Feature-Based Molecular Networks Identification of Bioactive Metabolites from Three Plants of the Polynesian Cosmetopoeia Targeting the Dermal Papilla Cells of the Hair Cycle
Abstract
:1. Introduction
2. Results and Discussion
2.1. Interspecies Correlations between the Cell Proliferative Activity of the Fractions/Extracts and Their Chemical Constituents
2.2. The Chemical Composition of the Bioactive Fractions
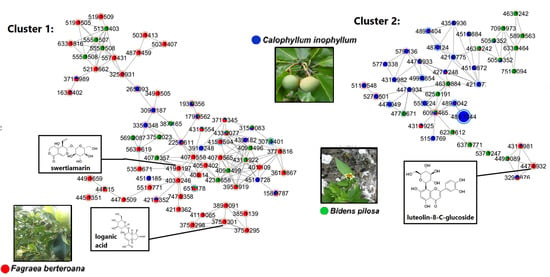
2.2.1. Chemical Constituents of BF4 from the Aerial Parts of Bidens pilosa
2.2.2. Chemical Constituents of CF5 from the Leaves of Calophyllum inophyllum
2.2.3. Chemical Constituents of FF1 from the Fruits of Fagraea berteroana
2.3. Bioactivity-Based Molecular Networking on Hair-Related Targets: Hair Follicle Dermal Papilla Cells Proliferation
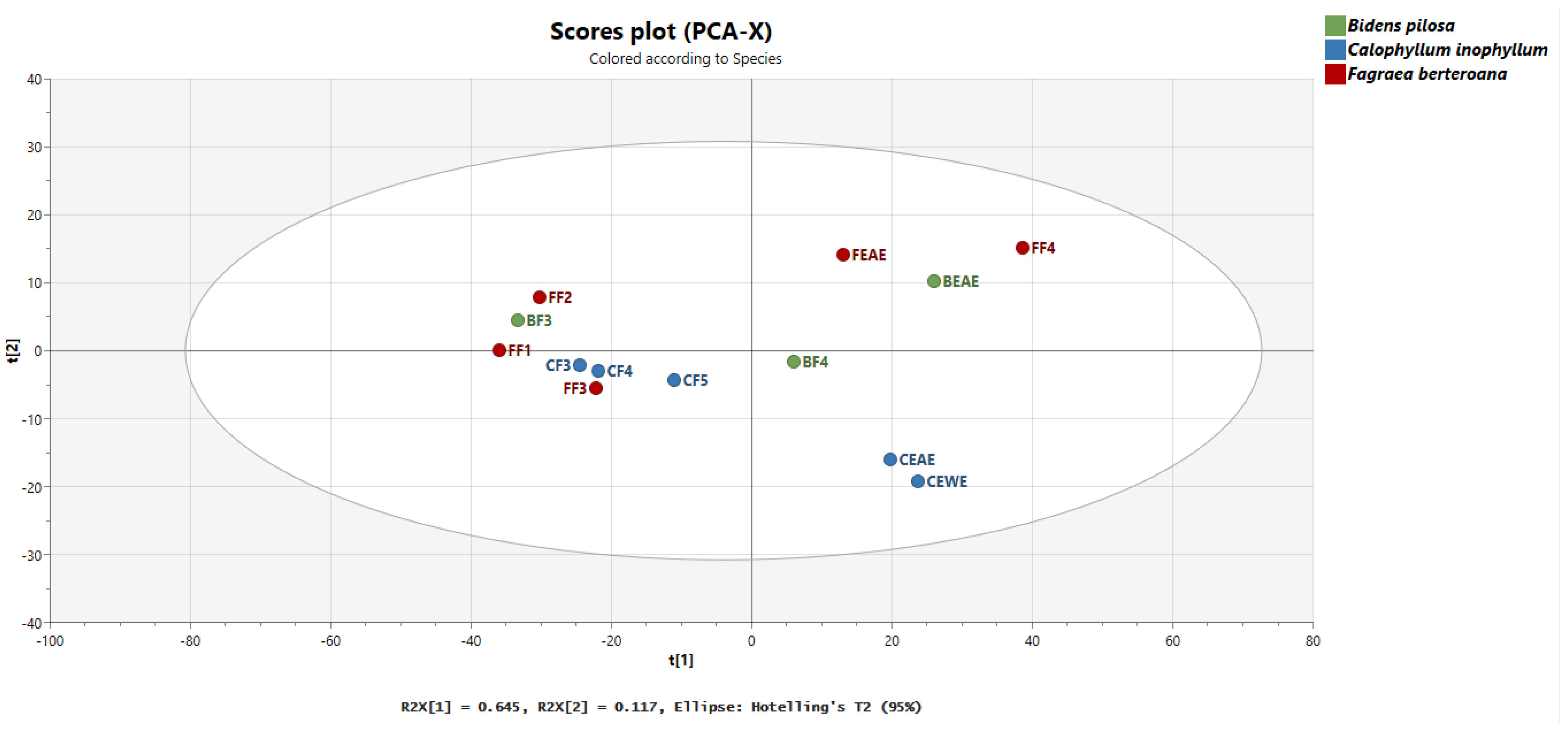
2.4. Multivariate Analysis of Interspecies Mapping with Principal Component Analysis
3. Materials and Methods
3.1. Plant Material
3.2. Plant Extraction and Compound Isolation
3.3. UHPLC-MS/MS Analysis
3.4. Construction of the Molecular Network
3.5. Proliferation Assay of Hair Follicle Dermal Papilla Cells
3.6. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Bai, Y.; Jia, Q.; Su, W.; Yan, Z.; Situ, W.; He, X.; Peng, W.; Yao, H. Integration of Molecular Networking and Fingerprint Analysis for Studying Constituents in Microctis Folium. PLoS ONE 2020, 15, e0235533. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking as a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Pétard, P. Plantes Utiles de Polynésie Française et Raau Tahiti; Editions Haere Po No Tahiti: Papeete, French Polynesia, 1986; ISBN 978-2-904171-06-2. [Google Scholar]
- Handy, E.S.C. The Native Culture in the Marquesas; Bernice, P., Ed.; Bishop Museum: Honolulu, HI, USA, 1923. [Google Scholar]
