Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms
Abstract
:1. Introduction
2. Methods
2.1. Data
2.2. Feature Computation
2.3. Model Construction
2.4. Illogical/Random Feature Models to Serve as Control
2.5. Evaluation Metrics
3. Results
3.1. Realistic Datasets for Benchmark Evaluations
3.2. Sequence-Based Predictors
3.3. Benchmark Evaluation of Sequence-Based Methods
| Dataset Creator | Species | Dataset-Referencing Paper | Positive Pairs | Random Pairs | Proteins in Positive Data | Proteins in Random Data |
|---|---|---|---|---|---|---|
| Du [56] | Yeast a | Du [56] | 17,257 | 48,594 | 4382 | 2521 |
| Guo [9] | Yeast b | Chen [66] | 5594 | 5594 | 2217 | 2421 |
| Guo [67] | Multi | Chen [66] | 32,959 | 32,959 | 11,527 | 1399 |
| Jia [57] | Yeast e,f | Jia [57] | 17,339 | 33,056 | 4436 | 3260 |
| Liu [68] | Fruit Fly | Liu [68] | 4156 | 4241 | 2463 | 4080 |
| Martin [69] | H.Pylori | Jia [53] | 1420 | 1458 | 1313 | 727 |
| Martin [69] | Human | Pan [10] | 937 | 938 | 828 | 740 |
| Pan [10] | Human | Pan [10] | 36,617 | 36,480 | 9473 | 2184 |
| Pan [10] | Human | Pan [10] | 3899 | 4262 | 2502 | 661 |
| Guo [9] | Yeast b | Tian [15] | 5594 | 5594 | 2521 | 1194 |
| Li [17] | Human c,d | Li [17] | 4096 | 4096 | 2805 | 1865 |
| Richoux [22] | Human c | Richoux [22] | 39,672 | 64,388 | 6676 | 15,869 |
| Algorithm | Dataset | Accuracy | AUC |
|---|---|---|---|
| Guo 2008 AC SVM [9] | Guo Tian Yeast | 87 (−2) | |
| Pan 2010 PSAAC SVM [10] | Martin Human | 68 (−12) | |
| Pan 2010 PSAAC SVM [10] | Pan Small | 91 (−18) | 95 (−17) |
| Pan 2010 PSAAC Rot [10] | Pan Small | 95 (+3) | 97 (+2) |
| Pan 2010 PSAAC Rand [10] | Pan Small | 96 (+2) | 97 (+2) |
| Pan 2010 LDA Rot [10] | Pan Large | 97 (+1) | 99 (+0) |
| Pan 2010 LDA Rot [10] | Pan Small | 96 (+2) | 98 (+1) |
| Pan 2010 LDA Rand [10] | Pan Large | 98 (+0) | 99 (+0) |
| Pan 2010 LDA Rand [10] | Pan Small | 96 (+2) | 98 (+1) |
| Pan 2010 LDA SVM [10] | Martin Human | 69 (−6) | |
| Pan 2010 LDA SVM [10] | Pan Large | 95 (+2) | 98 (+1) |
| Pan 2010 LDA SVM [10] | Pan Small | 91 (+4) | 95 (+3) |
| Pan 2010 AC SVM [10] | Martin Human | 51 (+15) | |
| Pan 2010 AC SVM [10] | Pan Small | 89 (+7) | 94 (+4) |
| Pan 2010 AC Rot [10] | Pan Small | 95 (+2) | 96 (+3) |
| Pan 2010 AC Rand [10] | Pan Small | 96 (+2) | 97 (+2) |
| Zhou 2011 SVM [70] | Guo Tian Yeast | 89 (+2) | 95 (+1) |
| Zhao 2012 SVM [11] | Liu Fruit Fly | 81 (−4) | |
| Zhao 2012 SVM [11] | Martin H Pylori | 89 (−3) | |
| Jia 2015 RF [57] | Jia Yeast Held | 87 (−5) | |
| Jia 2015 RF [57] | Jia Yeast Cross | 84 (−6) | |
| Jia 2015 RF [57] | Martin H Pylori | 91 (−3) | |
| You 2015 RF [61] | Guo Tian Yeast | 95 (−1) | |
| You 2015 RF [61] | Martin H Pylori | 88 (−2) | |
| Ding 2016 RF [63] | Guo Tian Yeast | 95 (−1) | |
| Ding 2016 RF [63] | Martin H Pylori | 88 (+2) | |
| Ding 2016 RF [63] | Pan Small | 98 (+0) | |
| Du 2017 Sep [56] | Du Yeast | 93 (+0) | 97 (−0) |
| Du 2017 Sep [56] | Guo Tian Yeast | 94 (+1) | |
| Du 2017 Sep [56] | Martin H Pylori | 86 (+2) | |
| Du 2017 Sep [56] | Pan Small | 98 (+1) | |
| Du 2017 Comb [56] | Du Yeast | 90 (+1) | 96 (+0) |
| Sun 2017 CT Auto [12] | Pan Large | 95 (−1) | |
| Sun 2017 AC Auto [12] | Pan Large | 97 (−0) | |
| Wang 2017 Rot [19] | Guo Tian Yeast | 90 (−0) | |
| Wang 2017 Rot [19] | Martin H Pylori | 88 (−12) | |
| Göktepe 2018 SVM [13] | Martin Human | 74 (−6) | 83 (−11) |
| Göktepe 2018 SVM [13] | Martin H Pylori | 89 (−5) | 94 (−3) |
| Göktepe 2018 SVM [13] | Pan Small | 94 (+4) | 93 (+6) |
| Gonzalez-Lopez 2018 [21] | Du Yeast | 93 (−1) | 97 (−0) |
| Gonzalez-Lopez 2018 [21] | Guo Tian Yeast | 95 (−1) | 98 (−0) |
| Gonzalez-Lopez 2018 [21] | Martin H Pylori | 85 (+1) | 92 (−0) |
| Gonzalez-Lopez 2018 [21] | Pan Small | 98 (+1) | 100 (−0) |
