Identification of Molecules from Coffee Silverskin That Suppresses Myostatin Activity and Improves Muscle Mass and Strength in Mice
Abstract
1. Introduction
2. Results
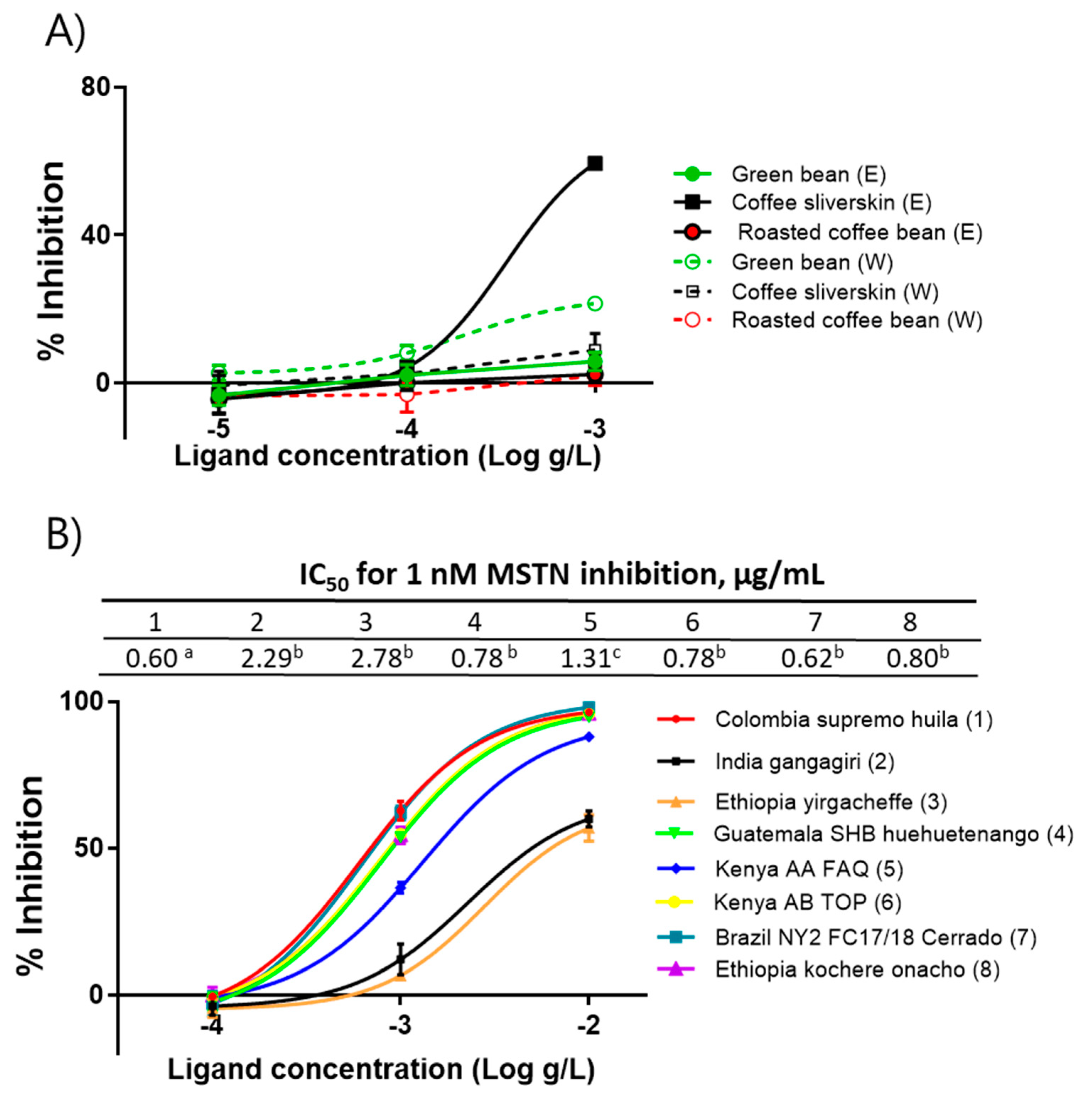
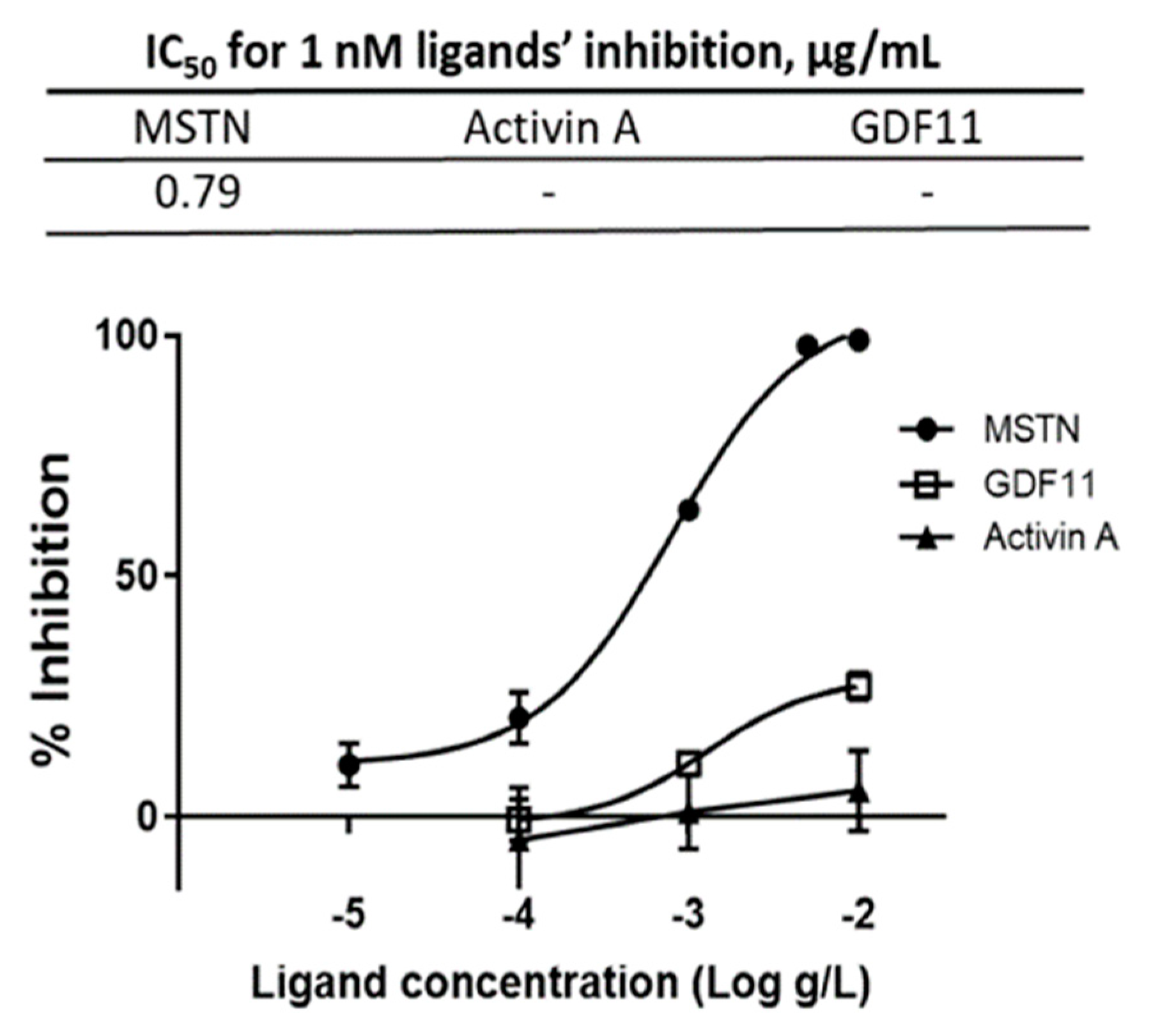
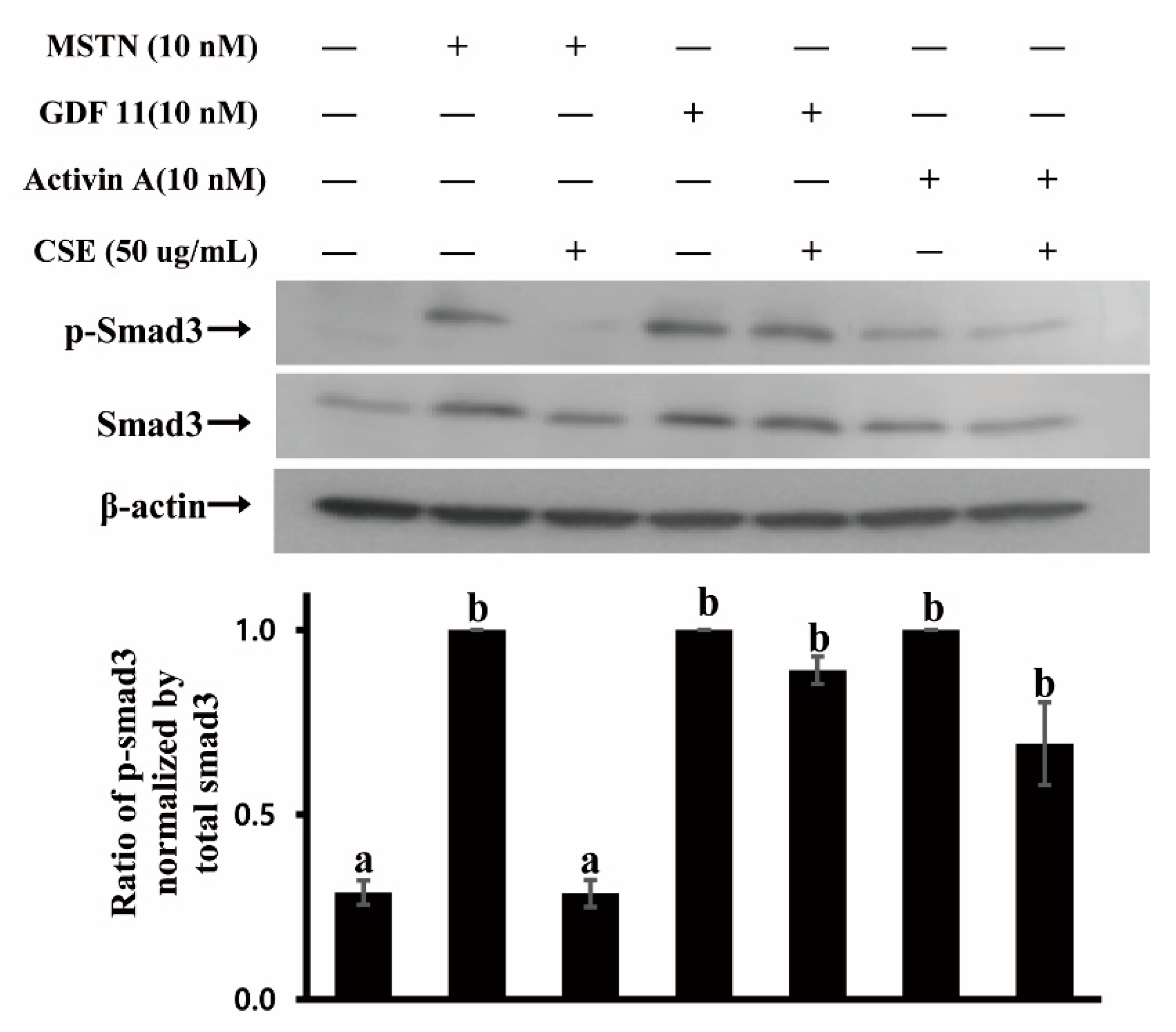
2.1. Coffee Silverskin Ethanol-Extract (CSE) Suppresses MSTN Activity
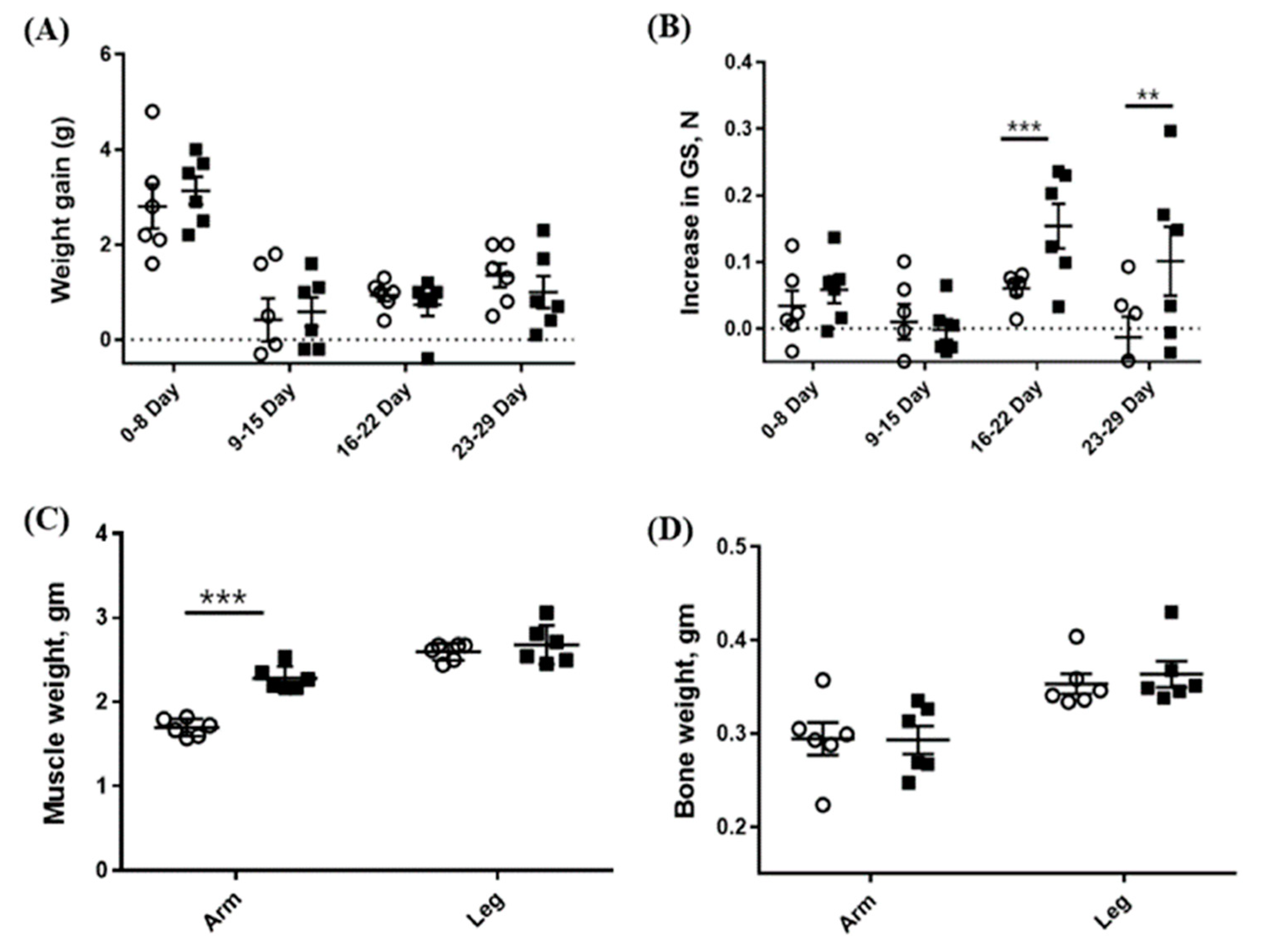
2.2. Oral Administration of CSE Increases the Muscle Mass and Grip Strength but Not Body Weight and Bone Mass in Mice
2.3. CSE Administration Decreases the Blood Free Fatty Acid Level
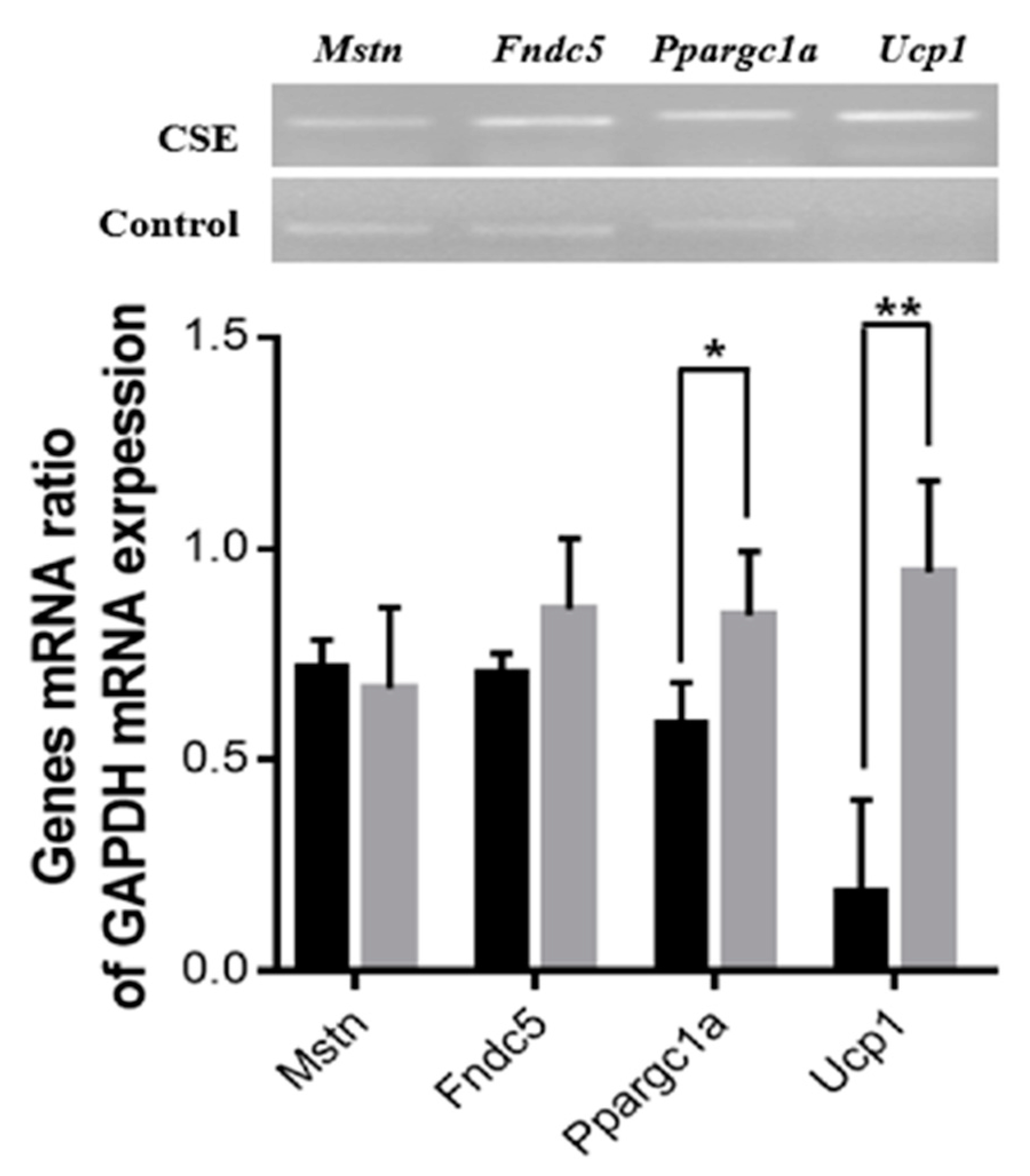
2.4. Administration of CSE Upregulated the mRNA Expression Level of Ppargc1a and Ucp1 in Mice
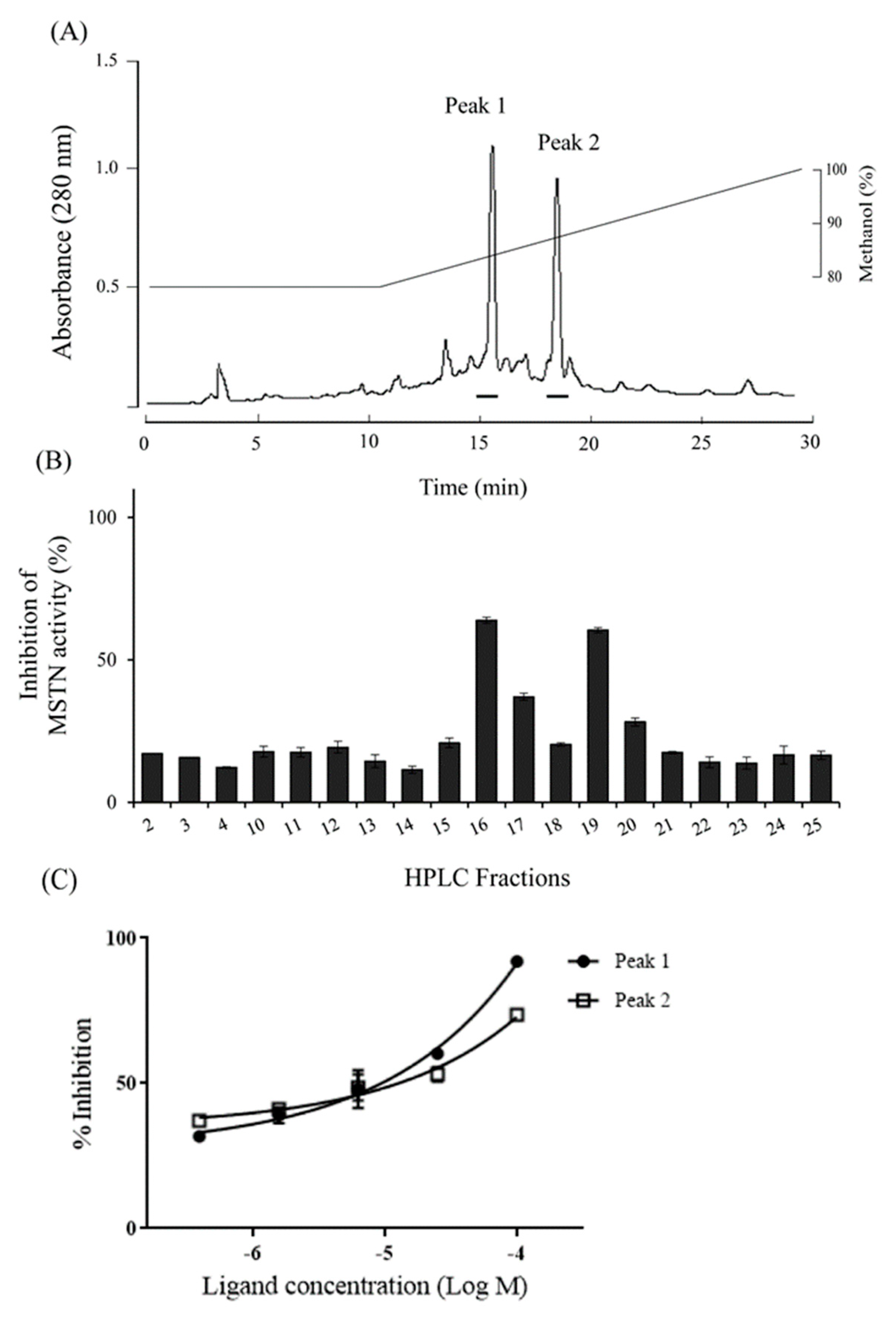
2.5. Purification and Structural Identification of Compounds with MSTN-Inhibitory Capacity
3. Discussion
4. Materials and Methods
4.1. Coffee Beans
4.2. Preparation of Ethanol and Water Extracts of Samples
4.3. pGL3-(CAGA)12-Luciferase Assay
4.4. Western Blot Analysis
4.5. Animal Experiment
4.5.1. Animals
4.5.2. Forelimb Grip Strength Measurement
4.5.3. Analysis of Blood Glucose, Cholesterol, Triglyceride, and Free Fatty Acids
4.5.4. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.5.5. Statistical Analysis
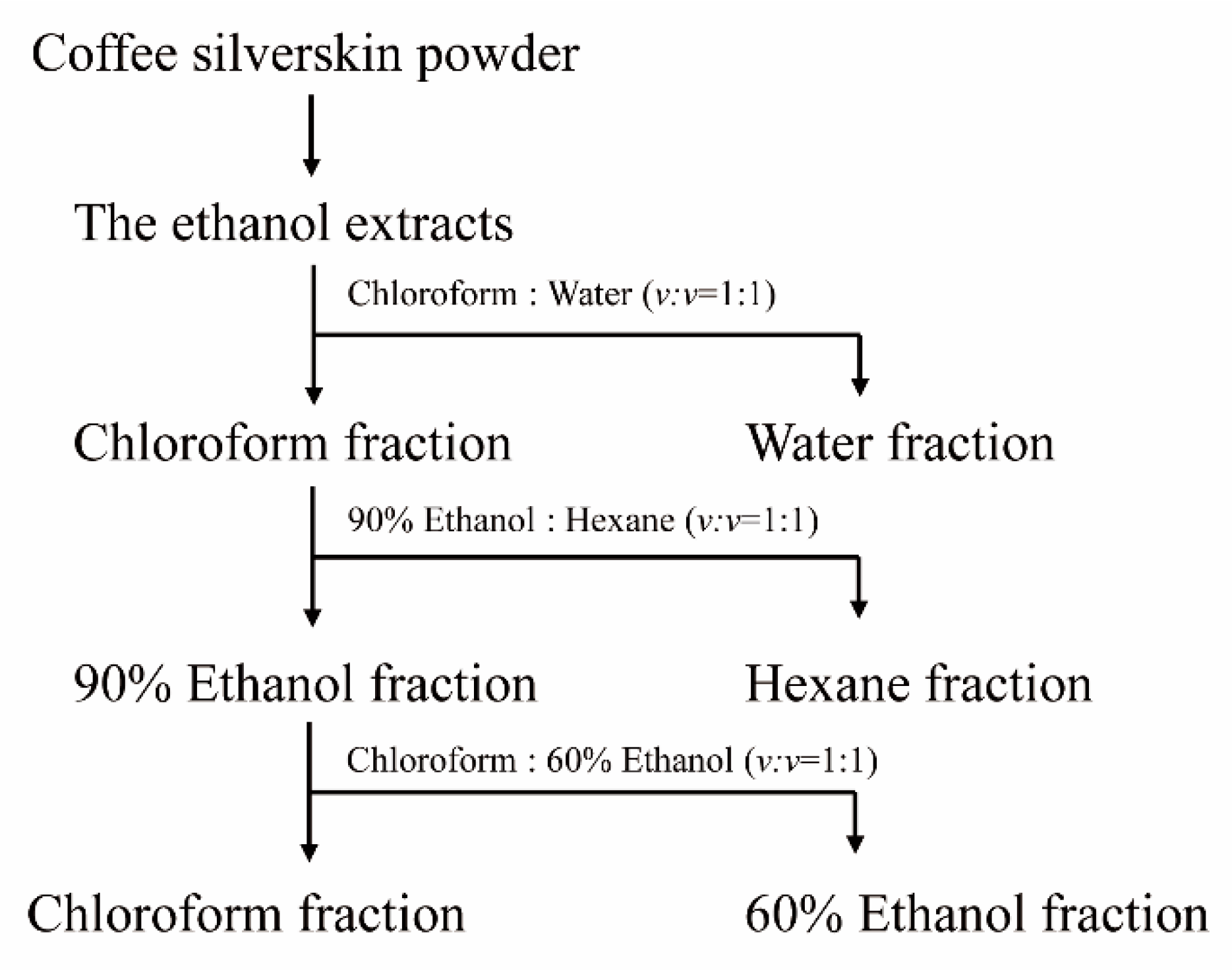
4.6. Isolation of Anti-MSTN Inhibitors from CSE
4.7. Identification of MSTN Antagonists
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nat. Cell Biol. 1997, 387, 83–90. [Google Scholar] [CrossRef]
- McFarland, D.C.; Velleman, S.G.; Pesall, J.E.; Liu, C. The role of myostatin in chicken (Gallus domesticus) myogenic satellite cell proliferation and differentiation. Gen. Comp. Endocrinol. 2007, 151, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Ríos, R.; Carneiro, I.; Arce, V.M.; Devesa, J. Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol. Physiol. 2002, 282, C993–C999. [Google Scholar] [CrossRef] [PubMed]