- Hughes, K.; Ho, R.; Butaud, J.-F.; Filaire, E.; Ranouille, E.; Berthon, J.-Y.; Raharivelomanana, P. A Selection of Eleven Plants Used as Traditional Polynesian Cosmetics and Their Development Potential as Anti-Aging Ingredients, Hair Growth Promoters and Whitening Products. J. Ethnopharmacol. 2019, 245, 112–159. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.; Ho, R.; Greff, S.; Filaire, E.; Ranouille, E.; Chazaud, C.; Herbette, G.; Butaud, J.-F.; Berthon, J.-Y.; Raharivelomanana, P. Hair Growth Activity of Three Plants of the Polynesian Cosmetopoeia and Their Regulatory Effect on Dermal Papilla Cells. Molecules 2020, 25, 4360. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.; Ho, R.; Chazaud, C.; Hermitte, S.; Greff, S.; Butaud, J.-F.; Filaire, E.; Ranouille, E.; Berthon, J.-Y.; Raharivelomanana, P. In Vitro Hair Dermal Papilla Cells Induction by Fagraea berteroana, a Tree of the Marquesan Cosmetopoeia (French Polynesia). Cosmetics 2021, 8, 13. [Google Scholar] [CrossRef]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Phelan, V.V. Feature-Based Molecular Networking for Metabolite Annotation. In Computational Methods and Data Analysis for Metabolomics; Li, S., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2104, pp. 227–243. ISBN 978-1-07-160238-6. [Google Scholar]
- Nothias, L.-F.; Nothias-Esposito, M.; da Silva, R.; Wang, M.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef] [Green Version]
- Olivon, F.; Allard, P.-M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.-F.; Cachet, X.; et al. Bioactive Natural Products Prioritization Using Massive Multi-Informational Molecular Networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef]
- Plikus, M.V. New Activators and Inhibitors in the Hair Cycle Clock: Targeting Stem Cells’ State of Competence. J. Investig. Dermatol. 2012, 132, 1321–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitagawa, T.; Matsuda, K.-I.; Inui, S.; Takenaka, H.; Katoh, N.; Itami, S.; Kishimoto, S.; Kawata, M. Keratinocyte Growth Inhibition through the Modification of Wnt Signaling by Androgen in Balding Dermal Papilla Cells. J. Clin. Endocrinol. Metab. 2009, 94, 1288–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, W.; Wu, E.; Morgan, B.A. Dermal Papilla Cell Number Specifies Hair Size, Shape and Cycling and Its Reduction Causes Follicular Decline. Development 2013, 140, 1676–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartolome, A.P.; Villaseñor, I.M.; Yang, W.