| Hashemifar 2018 CNN [20] | Guo Tian Yeast | 95 (+0) | |
| Li 2018 CNN/LSTM [23] | Pan Large | 99 (−0) | |
| Chen 2019 LGBM [14] | Guo Tian Yeast | 95 (−0) | |
| Chen 2019 LGBM [14] | Martin H Pylori | 89 (−1) | |
| Chen 2019 RNN [66] | Guo Chen Yeast | 97 (−1) | |
| Jia 2019 RF [53] | Jia Yeast C. Full | 88 (−4) | |
| Jia 2019 RF [53] | Martin H Pylori | 93 (−4) | |
| Richoux 2019 LSTM [22] | Richoux Strict | 78 (−1) | |
| Richoux 2019 Full [22] | Richoux Strict | 76 (−1) | |
| Tian 2019 SVM [15] a | Guo Tian Yeast | 96 (−12) | |
| Tian 2019 SVM [15] a | Martin H Pylori | 96 (−17) | |
| Yao 2019 Net [71] | Guo Tian Yeast | 95 (+1) | |
| Yao 2019 Net [71] | Pan Small | 99 (+0) | |
| Zhang 2019 Deep [16] | Du Yeast | 95 (−4) | 97 (−2) |
| Li 2020 Deep [17] | Li AD | 95 (+3) | 95 (+5) |
| Czibula 2021 Auto SS [18] | Guo Chen Multi | 97 (+0) | 97 (+2) |
| Czibula 2021 Auto SS [18] | Pan Large | 98 (−1) | 98 (+1) |
| Czibula 2021 Auto SJ [18] | Guo Chen Multi | 97 (−0) | 97 (+2) |
| Czibula 2021 Auto SJ [18] | Pan Large | 98 (−1) | 98 (+1) |
| Czibula 2021 Auto JJ [18] | Guo Chen Multi | 98 (−1) | 98 (+1) |
| Czibula 2021 Auto JJ [18] | Pan Large | 98 (−1) | 96 (+3) |
| Dataset Referred by Table 1 Column 1, Column 2, Column 3 | Count Bias | Seq Sim Bias | Rand Net | Rand RF | Number of Implementations | Results Reported in Publications | Results of Our Implementations | Max Improvement over Bias Methods |
|---|---|---|---|---|---|---|---|---|
| Du Yeast | 87.7 | 87.5 | 92.5 | 88.5 | 4 | 90–95.3 | 90.5–92.6 | 2.8% |
| Guo Yeast Chen | 81.5 | 81.4 | 84 | 74.4 | 1 | 97.1 | 96.3 | 13.1% |
| Guo Yeast Tian | 87 | 85.9 | 94.3 | 94 | 10 | 87.3–95.1 | 84.5–95.5 | 1.2% |
| Guo Multi Chen | 93.5 | 93.1 | 98.7 | 96.4 | 3 | 96.9–98.2 | 96.6–97.3 | −0.5% |
| Jia Yeast Cross | 78.7 | 78.4 | 75.2 | 76.5 | 1 | 84.4 | 77.9 | 5.7% |
| Jia Yeast Held | 82.9 | 82.5 | 81.3 | 80.7 | 1 | 86.5 | 82.0 | 3.6% |
| Jia Yeast C. Full | 83.8 | 83.1 | 85.4 | 81.1 | 1 | 88 | 84.3 | 2.6% |
| Li AD | 96.7 | 79.1 | 76.2 | 97.3 | 1 | 94.7 | 97.7 | 0.4% |
| Liu Fruit Fly | 84.1 | 95.7 | 96.6 | 84.2 | 1 | 80.9 | 76.8 | −15.7% |
| Martin Human | 61.2 | 83.1 | 81.1 | 62.2 | 4 | 51–73.8 | 55.7–67.8 | −9.3% |
| Martin H Pylori | 83.6 | 61 | 59.2 | 89.6 | 10 | 85.2–93 | 75.9–89.9 | 3.4% |
| Pan Human Large | 96.3 | 83.1 | 82.2 | 97.8 | 9 | 94.5–99 | 94.3–98.9 | 1.2% |
| Pan Human Small | 94.5 | 93.1 | 98.8 | 98.6 | 14 | 89.3–98.7 | 72.5–99.4 | 0.6% |
| Richoux Strict | 79.6 | 94.4 | 96.9 | 79.5 | 2 | 76.3–78.3 | 74.7–76.8 | −18.6% |
| Algorithm | 50% Pos Full | 10% Pos Full | 0.3% Pos Full | 50% Pos Held | 10% Pos Held | 0.3% Pos Held | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | AUC | Prec | Avg P | Prec | Avg P | Acc | AUC | Prec | Avg P | Prec | Avg P | |
| Control Methods | ||||||||||||
| Count Bias | 84.2 | 91.5 | 91.9 | 56.4 | 28.0 | 6.5 | 50.0 | 50.0 | 10.0 | 10.0 | 0.3 | 0.3 |
| Seq Sim Bias | 82.3 | 90.0 | 84.5 | 53.0 | 14.8 | 5.2 | 65.6 | 70.7 | 41.8 | 21.6 | 2.1 | 0.9 |
| Random Vec NNet | 84.7 | 92.0 | 92.5 | 60.4 | 29.1 | 7.4 | 51.0 | 50.2 | 11.1 | 10.0 | 0.4 | 0.3 |
| Random Vec RF | 78.1 | 85.9 | 87.3 | 45.9 | 17.9 | 3.8 | 50.9 | 50.5 | 10.5 | 10.1 | 0.3 | 0.3 |
| Sequence-Based Predictors | ||||||||||||
| Guo 2008 AC SVM [9] | 74.1 | 81.5 | 83.3 | 40.4 | 16.1 | 3.4 | 61.4 | 65.4 | 44.2 | 17.3 | 2.4 | 0.7 |
| Pan 2010 PSAAC SVM [10] | 64.2 | 68.4 | 46.7 | 21.3 | 2.7 | 1.4 | 63.2 | 67.1 | 43.5 | 19.2 | 2.2 | 1.1 |
| Pan 2010 PSAAC Rot [10] | 82.9 | 90.6 | 96.1 | 57.5 | 44.5 | 8.5 | 64.4 | 70.2 | 62.1 | 20.8 | 4.9 | 1.4 |
| Pan 2010 PSAAC Rand [10] | 83.7 | 91.4 | 96.8 | 59.9 | 46.9 | 9.3 | 66.4 | 72.5 | 65.7 | 22.6 | 5.4 | 1.5 |
| Pan 2010 LDA Rot [10] | 82.7 | 90.4 | 93.7 | 54.7 | 33.7 | 6.8 | 59.7 | 63.9 | 38.0 | 16.6 | 2.3 | 0.9 |