- Siriett, V.; Salerno, M.S.; Berry, C.; Nicholas, G.; Bower, R.; Kambadur, R.; Sharma, M. Antagonism of Myostatin Enhances Muscle Regeneration During Sarcopenia. Mol. Ther. 2007, 15, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, L.-A.; Song, K.; Li, X.; Aghajanian, J.; Davies, M.; Girgenrath, S.; Hill, J.J.; Jalenak, M.; Kelley, P.; Knight, A.; et al. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem. Biophys. Res. Commun. 2003, 300, 965–971. [Google Scholar] [CrossRef]
- Han, H.; Mitch, W.E. Targeting the myostatin signaling pathway to treat muscle wasting diseases. Curr. Opin. Support. Palliat. Care 2011, 5, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, H.; Enomoto, H.; Ishii, A.; Iwata, Y.; Miyamoto, Y.; Ishii, N.; Yuri, Y.; Hasegawa, K.; Nakano, C.; Nishimura, T.; et al. Elevated serum myostatin level is associated with worse survival in patients with liver cirrhosis. J. Cachex Sarcopenia Muscle 2017, 8, 915–925. [Google Scholar] [CrossRef]
- Plant, P.J.; Brooks, D.; Faughnan, M.; Bayley, T.; Bain, J.; Singer, L.; Correa, J.; Pearce, D.; Binnie, M.; Batt, J. Cellular Markers of Muscle Atrophy in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2010, 42, 461–471. [Google Scholar] [CrossRef]
- Shyu, K.G.; Lü, M.J.; Wang, B.W.; Sun, H.Y.; Chang, H. Myostatin expression in ventricular myocardium in a rat model of volume-overload heart failure. Eur. J. Clin. Investig. 2006, 36, 713–719. [Google Scholar] [CrossRef]
- Chen, P.R.; Lee, K. INVITED REVIEW: Inhibitors of myostatin as methods of enhancing muscle growth and development1. J. Anim. Sci. 2016, 94, 3125–3134. [Google Scholar] [CrossRef]