-C. Bidens pilosa L. (Asteraceae): Botanical Properties, Traditional Uses, Phytochemistry, and Pharmacology. Evid. Based Complementary Altern. Med. 2013, 2013, 1–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Matsuura, D.; Kanatani, H.; Yano, S.; Tsunakawa, M.; Matsuyama, S.; Shigemori, H. Inhibitory Effects of Polyacetylene Compounds from Panax ginseng on Neurotrophin Receptor-Mediated Hair Growth. Biol. Pharm. Bull. 2017, 40, 1784–1788. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, Z.; Hua, H.; Li, Z.; Liu, M. Studies on flavonoids from stems and leaves of Calophyllum inophyllum. Zhongguo Zhong Yao Za Zhi 2007, 32, 692–694. [Google Scholar]
- Shakila, K. Flavonoids of Calophyllum inophyllum Linnaeus. C. Von. J. Chem. Environ. Sci. Appl. 2016, 2, 135–144. [Google Scholar] [CrossRef]
- Aminudin, N.I.; Ahmad, F.; Taher, M.; Zulkifli, R.M. Incrassamarin A–D: Four New 4-Substituted Coumarins from Calophyllum incrassatum and Their Biological Activities. Phytochem. Lett. 2016, 16, 287–293. [Google Scholar] [CrossRef]
- Ferchichi, L.; Derbré, S.; Mahmood, K.; Touré, K.; Guilet, D.; Litaudon, M.; Awang, K.; Hadi, A.H.A.; Le Ray, A.M.; Richomme, P. Bioguided Fractionation and Isolation of Natural Inhibitors of Advanced Glycation End-Products (AGEs) from Calophyllum flavoramulum. Phytochemistry 2012, 78, 98–106. [Google Scholar] [CrossRef]
- Laure, F. Etude de la composition chimique et de la biodiversité du Calophyllum inophyllum de Polynésie française. Ph.D. Thesis, Université de la Polynésie Française, Papeete, French Polynesia, 2005. [Google Scholar]
- Laure, F.; Raharivelomanana, P.; Butaud, J.-F.; Bianchini, J.-P.; Gaydou, E.M. Screening of Anti-HIV-1 Inophyllums by HPLC–DAD of Calophyllum inophyllum Leaf Extracts from French Polynesia Islands. Anal. Chim. Acta 2008, 624, 147–153. [Google Scholar] [CrossRef]
- Prasad, J.; Shrivastava, A.; Khanna, A.K.; Bhatia, G.; Awasthi, S.K.; Narender, T. Antidyslipidemic and Antioxidant Activity of the Constituents Isolated from the Leaves of Calophyllum inophyllum. Phytomedicine 2012, 19, 1245–1249. [Google Scholar] [CrossRef]