| Pan 2010 LDA Rand [10] | 83.5 | 91.1 | 94.2 | 57 | 35.9 | 7.4 | 61.3 | 65.9 | 44.6 | 17.9 | 3.1 | 1.0 |
| Pan 2010 LDA SVM [10] | 77.8 | 85.3 | 88.4 | 45.9 | 19.2 | 4.2 | 58.8 | 61.9 | 27.4 | 14.9 | 1.3 | 0.5 |
| Pan 2010 AC SVM [10] | 80.2 | 87.2 | 85.5 | 47.9 | 13.6 | 3.8 | 59.9 | 64.5 | 41.8 | 18.2 | 2.0 | 0.7 |
| Pan 2010 AC Rot [10] | 83.2 | 91.0 | 94.3 | 55.2 | 37.7 | 6.8 | 57.8 | 61.3 | 30.7 | 14.5 | 1.3 | 0.7 |
| Pan 2010 AC Rand [10] | 83.9 | 91.7 | 94.1 | 57.0 | 39.0 | 7.4 | 59.8 | 64.3 | 37.0 | 15.4 | 2.1 | 0.8 |
| Zhou 2011 SVM [70] | 80.4 | 88.2 | 89.0 | 51.7 | 23.6 | 5.4 | 60.6 | 64.5 | 33.6 | 17.1 | 1.5 | 0.6 |
| Zhao 2012 SVM [11] | 77.9 | 83.5 | 86.6 | 39.6 | 14.9 | 2.9 | 64.4 | 68.5 | 35.0 | 19.1 | 1.7 | 0.7 |
| Jia 2015 RF [57] | 84.6 | 92.2 | 95.9 | 60.7 | 41.8 | 8.7 | 65.1 | 70.4 | 51.1 | 19.5 | 3.3 | 1.2 |
| You 2015 RF [61] | 83.1 | 90.8 | 95.8 | 56.5 | 43.9 | 7.7 | 61.2 | 65.9 | 42.9 | 17.4 | 2.5 | 1.0 |
| Ding 2016 RF [63] | 84.7 | 92.3 | 96.9 | 61.1 | 49.8 | 10.0 | 64.1 | 70.0 | 58.3 | 18.3 | 4.6 | 1.2 |
| Du 2017 Sep [56] | 85.5 | 92.8 | 94.9 | 64.5 | 39.5 | 9.8 | 67.0 | 73.3 | 56.3 | 24.9 | 3.9 | 1.2 |
| Du 2017 Comb [56] | 83.2 | 90.6 | 94.7 | 59.0 | 33.8 | 7.9 | 65.1 | 70.7 | 58.5 | 23.5 | 4.1 | 1.1 |
| Sun 2017 CT Auto [12] | 74.4 | 82.0 | 61.7 | 37.8 | 4.3 | 2.0 | 58.8 | 62.2 | 26.9 | 14.4 | 1.0 | 0.5 |
| Sun 2017 AC Auto [12] | 77.3 | 84.4 | 74.1 | 42.5 | 8.8 | 2.9 | 58.1 | 60.5 | 22.4 | 14.5 | 0.8 | 0.5 |
| Wang 2017 Rot [19] | 70.2 | 73.5 | 33.0 | 21.8 | 1.3 | 0.8 | 56.2 | 57.1 | 16.1 | 11.8 | 0.5 | 0.4 |
| Göktepe 2018 SVM [13] | 82.5 | 90.2 | 93.6 | 57.0 | 29.2 | 7.1 | 65.6 | 71.2 | 59.4 | 24.0 | 4.6 | 1.2 |
| Gonzalez-Lopez 2018 [21] | 83 | 90.5 | 89.9 | 55.6 | 23.9 | 5.9 | 54.1 | 55.4 | 17.8 | 11.9 | 0.7 | 0.4 |
| Hashemifar 2018 CNN [20] | 82.2 | 89.3 | 84.8 | 48.9 | 15.0 | 3.9 | 61.4 | 65.5 | 30.8 | 16.8 | 1.4 | 0.6 |
| Li 2018 CNN/LSTM [23] | 84.3 | 91.8 | 93.6 | 60.9 | 28.3 | 7.3 | 56.0 | 58.5 | 24.7 | 13.6 | 1.1 | 0.5 |
| Chen 2019 LGBM [14] | 81.9 | 89.7 | 96.0 | 57.6 | 45.2 | 8.4 | 62.7 | 67.7 | 43.6 | 19.8 | 2.4 | 1.0 |
| Chen 2019 RNN [66] | 83.9 | 90.4 | 75.4 | 54.7 | 7.7 | 4.0 | 59.8 | 63.2 | 27.1 | 15.7 | 1.2 | 0.5 |
| Jia 2019 RF [53] | 83.2 | 91.0 | 97.1 | 59.5 | 52.7 | 10.0 | 66.0 | 72.0 | 68.2 | 23.2 | 5.6 | 1.6 |
| Richoux 2019 LSTM [22] | 80.0 | 87.0 | 91.6 | 52.8 | 25.2 | 5.8 | 54.3 | 55.5 | 15.6 | 11.9 | 0.6 | 0.4 |
| Richoux 2019 Full [22] | 82.8 | 90.4 | 91.7 | 57.9 | 25.7 | 6.5 | 55.2 | 56.8 | 15.1 | 12.2 | 0.6 | 0.4 |
| Tian 2019 SVM [15] | 76.0 | 83.6 | 86.4 | 44.3 | 18.8 | 4.1 | 65.3 | 70.8 | 57.7 | 22.0 | 4.3 | 1.0 |
| Yao 2019 Net [71] | 83.5 | 90.7 | 89.3 | 55.9 | 20.1 | 5.6 | 57.7 | 60.7 | 27.1 | 14.7 | 1.2 | 0.5 |
| Zhang 2019 Deep [16] | 81.4 | 88.1 | 74.8 | 51.0 | 8.0 | 3.7 | 59.5 | 61.7 | 38.2 | 17.1 | 1.9 | 0.6 |
| Li 2020 Deep [17] | 86.4 | 93.4 | 96.6 | 67.7 | 50.7 | 12.4 | 67.3 | 73.8 | 65.9 | 26.8 | 6.1 | 1.7 |
| Czibula 2021 Auto SS [18] | 76.5 | 84.7 | 66.8 | 36.5 | 5.7 | 2.1 | 53.1 | 54.0 | 11.3 | 11.3 | 0.7 | 0.4 |
| Czibula 2021 Auto SJ [18] | 66.6 | 74.8 | 66.6 | 33.3 | 5.9 | 1.8 | 53.0 | 54.4 | 18.2 | 11.6 | 0.8 | 0.4 |
| Czibula 2021 Auto JJ [18] | 74.7 | 82.6 | 67.9 | 35.8 | 5.8 | 2.1 | 55.7 | 57.2 | 40 | 12.9 | 1.7 | 0.5 |
3.4. De-Biasing Annotation-Based Predictors
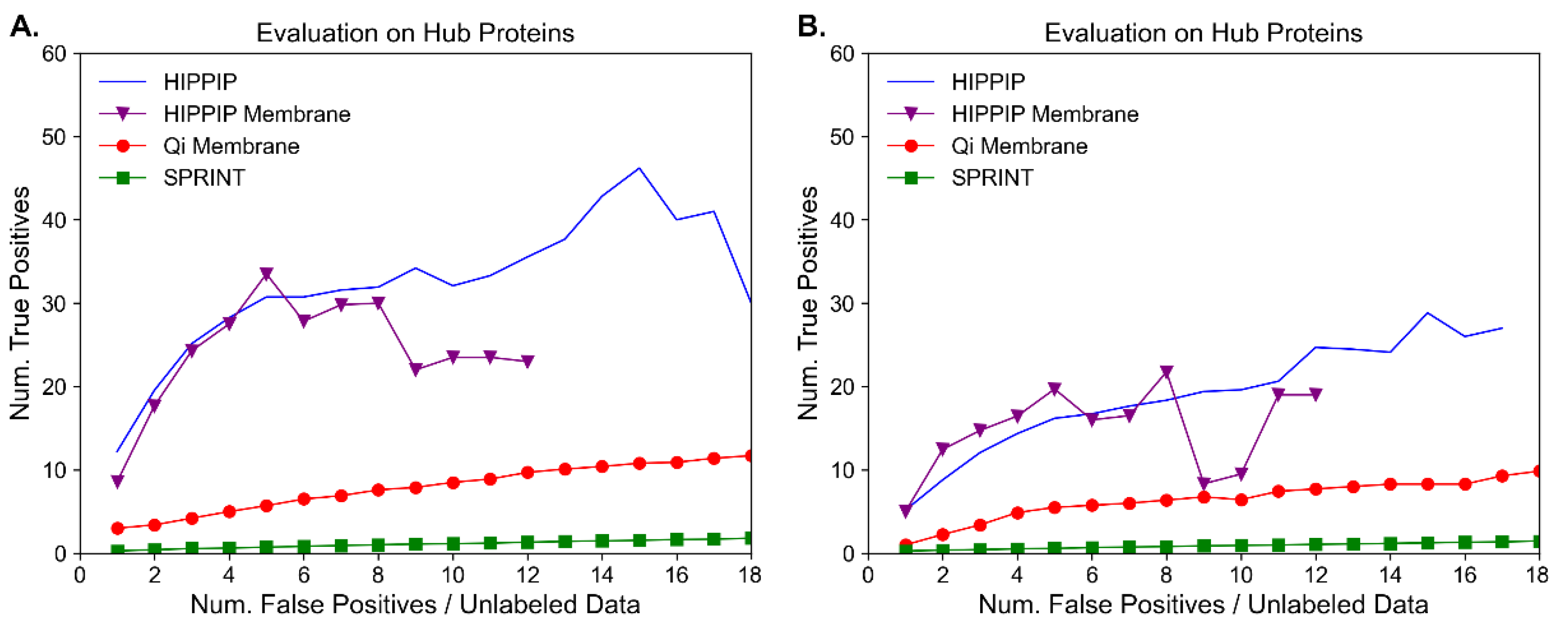
3.5. Benchmark Evaluation of Annotation-Based Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Tang, Z.; Takahashi, Y. Analysis of Protein–Protein Interaction by Co-IP in Human Cells. In Two-Hybrid Systems; Springer: New York, NY, USA, 2018; pp. 289–296. [Google Scholar]