- Lebrasseur, N.K.; Schelhorn, T.M.; Bernardo, B.L.; Cosgrove, P.G.; Loria, P.M.; Brown, T.A. Myostatin Inhibition Enhances the Effects of Exercise on Performance and Metabolic Outcomes in Aged Mice. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2009, 64, 940–948. [Google Scholar] [CrossRef]
- Murphy, K.T.; Ryall, J.G.; Snell, S.M.; Nair, L.; Koopman, R.; Krasney, P.A.; Ibebunjo, C.; Holden, K.S.; Loria, P.M.; Salatto, C.T.; et al. Antibody-Directed Myostatin Inhibition Improves Diaphragm Pathology in Young but not Adult Dystrophic mdx Mice. Am. J. Pathol. 2010, 176, 2425–2434. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Lord, S.R.; A Studenski, S.; Warden, S.J.; A Fielding, R.; Recknor, C.P.; Hochberg, M.C.; Ferrari, S.L.; Blain, H.; Binder, E.F.; et al. Myostatin antibody (LY2495655) in older weak fallers: A proof-of-concept, randomised, phase 2 trial. Lancet Diabetes Endocrinol. 2015, 3, 948–957. [Google Scholar] [CrossRef]
- Lach-Trifilieff, E.; Minetti, G.C.; Sheppard, K.; Ibebunjo, C.; Feige, J.N.; Hartmann, S.; Brachat, S.; Rivet, H.; Koelbing, C.; Morvan, F.; et al. An Antibody Blocking Activin Type II Receptors Induces Strong Skeletal Muscle Hypertrophy and Protects from Atrophy. Mol. Cell. Biol. 2014, 34, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Morvan, F.; Rondeau, J.-M.; Zou, C.; Minetti, G.; Scheufler, C.; Scharenberg, M.; Jacobi, C.; Brebbia, P.; Ritter, V.; Toussaint, G.; et al. Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy. Proc. Natl. Acad. Sci.USA 2017, 114, 12448–12453. [Google Scholar] [CrossRef] [PubMed]