- Ghosal, S.; Singh, A.K.; Sharma, P.V.; Chaudhuri, R.K. Chemical Constituents of Gentianaceae IX: Natural Occurrence of Erythrocentaurin in Enicostemma hyssopifolium and Swertia lawii. J. Pharm. Sci. 1974, 63, 944–945. [Google Scholar] [CrossRef]
- Li, L.; Li, M.H.; Zhang, N.; Huang, L.Q. Chemical Constituents from Lomatogonium Carinthiacum (Gentianaceae). Biochem. Syst. Ecol. 2011, 39, 766–768. [Google Scholar] [CrossRef]
- Lv, H.; Chen, J.; Li, W.-L.; Zhang, H.-Q. Studies on the triterpenes from loquat leaf (Eriobotrya japonica). Zhong Yao Cai 2008, 31, 1351–1354. [Google Scholar] [PubMed]
- Klein, G.; Kim, J.; Himmeldirk, K.; Cao, Y.; Chen, X. Antidiabetes and Anti-Obesity Activity of Lagerstroemia Speciosa. Evid. Based Complement. Altern. Med. 2007, 4, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, W.; Xu, C.; Li, X. Biological Activities of Extracts from Loquat (Eriobotrya japonica Lindl.): A Review. Int. J. Mol. Sci. 2016, 17, 1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stohs, S.J.; Miller, H.; Kaats, G.R. A Review of the Efficacy and Safety of Banaba (Lagerstroemia speciosa L.) and Corosolic Acid. Phytother. Res. 2012, 26, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Zubair, S.; Mujtaba, G. Hair—A Mirror of Diabetes. PakMediNet 2016, 19, 4. [Google Scholar]
- Coogan, P.F.; Bethea, T.N.; Cozier, Y.C.; Bertrand, K.A.; Palmer, J.R.; Rosenberg, L.; Lenzy, Y. Association of Type 2 Diabetes with Central-Scalp Hair Loss in a Large Cohort Study of African American Women. Int. J. Women Dermatol. 2019, 5, 261–266. [Google Scholar] [CrossRef]
- Labo Cosprophar AG. A Composition for Activating Hair Follicle Stem Cells to Stimulate Hair Growth. Patent EP2561858A3, 4 June 2016. [Google Scholar]