- Johnson, K.L.; Qi, Z.; Yan, Z.; Wen, X.; Nguyen, T.C.; Zaleta-Rivera, K.; Chen, C.J.; Fan, X.; Sriram, K.; Wan, X.; et al. Revealing protein-protein interactions at the transcriptome scale by sequencing. Mol. Cell 2021, 81, 4091–4103. [Google Scholar] [CrossRef]
- Huang, H.; Jedynak, B.M.; Bader, J.S. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 2007, 3, e214. [Google Scholar] [CrossRef] [Green Version]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lund-Johansen, F.; Tran, T.; Mehta, A. Towards reproducibility in large-scale analysis of protein–protein interactions. Nat. Methods 2021, 18, 720–721. [Google Scholar] [CrossRef]
- Hart, G.T.; Ramani, A.K.; Marcotte, E.M. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006, 7, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpf, M.P.; Thorne, T.; de Silva, E.; Stewart, R.; An, H.J.; Lappe, M.; Wiuf, C. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. USA 2008, 105, 6959–6964. [Google Scholar] [CrossRef] [Green Version]
- Rual, J.F.; Hirozane-Kishikawa, T.; Hao, T.; Bertin, N.; Li, S.; Dricot, A.; Li, N.; Rosenberg, J.; Lamesch, P.; Vidalain, P.O.; et al. Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 2004, 14, 2128–2135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Yu, L.; Wen, Z.; Li, M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res. 2008, 36, 3025–3030. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.-Y.; Zhang, Y.-N.; Shen, H.-B. Large-Scale prediction of human protein− protein interactions from amino acid sequence based on latent topic features. J. Proteome Res. 2010, 9, 4992–5001. [Google Scholar] [CrossRef]
- Zhao, X.-W.; Ma, Z.-Q.; Yin, M.-H. Predicting protein-protein interactions by combing various sequence-derived features into the general form of Chou’s Pseudo amino acid composition. Protein Pept. Lett. 2012, 19, 492–500. [Google Scholar] [CrossRef]
- Sun, T.; Zhou, B.; Lai, L.; Pei, J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinform. 2017, 18, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Göktepe, Y.E.; Kodaz, H. Prediction of protein-protein interactions using an effective sequence based combined method. Neurocomputing 2018, 303, 68–74. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Q.; Ma, Q.; Yu, B. LightGBM-PPI: Predicting protein-protein interactions through LightGBM with multi-information fusion. Chemom. Intell. Lab. Syst. 2019, 191, 54–64. [Google Scholar] [CrossRef]
- Tian, B.; Wu, X.; Chen, C.; Qiu, W.; Ma, Q.; Yu, B. Predicting protein–protein interactions by fusing various Chou’s pseudo components and using wavelet denoising approach. J. Theor. Biol. 2019, 462, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, G.; Xia, D.; Wang, J. Protein–protein interactions prediction based on ensemble deep neural networks. Neurocomputing 2019, 324, 10–19. [Google Scholar] [CrossRef]
- Li, F.; Zhu, F.; Ling, X.; Liu, Q. Protein Interaction Network Reconstruction Through Ensemble Deep Learning with Attention Mechanism. Front. Bioeng. Biotechnol. 2020, 8, 390. [Google Scholar] [CrossRef]
- Czibula, G.; Albu, A.-I.; Bocicor, M.I.; Chira, C. AutoPPI: An Ensemble of Deep Autoencoders for Protein–Protein Interaction Prediction. Entropy 2021, 23, 643. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; You, Z.-H.; Xia, S.-X.; Liu, F.; Chen, X.; Yan, X.; Zhou, Y. Advancing the prediction accuracy of protein-protein interactions by utilizing evolutionary information from position-specific scoring matrix and ensemble classifier. J. Theor. Biol. 2017, 418, 105–110. [Google Scholar] [CrossRef]
- Hashemifar, S.; Neyshabur, B.; Khan, A.A.; Xu, J. Predicting protein–protein interactions through sequence-based deep learning. Bioinformatics 2018, 34, i802–i810. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Lopez, F.; Morales-Cordovilla, J.A.; Villegas-Morcillo, A.; Gomez, A.M.; Sanchez, V. End-to-end prediction of protein-protein interaction based on embedding and recurrent neural networks. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2344–2350. [Google Scholar]