- Rooks, D.; Praestgaard, J.; Hariry, S.; Laurent, D.; Petricoul, O.; Perry, R.G.; Lach-Trifilieff, E.; Roubenoff, R. Treatment of Sarcopenia with Bimagrumab: Results from a Phase II, Randomized, Controlled, Proof-of-Concept Study. J. Am. Geriatr. Soc. 2017, 65, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Dong, Y.; Chen, F.; E Mitch, W.; Zhang, L. Inhibition of myostatin in mice improves insulin sensitivity via irisin-mediated cross talk between muscle and adipose tissues. Int. J. Obes. 2016, 40, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Bond, N.D.; Jou, W.; Gavrilova, O.; Portas, J.; McPherron, A.C. Myostatin Inhibition Prevents Diabetes and Hyperphagia in a Mouse Model of Lipodystrophy. Diabetes 2012, 61, 2414–2423. [Google Scholar] [CrossRef]
- Guo, T.; Jou, W.; Chanturiya, T.; Portas, J.; Gavrilova, O.; McPherron, A.C. Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS ONE 2009, 4, e4937. [Google Scholar] [CrossRef]
- Cai, C.; Qian, L.; Jiang, S.; Sun, Y.; Wang, Q.; Ma, D.; Xiao, G.; Li, B.; Xie, S.; Gao, T.; et al. Loss-of-function myostatin mutation increases insulin sensitivity and browning of white fat in Meishan pigs. Oncotarget 2017, 8, 34911–34922. [Google Scholar] [CrossRef]
- Shen, S.; Yu, H.; Gan, L.; Ye, Y.; Lin, L. Natural constituents from food sources: Potential therapeutic agents against muscle wasting. Food Funct. 2019, 10, 6967–6986. [Google Scholar] [CrossRef]
- Otsuka, Y.; Egawa, K.; Kanzaki, N.; Izumo, T.; Rogi, T.; Shibata, H. Quercetin glycosides prevent dexamethasone-induced muscle atrophy in mice. Biochem. Biophys. Rep. 2019, 18, 100618. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Niu, K.; Okazaki, T.; Wu, H.; Yoshikawa, T.; Ohrui, T.; Furukawa, K.; Ichinose, M.; Yanai, K.; Arai, H.; et al. Coffee treatment prevents the progression of sarcopenia in aged mice in vivo and in vitro. Exp. Gerontol. 2014, 50, 1–8. [Google Scholar] [CrossRef]