- Buonocore, D.; Nobile, V.; Michelotti, A.; Marzatico, F. Clinical Efficacy of a Cosmetic Treatment by Crescina® Human Follicle Stem Cell on Healthy Males with Androgenetic Alopecia. Dermatol. Ther. 2013, 3, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Baba, K.; Hiramatsu, R.; Suradej, B.; Tanigaki, R.; Koeda, S.; Waku, T.; Kataoka, T. Asiatic Acid, Corosolic Acid, and Maslinic Acid Interfere with Intracellular Trafficking and N-Linked Glycosylation of Intercellular Adhesion Molecule-1. Biol. Pharm. Bull. 2018, 41, 1757–1768. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Mena, G.; Sánchez-González, M.; Juan, M.E.; Planas, J.M. Maslinic Acid, a Natural Phytoalexin-Type Triterpene from Olives—A Promising Nutraceutical? Molecules 2014, 19, 11538–11559. [Google Scholar] [CrossRef] [Green Version]
- Mokhtari, K.; Rufino-Palomares, E.E.; Pérez-Jiménez, A.; Reyes-Zurita, F.J.; Figuera, C.; García-Salguero, L.; Medina, P.P.; Peragón, J.; Lupiáñez, J.A. Maslinic Acid, a Triterpene from Olive, Affects the Antioxidant and Mitochondrial Status of B16F10 Melanoma Cells Grown under Stressful Conditions. Evid. Based Complementary Altern. Med. 2015, 2015, e272457. [Google Scholar] [CrossRef] [Green Version]
- Bangprapai, A.; Thongphasuk, P.; Songsak, T. Determination of Swertiamarin Content by Tlc-Densitometer in Fagraea Fragrans Roxb. Leaves. Bull. Health Sci. Technol. 2016, 14, 13–18. [Google Scholar]
- Cuendet, M.; Hostettmann, K.; Potterat, O.; Dyatmiko, W. Iridoid Glucosides with Free Radical Scavenging Properties from Fagraea Blumei. Helv. Chim. Acta 1997, 80, 1144–1152. [Google Scholar] [CrossRef]
- Cambie, R.C.; Lal, A.R.; Rickard, C.E.F.; Tanaka, N. Chemistry of Fijian Plants. V.: Constituents of Fagraea gracilipes, A. Gray. Chem. Pharm. Bull. 1990, 38, 1857–1861. [Google Scholar] [CrossRef] [Green Version]
- Jonville, M.-C.; Capel, M.; Frédérich, M.; Angenot, L.; Dive, G.; Faure, R.; Azas, N.; Ollivier, E. Fagraldehyde, a Secoiridoid Isolated from Fagraea fragrans. J. Nat. Prod. 2008, 71, 2038–2040. [Google Scholar] [CrossRef]
- Madmanang, S.; Cheyeng, N.; Heembenmad, S.; Mahabusarakam, W.; Saising, J.; Seeger, M.; Chusri, S.; Chakthong, S. Constituents of Fagraea fragrans with Antimycobacterial Activity in Combination with Erythromycin. J. Nat. Prod. 2016, 79, 767–774. [Google Scholar] [CrossRef]