- Richoux, F.; Servantie, C.; Borès, C.; Téletchéa, S. Comparing two deep learning sequence-based models for protein-protein interaction prediction. arXiv 2019, arXiv:1901.06268. [Google Scholar]
- Li, H.; Gong, X.-J.; Yu, H.; Zhou, C. Deep neural network based predictions of protein interactions using primary sequences. Molecules 2018, 23, 1923. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Liu, R.; Shriver, C.D.; Hu, H.; Liebman, M.N. Assessing semantic similarity measures for the characterization of human regulatory pathways. Bioinformatics 2006, 22, 967–973. [Google Scholar] [CrossRef]
- Zhang, S.-B.; Tang, Q.-R. Protein–protein interaction inference based on semantic similarity of gene ontology terms. J. Theor. Biol. 2016, 401, 30–37. [Google Scholar] [CrossRef]
- Chen, X.-W.; Liu, M. Prediction of protein–protein interactions using random decision forest framework. Bioinformatics 2005, 21, 4394–4400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiao, X.; Song, J.; Chang, S. Prediction of human protein–protein interaction by a domain-based approach. J. Theor. Biol. 2016, 396, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Maetschke, S.R.; Simonsen, M.; Davis, M.J.; Ragan, M.A. Gene Ontology-driven inference of protein–protein interactions using inducers. Bioinformatics 2012, 28, 69–75. [Google Scholar] [CrossRef]
- Qi, Y.; Dhiman, H.K.; Bhola, N.; Budyak, I.; Kar, S.; Man, D.; Dutta, A.; Tirupula, K.; Carr, B.I.; Grandis, J. Systematic prediction of human membrane receptor interactions. Proteomics 2009, 9, 5243–5255. [Google Scholar] [CrossRef] [Green Version]
- Thahir, M.; Sharma, T.; Ganapathiraju, M.K. An efficient heuristic method for active feature acquisition and its application to protein-protein interaction prediction. In Proceedings of the Great Lakes Bioinformatics Conference 2012, Ann Arbor, MI, USA, 15–17 May 2012; pp. 1–9. [Google Scholar]
- Goldberg, D.S.; Roth, F.P. Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. USA 2003, 100, 4372–4376. [Google Scholar] [CrossRef] [Green Version]
- Stark, C.; Breitkreutz, B.-J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Marcotte, E.M. Flaws in evaluation schemes for pair-input computational predictions. Nat. Methods 2012, 9, 1134. [Google Scholar] [CrossRef]
- Yu, J.; Guo, M.; Needham, C.J.; Huang, Y.; Cai, L.; Westhead, D.R. Simple sequence-based kernels do not predict protein–protein interactions. Bioinformatics 2010, 26, 2610–2614. [Google Scholar] [CrossRef] [Green Version]
- Pinker, E. Reporting accuracy of rare event classifiers. NPJ Digit. Med. 2018, 1, 56. [Google Scholar] [CrossRef]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganapathiraju, M.K.; Thahir, M.; Handen, A.; Sarkar, S.N.; Sweet, R.A.; Nimgaonkar, V.L.; Loscher, C.E.; Bauer, E.M.; Chaparala, S. Schizophrenia interactome with 504 novel protein-protein interactions. NPJ Schizophr. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Consortium, G.O. Gene ontology consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Huntley, R.P.; Sawford, T.; Mutowo-Meullenet, P.; Shypitsyna, A.; Bonilla, C.; Martin, M.J.; O’Donovan, C. The GOA database: Gene ontology annotation updates for 2015. Nucleic Acids Res. 2015, 43, D1057–D1063. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [Green Version]
- Hulo, N.; Bairoch, A.; Bulliard, V.; Cerutti, L.; De Castro, E.; Langendijk-Genevaux, P.S.; Pagni, M.; Sigrist, C.J. The PROSITE database. Nucleic Acids Res. 2006, 34, D227–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 22 November 2021).