- Lee, S.-J.; McPherron, A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA 2001, 98, 9306–9311. [Google Scholar] [CrossRef]
- Rebbapragada, A.; Benchabane, H.; Wrana, J.L.; Celeste, A.J.; Attisano, L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 2003, 23, 7230–7242. [Google Scholar] [CrossRef] [PubMed]
- Bogdanovich, S.; Krag, T.O.B.; Barton, E.R.; Morris, L.D.; Whittemore, L.-A.; Ahima, R.S.; Khurana, T.S. Functional improvement of dystrophic muscle by myostatin blockade. Nat. Cell Biol. 2002, 420, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Langley, B.; Thomas, M.; Bishop, A.; Sharma, M.; Gilmour, S.; Kambadur, R. Myostatin Inhibits Myoblast Differentiation by Down-regulating MyoD Expression. J. Biol. Chem. 2002, 277, 49831–49840. [Google Scholar] [CrossRef]
- Zhu, X.; Topouzis, S.; Liang, L.-F.; Stotish, R.L. Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine 2004, 26, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.P.; Yeo, C.-Y.; Lee, Y.; Schrewe, H.; Whitman, M.; Li, E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002, 16, 2749–2754. [Google Scholar] [CrossRef]
- A Pangas, S.; Woodruff, T.K. Activin Signal Transduction Pathways. Trends Endocrinol. Metab. 2000, 11, 309–314. [Google Scholar] [CrossRef]
- Lee, S.-J. Extracellular Regulation of Myostatin: A Molecular Rheostat for Muscle Mass. Immunol. Endocr. Metab. Agents Med. Chem. 2010, 10, 183–194. [Google Scholar] [CrossRef]