- Suciati; Lambert, L.K.; Ross, B.P.; Deseo, M.A.; Garson, M.J. Phytochemical Study of Fagraea spp. Uncovers a New Terpene Alkaloid with Anti-Inflammatory Properties. Aust. J. Chem. 2011, 64, 489. [Google Scholar] [CrossRef]
- Suzuki, M.; Ninagawa, Y.; Hosokawa, K.; Matsunaga, K.; Hayakawa, R.; Fukushima, E.; Ueda, H. Clinical Evaluation of Hair Regrowth Tonics Containing Oleanolic Acid. Skin Res. 1988, 31, 136–143. (In Japanese) [Google Scholar] [CrossRef]
- Bassino, E.; Gasparri, F.; Munaron, L. Protective Role of Nutritional Plants Containing Flavonoids in Hair Follicle Disruption: A Review. Int. J. Mol. Sci. 2020, 21, 523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambwawasvika, H.; Dzomba, P.; Gwatidzo, L. Hair Growth Promoting Effect of Dicerocaryum senecioides Phytochemicals. Int. J. Med. Chem. 2019, 2019, e7105834. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, A.; Takahashi, T. Procyanidin B-2, Extracted from Apples, Promotes Hair Growth: A Laboratory Study: Procyanidin B-2 Modulates PKC Expression. Br. J. Dermatol. 2002, 146, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Sashida, Y.; Ogawa, K.; Kitada, M.; Karikome, H.; Mimaki, Y.; Shimomura, H. New Aurone Glucosides and New Phenylpropanoid Glucosides from Bidens pilosa. Chem. Pharm. Bull. 1991, 39, 709–711. [Google Scholar] [CrossRef] [Green Version]
- Rücker, G.; Kehrbaum, S.; Sakulas, H.; Lawong, B.; Goeltenboth, F. Acetylenic Glucosides from Microglossa pyrifolia. Planta Med. 1992, 58, 266–269. [Google Scholar] [CrossRef]
- Miranda, M.L.D.; Souza, A.F.; Rodrigues, E.D.; Garcez, F.R.; Garcez, W.S.; Abot, A. Constituintes químicos das folhas de Riedeliella graciliflora Harms (Leguminosae). Quím. Nova 2012, 35, 1306–1311. [Google Scholar] [CrossRef] [Green Version]
- Valentão, P.; Andrade, P.B.; Silva, A.M.S.; Moreira, M.M.; Seabra, R.M. Isolation and Structural Elucidation of 5-Formyl-2,3-Dihydroisocoumarin from Centaurium erythraea Aerial Parts. Nat. Prod. Res. 2003, 17, 361–364. [Google Scholar] [CrossRef]