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Shi, J.; Li, Q.; He, B.; Chen, J. ThunderSVM: A fast SVM library on GPUs and CPUs. J. Mach. Learn. Res. 2018, 19, 797–801. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Rodriguez, J.J.; Kuncheva, L.I.; Alonso, C.J. Rotation forest: A new classifier ensemble method. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 1619–1630. [Google Scholar] [CrossRef]
- Li, Y.; Ilie, L. SPRINT: Ultrafast protein-protein interaction prediction of the entire human interactome. BMC Bioinform. 2017, 18, 485. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Li, X.; Qiu, W.; Xiao, X.; Chou, K.-C. iPPI-PseAAC (CGR): Identify protein-protein interactions by incorporating chaos game representation into PseAAC. J. Theor. Biol. 2019, 460, 195–203. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubchak, I.; Muchnik, I.; Holbrook, S.R.; Kim, S.-H. Prediction of protein folding class using global description of amino acid sequence. Proc. Natl. Acad. Sci. USA 1995, 92, 8700–8704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, X.; Sun, S.; Hu, C.; Yao, Y.; Yan, Y.; Zhang, Y. DeepPPI: Boosting prediction of protein–protein interactions with deep neural networks. J. Chem. Inf. Model. 2017, 57, 1499–1510. [Google Scholar] [CrossRef]
- Jia, J.; Liu, Z.; Xiao, X.; Liu, B.; Chou, K.-C. iPPI-Esml: An ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J. Theor. Biol. 2015, 377, 47–56. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Wang, Z.-H.; Wang, Y.-X. A new encoding scheme to improve the performance of protein structural class prediction. In Proceedings of the International Conference on Natural Computation, Changsha, China, 27–29 August 2005; pp. 1164–1173. [Google Scholar]
- Yu, B.; Chen, C.; Wang, X.; Yu, Z.; Ma, A.; Liu, B. Prediction of protein–protein interactions based on elastic net and deep forest. Expert Syst. Appl. 2021, 176, 114876. [Google Scholar] [CrossRef]
- Yang, L.; Xia, J.-F.; Gui, J. Prediction of protein-protein interactions from protein sequence using local descriptors. Protein Pept. Lett. 2010, 17, 1085–1090. [Google Scholar] [CrossRef]
- You, Z.-H.; Chan, K.C.; Hu, P. Predicting protein-protein interactions from primary protein sequences using a novel multi-scale local feature representation scheme and the random forest. PLoS ONE 2015, 10, e0125811. [Google Scholar] [CrossRef]
- You, Z.-H.; Zhu, L.; Zheng, C.-H.; Yu, H.-J.; Deng, S.-P.; Ji, Z. Prediction of protein-protein interactions from amino acid sequences using a novel multi-scale continuous and discontinuous feature set. BMC Bioinform. 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Tang, J.; Guo, F. Predicting protein-protein interactions via multivariate mutual information of protein sequences. BMC Bioinform. 2016, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.C. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct. Funct. Bioinform. 2001, 43, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C. Prediction of protein subcellular locations by incorporating quasi-sequence-order effect. Biochem. Biophys. Res. Commun. 2000, 278, 477–483. [Google Scholar] [CrossRef]
- Chen, M.; Ju, C.J.-T.; Zhou, G.; Chen, X.; Zhang, T.; Chang, K.-W.; Zaniolo, C.; Wang, W. Multifaceted protein–protein interaction prediction based on Siamese residual RCNN. Bioinformatics 2019, 35, i305–i314. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Li, M.; Pu, X.; Li, G.; Guang, X.; Xiong, W.; Li, J. PRED_PPI: A server for predicting protein-protein interactions based on sequence data with probability assignment. BMC Res. Notes 2010, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Cai, Y.; Lu, W.; Feng, K.; Peng, C.; Niu, B. Prediction of protein–protein interactions based on PseAA composition and hybrid feature selection. Biochem. Biophys. Res. Commun. 2009, 380, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Roe, D.; Faulon, J.-L. Predicting protein–protein interactions using signature products. Bioinformatics 2005, 21, 218–226. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Gao, Y.; Zheng, Y.Y. Prediction of protein-protein interactions using local description of amino acid sequence. In Advances in Computer Science and Education Applications; Springer: Qingdao, China, 2011; pp. 254–262. [Google Scholar]