- Kim, J.H.; Sutikno, L.A.; Lee, S.B.; Jin, D.-H.; Hong, Y.-K.; Kim, Y.S.; Jin, H.-J.; Kim, J.H. Identification of the minimum region of flatfish myostatin propeptide (Pep45-65) for myostatin inhibition and its potential to enhance muscle growth and performance in animals. PLoS ONE 2019, 14, e0215298. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Pan, J.S.; Zhang, L. Myostatin Suppression of Akirin1 Mediates Glucocorticoid-Induced Satellite Cell Dysfunction. PLoS ONE 2013, 8, e58554. [Google Scholar] [CrossRef] [PubMed]
- Calvo, J.A.; Daniels, T.G.; Wang, X.; Paul, A.; Lin, J.; Spiegelman, B.M.; Stevenson, S.C.; Rangwala, S.M. Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J. Appl. Physiol. 2008, 104, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Hatazawa, Y.; Senoo, N.; Tadaishi, M.; Ogawa, Y.; Ezaki, O.; Kamei, Y.; Miura, S. Metabolomic Analysis of the Skeletal Muscle of Mice Overexpressing PGC-1α. PLoS ONE 2015, 10, e0129084. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-Y.; Zheng, D.; Houmard, J.A.; Brault, J.J.; Hickner, R.C.; Cortright, R.N. Overexpression of PGC-1α increases peroxisomal activity and mitochondrial fatty acid oxidation in human primary myotubes. Am. J. Physiol. Metab. 2017, 312, E253–E263. [Google Scholar] [CrossRef]
- Boström, P.A.; Fernández-Real, J.M. Irisin, the metabolic syndrome and follistatin in humans. Nat. Rev. Endocrinol. 2013, 10, 11–12. [Google Scholar] [CrossRef]
- Whittle, A. Searching for ways to switch on brown fat: Are we getting warmer? J. Mol. Endocrinol. 2012, 49, R79–R87. [Google Scholar] [CrossRef]
- Riedel, A.; Hochkogler, C.M.; Lang, R.; Bytof, G.; Lantz, I.; Hofmann, T.; Somoza, V. N-Methylpyridinium, a degradation product of trigonelline upon coffee roasting, stimulates respiratory activity and promotes glucose utilization in HepG2 cells. Food Funct. 2014, 5, 454–462. [Google Scholar] [CrossRef]