- Yagi, A.; Okamura, N.; Haraguchi, Y.; Noda, K.; Nishioka, I. Studies on the Constituents of Zizyphi fructus II: Structure of New p-Coumaroylates of Maslinic Acid. Chem. Pharm. Bull. 1978, 26, 3075–3079. [Google Scholar] [CrossRef] [Green Version]
- Häberlein, H.; Tschiersch, K.-P. Triterpenoids and Flavonoids from Leptospermum scoparium. Phytochemistry 1994, 35, 765–768. [Google Scholar] [CrossRef]
- Numata, A.; Yang, P.; Takahashi, C.; Fujiki, R.; Nabae, M.; Fujita, E. Cytotoxic Triterpenes from a Chinese Medicine, Goreishi. Chem. Pharm. Bull. 1989, 37, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with GNPS. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
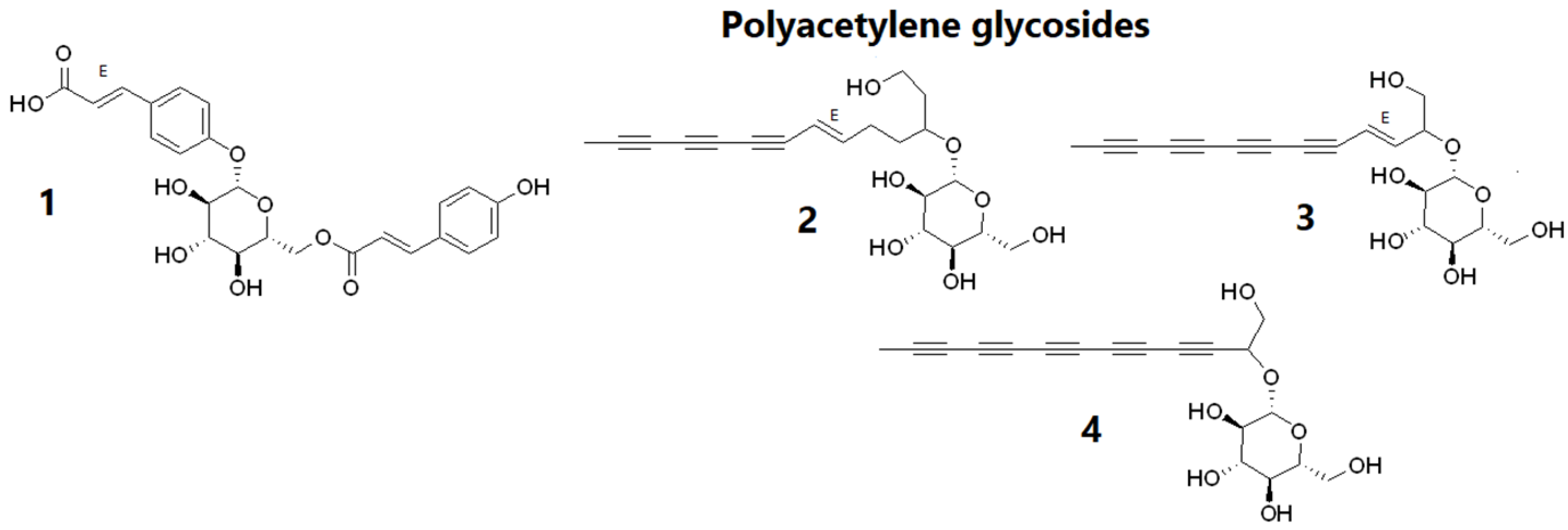
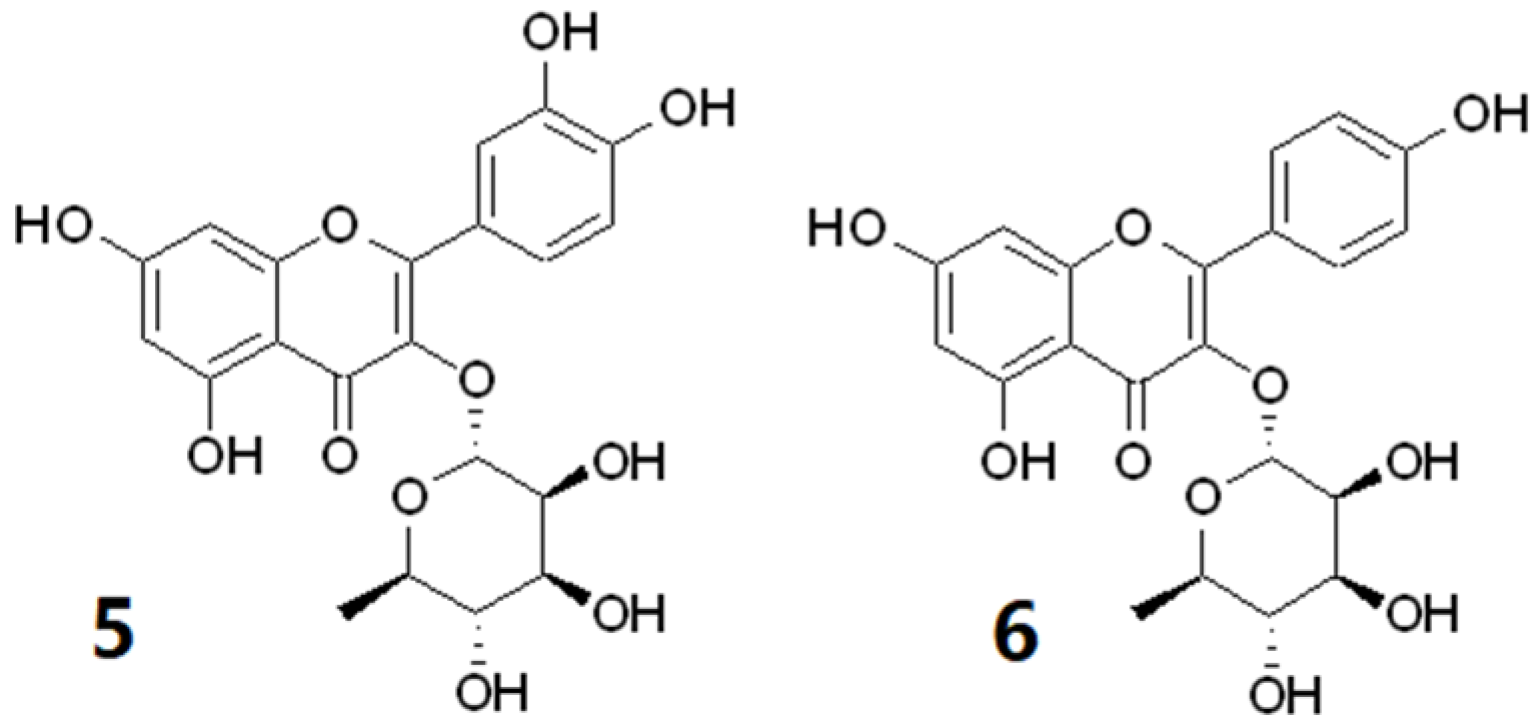
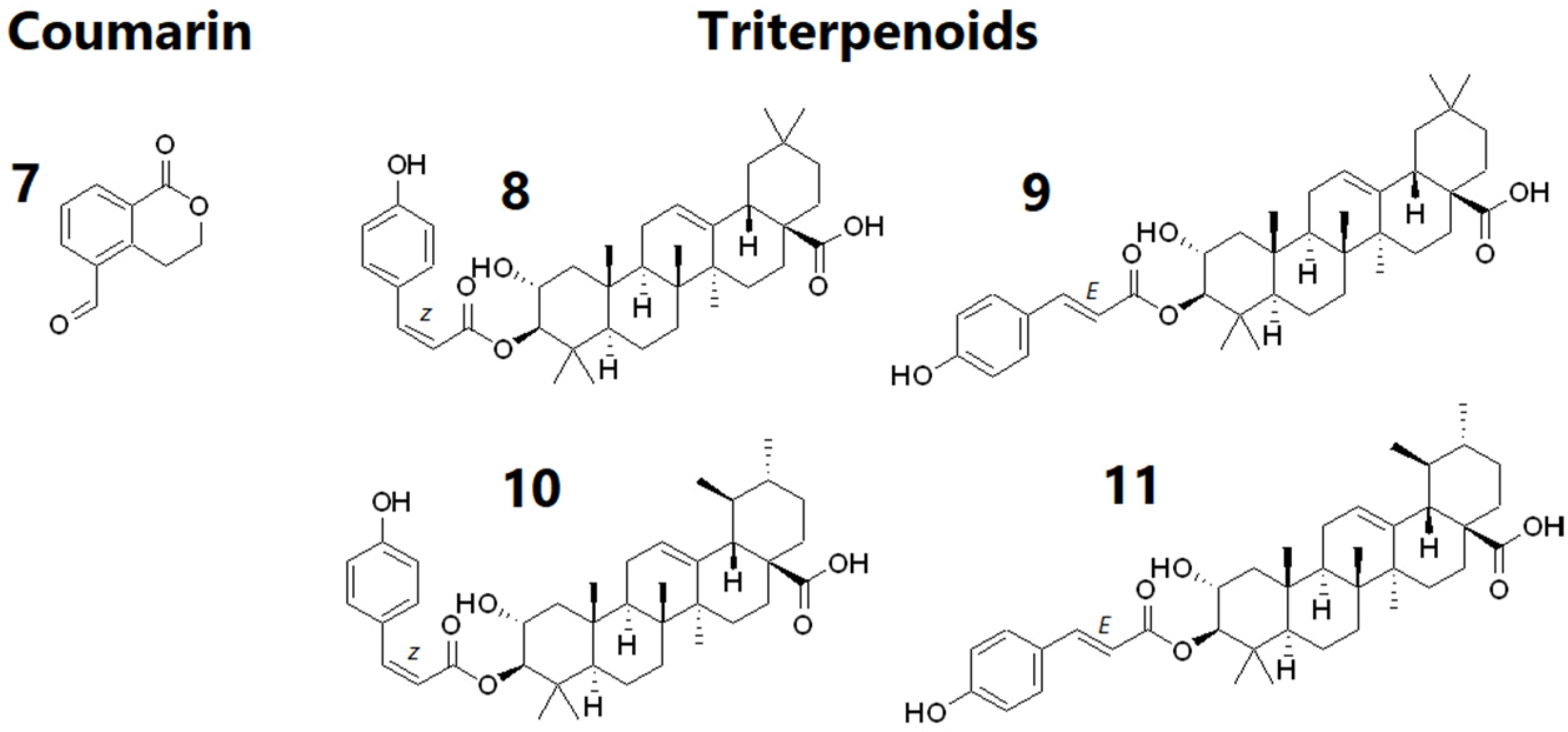
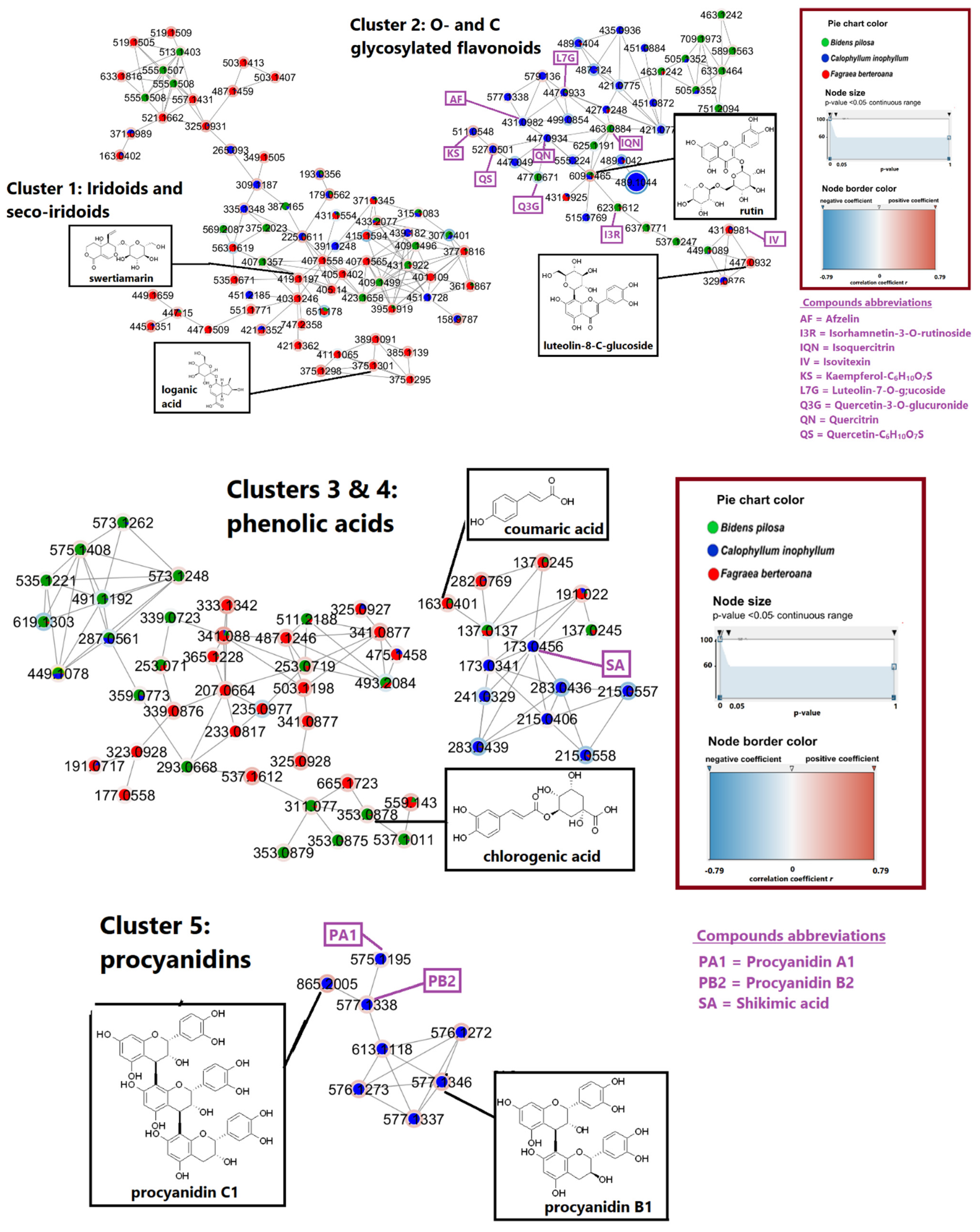
| Exp. m/z | Ion Type | Molecular Formula (Δppm) | Annotation | Comparison Source | Correlation to Cell Proliferation |
|---|---|---|---|---|---|
| 111.0088 | [M − H]− | C5H4O3 (0.3) | + | ||
| 163.0401 | [M − H]− | C9H8O3 (0.2) | p-coumaric acid | Massbank:RP016813 | + |
| 173.0456 | [M − H]− | C7H10O5 (0.3) | shikimic acid | Massbank: RP017513 | + |
| 339.0722 | [M − H]− | C15H16O9 (0.3) | + | ||
| 353.0878 | [M − H]− | C16H18O9 (0.0) | chlorogenic acid | MoNA:VF-NPL-QEHF011308 | + |
| 375.1301 | [M − H]− | C16H24O10 (1.1) | loganic acid | MoNA:VF-NPL-QTOF002452 | + |
| 419.1197 | [M + HCOO]− | C17H24O12 (0.5) | swertiamarin | Massbank:PR307657 | + |
| 431.0981 | [M − H]− | C21H20O10 (−0.6) | isovitexin | Massbank:PR307135 | + |
| 431.0982 | [M − H]− | C21H20O10 (−0.4) | afzelin | MoNA:VF-NPL-QEHF015100 | − |
| 447.0932 | [M − H]− | C21H20O11 (−0.2) | luteolin-8-C glucoside | Massbank:PR305779 | + |
| 447.0933 | [M − H]− | C21H20O11 (0.0) | luteolin-7-O-glucoside | Massbank:PR305563 | + |
| 447.0934 | [M − H]− | C21H20O11 (0.3) | quercitrin | Massbank:FIO00585 | − |
| 463.0884 | [M − H]− | C21H20O12 (0.4) | isoquercitrin | Massbank:FIO00167 | + |
| 477.0671 | [M − H]− | C21H18O13 (−0.8) | quercetin 3-O glucuronide | MoNA:VF-NPL-QEHF015166 | − |
| 511.0548 | [M − H]− | C21H20O13S (−0.8) | kaempferol-C6H10O7S | [7] | + |
| 515.1769 | [M − H]− | C23H32O13 (−0.2) | − | ||