- Yao, Y.; Du, X.; Diao, Y.; Zhu, H. An integration of deep learning with feature embedding for protein–protein interaction prediction. PeerJ 2019, 7, e7126. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.P.; Carbonell, J.G.; Ganapathiraju, M.K. Active learning for human protein-protein interaction prediction. BMC Bioinform. 2010, 11 (Suppl. 1), S57. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Pos Ratio | Usage | Instance Sampling | Number of Datasets | Positive Pairs | Negative Pairs | Proteins in Positive Data | Proteins in Negative Data |
|---|---|---|---|---|---|---|---|
| 1:1 (50%) | Both | Full | 1 (5-fold CV) | 62,500 | 62,500 | 12,895 | 19,082 |
| 1:4 (20%) | Train | Full | 5 | 20,000 | 80,000 | 9250–9345 | 19,105–19,110 |
| 1:9 (10%) | Test | Full | 5 | 10,000 | 90,000 | 6868–6987 | 19,109–19,111 |
| 1:332 (0.3%) | Test | Full | 5 | 1500 | 498,500 | 2099–2170 | 19,112 |
| 1:1 | Train | Held Out | 21 | 50,000 | 50,000 | 8849–10,762 | 12,734–15,899 |
| 1:1 | Test | Held Out | 21 | 3170–7129 | 3170–7129 | 1530–3233 | 2770–5707 |
| 1:4 | Train | Held Out | 21 | 20,000 | 80,000 | 6978–8297 | 12,749–15,927 |
| 1:9 | Test | Held Out | 21 | 3170–7129 | 28,530–64,161 | 1530–3233 | 3185–6372 |
| 1:332 | Test | Held Out | 21 | 300–600 | 99,700–199,400 | 398–846 | 3185–6372 |
| Amino Acid Composition (AAC) | Auto Covariance (AC) [9] | Chaos Game Representation [53] | Conjoint Triad (CT) [54] | Composition-Transition-Distribution (CTD) [55] |
|---|---|---|---|---|
| Dipeptide Composition [56] | Discrete Wavelet Transform Physicochemical [57] | Encoding Based on Grouped Weight (EBGW) [58] | Geary Autocorrelation [59] | Local Descriptor (LD) [60] |
| Multi-scale Local Descriptor [61] | Multi-scale Continuous and Discontinuous [62] | Multivariate Mutual Information [63] | Moran Autocorrelation [59] | Normalized Moreau-Broto Autocorrelation [59] |
| Numeric/One Hot encoding | Pseudo Amino Acid Composition [64] | PSSM ([46]) | PSSM(DPC)/PSSM(Bi-gram) [13] | PSSM Discrete Cosine Transform [19] |
| Quasi Sequence Order Descriptor [65] | Sequence Order [11] | Skip Gram [45] | Weighted Skip-Sequential Conjoint Triad [13] |
| Dataset | Pos% | Data Type | Class | GO CC | GO BP | GO MF | GO Any | Pfam | Prosite | InterPro |
|---|---|---|---|---|---|---|---|---|---|---|
| Full | 50% | Train | Positive | 7.3 | 13.3 | 2.9 | 0.7 | 8.2 | 47.8 | 1.2 |
| Full | 50% | Train | Negative | 13.6 | 22.2 | 17.9 | 5.9 | 11.9 | 57.7 | 2.3 |
| Full | 50% | Test | Positive | 7.3 | 13.3 | 2.9 | 0.7 | 8.2 | 47.8 | 1.2 |
| Full | 50% | Test | Negative | 13.6 | 22.2 | 17.9 | 5.9 | 11.9 | 57.7 | 2.3 |
| Full | 20% | Train | Positive | 7.2 | 13.2 | 2.8 | 0.7 | 8.2 | 47.6 | 1.2 |
| Full | 20% | Train | Negative | 13.8 | 22.2 | 17.9 | 6.0 | 12.0 | 57.8 | 2.4 |
| Full | 10% | Test | Positive | 7.2 | 13.2 | 2.9 | 0.7 | 8.2 | 48.0 | 1.2 |
| Full | 10% | Test | Negative | 13.7 | 22.0 | 17.9 | 6.0 | 12.1 | 57.8 | 2.4 |
| Full | 0.3% | Test | Positive | 7.3 | 13.3 | 2.7 | 0.7 | 7.9 | 48.6 | 1.1 |
| Full | 0.3% | Test | Negative | 13.7 | 22.1 | 17.9 | 6.0 | 12.0 | 57.8 | 2.4 |
| Held Out | 50% | Train | Positive | 7.2 | 13.3 | 2.8 | 0.7 | 8.2 | 47.9 | 1.2 |
| Held Out | 50% | Train | Negative | 13.7 | 22.0 | 17.8 | 6.0 | 12.0 | 57.8 | 2.4 |
| Held Out | 50% | Test | Positive | 7.2 | 13.2 | 2.8 | 0.7 | 8.1 | 47.9 | 1.2 |
| Held Out | 50% | Test | Negative | 13.9 | 22.1 | 17.9 | 6.1 | 12.0 | 57.6 | 2.3 |
| Held Out | 20% | Train | Positive | 7.2 | 13.3 | 2.8 | 0.7 | 8.1 | 48.0 | 1.2 |
| Held Out | 20% | Train | Negative | 13.7 | 22.0 | 17.9 | 6.0 | 12.0 | 57.8 | 2.4 |
| Held Out | 10% | Test | Positive | 7.2 | 13.2 | 2.8 | 0.7 | 8.1 | 47.9 | 1.2 |
| Held Out | 10% | Test | Negative | 13.6 | 22.0 | 17.9 | 5.9 | 12.0 | 57.8 | 2.4 |