- Folstar, P.; Pilnik, W.; de Heus, J.C.; Schols, H.A.; Melger, P.J. Liquid chromatographic analysis of N-alkanoyl-S-hydroxytryptamine (C-5-HT) in green coffee beans. J. Agric. Food Chem. 1979, 27, 12–15. [Google Scholar] [CrossRef]
- Folstar, P.; Schols, H.A.; Van Der Plas, H.C.; Pilnik, W.; Landheer, C.A.; Van Veldhuizen, A. New tryptamine derivatives isolated from wax of green coffee beans. J. Agric. Food Chem. 1980, 28, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Lang, R.; Bardelmeier, I.; Weiss, C.; Rubach, M.; Somoza, V.; Hofmann, T. Quantitation ofβN-Alkanoyl-5-hydroxytryptamides in Coffee by Means of LC-MS/MS-SIDA and Assessment of Their Gastric Acid Secretion Potential Using the HGT-1 Cell Assay. J. Agric. Food Chem. 2010, 58, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.-J.; Kim, J.; Sohn, C.H.; DeWreede, R.; Choi, T.; Towers, G.; Hudson, J.; Hong, Y. Inhibition of Taq DNA polymerase by seaweed extracts from British Columbia, Canada and Korea. Environ. Boil. Fishes 1997, 9, 383–388. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, J.H.; Jin, D.-H.; Jin, H.-J.; Kim, Y.S. Myostatin inhibitory region of fish (Paralichthys olivaceus) myostatin-1 propeptide. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 194-195, 65–70. [Google Scholar] [CrossRef]
- Cash, J.N.; Angerman, E.B.; Kattamuri, C.; Nolan, K.; Zhao, H.; Sidis, Y.; Keutmann, H.T.; Thompson, T.B. Structure of Myostatin·Follistatin-like 3. J. Biol. Chem. 2012, 287, 1043–1053. [Google Scholar] [CrossRef] [PubMed]


| Group | Glucose (mg/dL) | Total Cholesterol (mg/dL) | Triglyceride (mg/dL) | Free Fatty Acid (µEq/L) |
|---|---|---|---|---|
| Control | 286.0 ± 8.07 | 127.2 ± 5.74 | 129.8 ± 11.21 | 745.8 ± 119.59 |
| CSE | 291.8 ± 13.48 | 132.5 ± 8.12 | 129.3 ± 11.58 | 588.6 ± 127.37 + |