| 527.0501 | [M − H]− | C21H20O14S (0.0) | quercetin-C6H10O7S | [7] | + |
| 575.1195 | [M − H]− | C30H24O12 (0.0) | procyanidin A1 | + | |
| 577.1338 | [M − H]− | C30H26O12 (−2.3) | procyanidin B2 | MassBank: BS003942 | + |
| 577.1346 | [M − H]− | C30H26O12 (−1.0) | procyanidin B1 | MassBank:BS003943 | + |
| 609.1465 | [M − H]− | C27H30O16 (0.6) | rutin | Massbank:FIO00596 | + |
| 623.1612 | [M − H]− | C28H32O16 (−0.9) | isorhamnetin-3-O-rutinoside | Massbank:PR305498 | + |
| 865.2005 | [M − H]− | C45H38O18 (2.3) | procyanidin C1 | MassBank: PR101005 | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hughes, K.; Ho, R.; Greff, S.; Herbette, G.; Filaire, E.; Ranouille, E.; Berthon, J.-Y.; Raharivelomanana, P. Feature-Based Molecular Networks Identification of Bioactive Metabolites from Three Plants of the Polynesian Cosmetopoeia Targeting the Dermal Papilla Cells of the Hair Cycle. Molecules 2022, 27, 105. https://doi.org/10.3390/molecules27010105
Hughes K, Ho R, Greff S, Herbette G, Filaire E, Ranouille E, Berthon J-Y, Raharivelomanana P. Feature-Based Molecular Networks Identification of Bioactive Metabolites from Three Plants of the Polynesian Cosmetopoeia Targeting the Dermal Papilla Cells of the Hair Cycle. Molecules. 2022; 27(1):105. https://doi.org/10.3390/molecules27010105
Chicago/Turabian StyleHughes, Kristelle, Raimana Ho, Stéphane Greff, Gaëtan Herbette, Edith Filaire, Edwige Ranouille, Jean-Yves Berthon, and Phila Raharivelomanana. 2022. "Feature-Based Molecular Networks Identification of Bioactive Metabolites from Three Plants of the Polynesian Cosmetopoeia Targeting the Dermal Papilla Cells of the Hair Cycle" Molecules 27, no. 1: 105. https://doi.org/10.3390/molecules27010105
APA StyleHughes, K., Ho, R., Greff, S., Herbette, G., Filaire, E., Ranouille, E., Berthon, J. -Y., & Raharivelomanana, P. (2022). Feature-Based Molecular Networks Identification of Bioactive Metabolites from Three Plants of the Polynesian Cosmetopoeia Targeting the Dermal Papilla Cells of the Hair Cycle. Molecules, 27(1), 105. https://doi.org/10.3390/molecules27010105