| Held Out | 0.3% | Test | Positive | 6.5 | 12.9 | 2.9 | 0.7 | 7.7 | 47.3 | 1.2 |
| Held Out | 0.3% | Test | Negative | 13.7 | 22.0 | 17.9 | 6.0 | 12.0 | 57.7 | 2.4 |
| Algorithm | 50% Pos Rand | 10% Pos Rand | 0.3% Pos Rand | 50% Pos Held | 10% Pos Held | 0.3% Pos Held | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | AUC | Prec | Avg P | Prec | Avg P | Acc | AUC | Prec | Avg P | Prec | Avg P | |
| Dom Var All | 92.0 | 97.3 | 99.0 | 88.4 | 72.9 | 42.4 | 92.1 | 97.3 | 99.1 | 88.3 | 76.2 | 43.7 |
| Dom Var NonTest | 74.2 | 77.4 | 96.7 | 51.5 | 41.3 | 9.0 | 73.8 | 76.9 | 96.4 | 49.9 | 46.4 | 9.6 |
| Dom Var HeldOut | 63.3 | 64.2 | 91.8 | 28.7 | 25.6 | 2.8 | ||||||
| Ensemble All | 94.3 | 98.3 | 97.5 | 90.9 | 51.3 | 38.3 | 93.8 | 98.0 | 97.5 | 90.1 | 52.2 | 38.0 |
| Ensemble NonTest | 76.4 | 82.2 | 92.2 | 54.1 | 25.4 | 8.8 | 76.0 | 81.4 | 92.1 | 51.9 | 23.9 | 8.0 |
| Ensemble HeldOut | 64.7 | 67.2 | 88.4 | 27.9 | 16.4 | 2.3 | ||||||
| Algorithm | 50% Pos Full | 10% Pos Full | 0.3% Pos Full | 50% Pos Held | 10% Pos Held | 0.3% Pos Held | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | AUC | Prec | Avg P | Prec | Avg P | Acc | AUC | Prec | Avg P | Prec | Avg P | |
| Annotation-Based Methods | ||||||||||||
| Chen 2005 Dom RF [26] | 75.9 | 82.7 | 77.1 | 41.8 | 8.8 | 2.8 | 65.1 | 69.6 | 58.8 | 23.1 | 4.3 | 1.0 |
| Gou 2006 Sem [24] | 66.3 | 72.1 | 90.1 | 33.7 | 15.7 | 2.9 | 66.3 | 72.2 | 89.5 | 33.1 | 22.1 | 3.3 |
| Maetschke 2012 ULCA [28] | 71.0 | 77.4 | 46.2 | 28.6 | 2.3 | 1.2 | 69.5 | 75.2 | 42.0 | 25.5 | 2.0 | 1.0 |
| Dom Variant | 74.2 | 77.4 | 96.7 | 51.5 | 41.3 | 9.0 | 63.3 | 64.2 | 91.8 | 28.7 | 25.6 | 2.8 |
| Zhang 2016 Sem [25] | 73.5 | 80.4 | 86.0 | 35.7 | 13.5 | 2.7 | 73.0 | 79.5 | 85.4 | 33.4 | 19.7 | 2.9 |
| Simple Ensemble | 76.4 | 82.2 | 92.2 | 54.1 | 25.4 | 8.8 | 64.7 | 67.2 | 88.4 | 27.9 | 16.4 | 2.3 |
| Control Methods | ||||||||||||
| Count Bias | 84.2 | 91.5 | 91.9 | 56.4 | 28.0 | 6.5 | 50.0 | 50.0 | 10.0 | 10.0 | 0.3 | 0.3 |
| Seq Sim Bias | 82.3 | 90.0 | 84.5 | 53.0 | 14.8 | 5.2 | 65.6 | 70.7 | 41.8 | 21.6 | 2.1 | 0.9 |
| Seq Sim Bias + Protein Bias | 82.3 | 89.9 | 84.5 | 52.9 | 14.8 | 5.2 | 65.6 | 70.8 | 41.6 | 21.6 | 2.1 | 0.9 |
| Rand Net | 84.7 | 92.0 | 92.5 | 60.4 | 29.1 | 7.4 | 51.0 | 50.2 | 11.1 | 10.0 | 0.4 | 0.3 |
| Rand RF | 78.1 | 85.9 | 87.3 | 45.9 | 17.9 | 3.8 | 50.9 | 50.5 | 10.5 | 10.1 | 0.3 | 0.3 |
| Selected Best-Performing Sequence-Based Methods | ||||||||||||
| Pan 2010 PSAAC Rand [10] | 83.7 | 91.4 | 96.8 | 59.9 | 46.9 | 9.3 | 66.4 | 72.5 | 65.7 | 22.6 | 5.4 | 1.5 |
| Jia 2015 RF [57] | 84.6 | 92.2 | 95.9 | 60.7 | 41.8 | 8.7 | 65.1 | 70.4 | 51.1 | 19.5 | 3.3 | 1.2 |
| Ding 2016 RF [63] | 84.7 | 92.3 | 96.9 | 61.1 | 49.8 | 10.0 | 64.1 | 70.0 | 58.3 | 18.3 | 4.6 | 1.2 |
| Du 2017 Sep [56] | 85.5 | 92.8 | 94.9 | 64.5 | 39.5 | 9.8 | 67.0 | 73.3 | 56.3 | 24.9 | 3.9 | 1.2 |
| Göktepe 2018 SVM [13] | 82.5 | 90.2 | 93.6 | 57.0 | 29.2 | 7.1 | 65.6 | 71.2 | 59.4 | 24.0 | 4.6 | 1.2 |
| Li 2018 CNN/LSTM [23] | 84.3 | 91.8 | 93.6 | 60.9 | 28.3 | 7.3 | 56.0 | 58.5 | 24.7 | 13.6 | 1.1 | 0.5 |
| Jia 2019 RF [53] | 83.2 | 91.0 | 97.1 | 59.5 | 52.7 | 10.0 | 66.0 | 72.0 | 68.2 | 23.2 | 5.6 | 1.6 |
| Li 2020 Deep [17] | 86.4 | 93.4 | 96.6 | 67.7 | 50.7 | 12.4 | 67.3 | 73.8 | 65.9 | 26.8 | 6.1 | 1.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dunham, B.; Ganapathiraju, M.K. Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms. Molecules 2022, 27, 41. https://doi.org/10.3390/molecules27010041
Dunham B, Ganapathiraju MK. Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms. Molecules. 2022; 27(1):41. https://doi.org/10.3390/molecules27010041
Chicago/Turabian StyleDunham, Brandan, and Madhavi K. Ganapathiraju. 2022. "Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms" Molecules 27, no. 1: 41. https://doi.org/10.3390/molecules27010041
APA StyleDunham, B., & Ganapathiraju, M. K. (2022). Benchmark Evaluation of Protein–Protein Interaction Prediction Algorithms. Molecules, 27(1), 41. https://doi.org/10.3390/molecules27010041