| Relative level (%) | 102.0 | 104.1 | 99.6 | 78.9 |
| Gene | Forward Primer (5′–3′) | Reverse Primer (5´–3´) | Size (bp) | Accession |
|---|---|---|---|---|
| Mstn | GCTTGACTGCGATGAGCACT | GCACCCACAGCGGTCTACTA | 316 | NM 010834 |
| Fndc5 | TTGATGTGGCAGCAGACTTC | CTTATCCGTCCCTCCTCTCC | 312 | NM_027402 |
| Ppargc1a | ATGTGTCGCCTTCTTGCTCT | GCGGTATTCATCCCTCTTGA | 350 | NM_008904 |
| Ucp1 | CTTCTCAGCCGGAGTTTCAG | TGCCACACCTCCAGTCATTA | 331 | NM_009463 |
| Gapdh | CGTCCCGTAGACAAAATGGT | TCTCCATGGTGGTGAAGACA | 328 | NM_001289726 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Kim, J.H.; Jang, J.-P.; Jang, J.-H.; Jin, D.-H.; Kim, Y.S.; Jin, H.-J. Identification of Molecules from Coffee Silverskin That Suppresses Myostatin Activity and Improves Muscle Mass and Strength in Mice. Molecules 2021, 26, 2676. https://doi.org/10.3390/molecules26092676
Kim JH, Kim JH, Jang J-P, Jang J-H, Jin D-H, Kim YS, Jin H-J. Identification of Molecules from Coffee Silverskin That Suppresses Myostatin Activity and Improves Muscle Mass and Strength in Mice. Molecules. 2021; 26(9):2676. https://doi.org/10.3390/molecules26092676
Chicago/Turabian StyleKim, Jeong Han, Jae Hong Kim, Jun-Pil Jang, Jae-Hyuk Jang, Deuk-Hee Jin, Yong Soo Kim, and Hyung-Joo Jin. 2021. "Identification of Molecules from Coffee Silverskin That Suppresses Myostatin Activity and Improves Muscle Mass and Strength in Mice" Molecules 26, no. 9: 2676. https://doi.org/10.3390/molecules26092676
APA StyleKim, J. H., Kim, J. H., Jang, J.-P., Jang, J.-H., Jin, D.-H., Kim, Y. S., & Jin, H.-J. (2021). Identification of Molecules from Coffee Silverskin That Suppresses Myostatin Activity and Improves Muscle Mass and Strength in Mice. Molecules, 26(9), 2676. https://doi.org/10.3390/molecules26092676






