.; Lee, C.K.; Chang, S.T.; Kuo, C.J.; Lee, S.S.; et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J. Med. Chem. 2007, 50, 4087–4095. [Google Scholar] [CrossRef] [PubMed]
- Shady, H.N.; Youssif, K.A.; Sayed, A.M.; Belbahri, L.; Oszako, T.; Hassan, H.M.; Abdelmohsen, U.R. Sterols and triterpenes: Antiviral potential supported by in-silico analysis. Plants 2021, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Su, H.X.; Yao, S.; Zhao, W.F.; Li, M.J.; Liu, J.; Shang, W.J.; Xie, H.; Ke, C.-Q.; Hu, H.-C.; Gao, M.-N.; et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020, 41, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velazquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; El-Halawany, A.M.; Sirwi, A.; El-Araby, A.M.; Mohamed, G.A.; Ibrahim, S.R.; Elfaky, M.A. Repurposing of some natural product isolates as SARS-COV-2 main protease inhibitors via in vitro cell free and cell-based antiviral assessments and molecular modeling approaches. Pharmaceuticals 2021, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.I.; Higashi-Kuwata, N.; Hayashi, H.; Allu, S.R.; Raghavaiah, J.; Bulut, H.; Debananda, D.; Anson, B.J.; Lendy, E.K.; Takamatsu, Y. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef]
- Guinda, A.; Rada, M.; Delgado, T.; Gutierrez-Adanez, P.; Castellano, J.M. Pentacyclic triterpenoids from olive fruit and leaf. J. Agric. Food Chem. 2010, 58, 9685–9691. [Google Scholar] [CrossRef]
- Kong, L.; Li, S.; Liao, Q.; Zhang, Y.; Sun, R.; Zhu, X.; Zhang, Q.; Wang, J.; Wu, X.; Fang, X.; et al. Oleanolic acid and ursolic acid: Novel hepatitis C virus antivirals that inhibit NS5B activity. Antivir. Res. 2013, 98, 44–53. [Google Scholar] [CrossRef]
- Tohmé, M.J.; Giménez, M.C.; Peralta, A.; Colombo, M.I.; Delgui, L.R. Ursolic acid: A novel antiviral compound inhibiting rotavirus infection in vitro. Int. J. Antimicrob. Agents 2019, 54, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.; Rivas, F.; Lopez, P.E.; Garcia-Granados, A.; Martinez, A.; Albericio, F.; Muñoz, E. Solution-and solid-phase synthesis and anti-HIV activity of maslinic acid derivatives containing amino acids and peptides. Bioorganic Med. Chem. 2009, 17, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Parra, A.; Martin-Fonseca, S.; Rivas, F.; Reyes-Zurita, F.J.; Medina-O’Donnell, M.; Martinez, A.; Lupiañez, J.A.; Albericio, F. Semi-synthesis of acylated triterpenes from olive-oil industry wastes for the development of anticancer and anti-HIV agents. Eur. J. Med. Chem. 2014, 74, 278–301. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Salmon, J.K. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Desmond Molecular Dynamics System. Available online: https://www.schrodinger.com/products/desmond (accessed on 29 April 2021).
- Schrodinger LLC Maestro, version 9.0; Schrodinger: New York, NY, USA, 2009.
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhadrami, H.A.; Sayed, A.M.; Sharif, A.M.; Azhar, E.I.; Rateb, M.E. Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics. Molecules 2021, 26, 2654. https://doi.org/10.3390/molecules26092654
Alhadrami HA, Sayed AM, Sharif AM, Azhar EI, Rateb ME. Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics. Molecules. 2021; 26(9):2654. https://doi.org/10.3390/molecules26092654
Chicago/Turabian StyleAlhadrami, Hani A., Ahmed M. Sayed, Ahmed M. Sharif, Esam I. Azhar, and Mostafa E. Rateb. 2021. "Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics" Molecules 26, no. 9: 2654. https://doi.org/10.3390/molecules26092654
APA StyleAlhadrami, H. A., Sayed, A. M., Sharif, A. M., Azhar, E. I., & Rateb, M. E. (2021). Olive-Derived Triterpenes Suppress SARS COV-2 Main Protease: A Promising Scaffold for Future Therapeutics. Molecules, 26(9), 2654. https://doi.org/10.3390/molecules26092654








