The Donor-Dependent and Colon-Region-Dependent Metabolism of (+)-Catechin by Colonic Microbiota in the Simulator of the Human Intestinal Microbial Ecosystem
Abstract
:1. Introduction
2. Results
2.1. Interindividual Differences in Metabolic Rate of (+)-Catechin
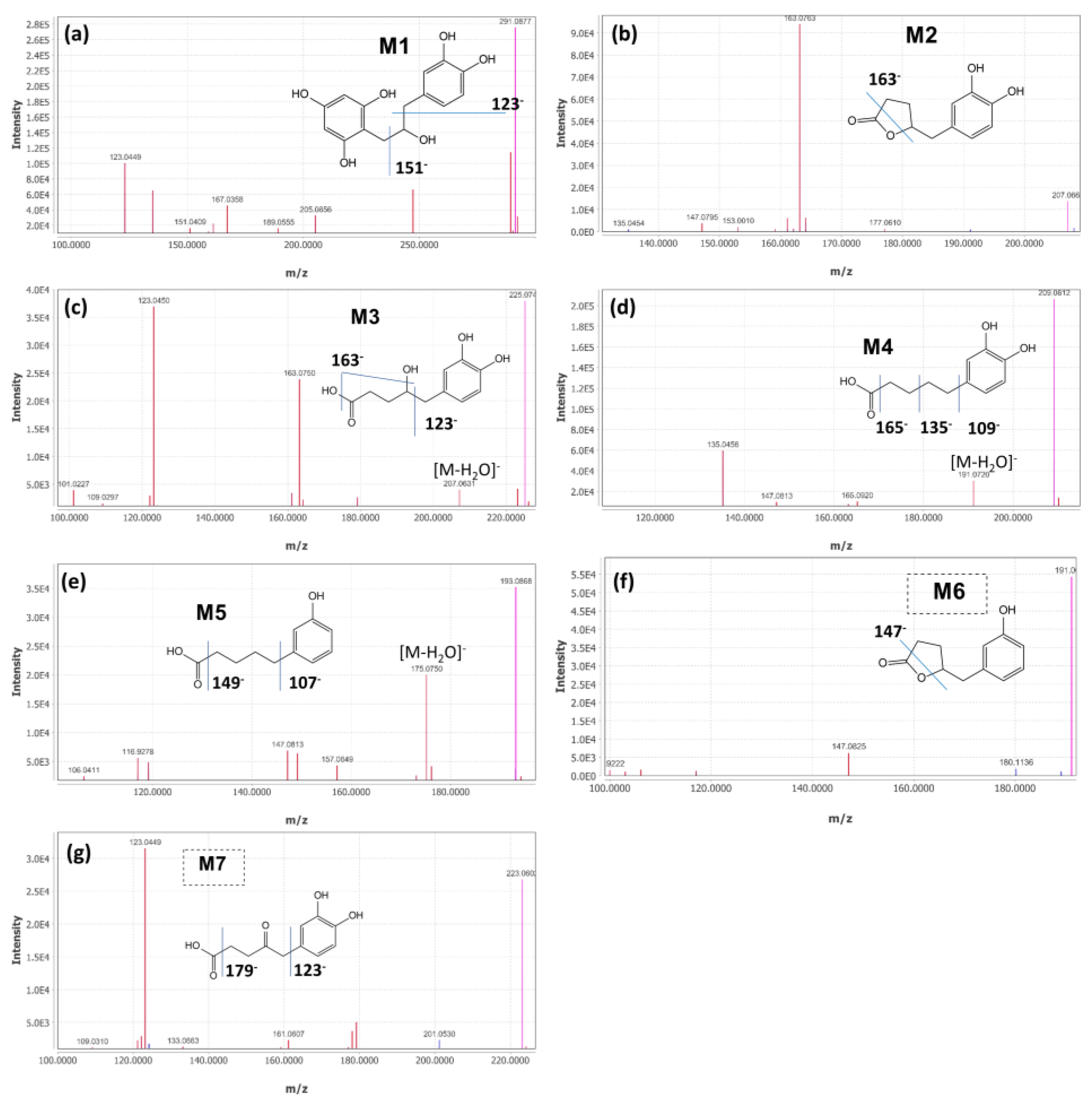
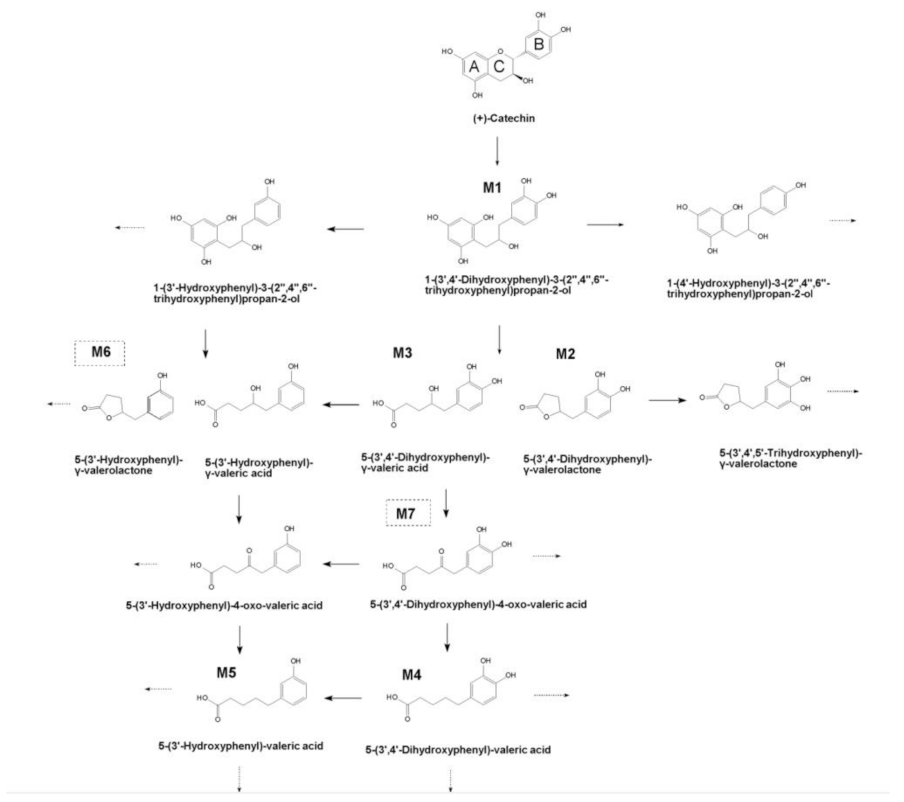
2.2. Identification of (+)-Catechin Metabolites during Simulated Colon Digestion in SHIME
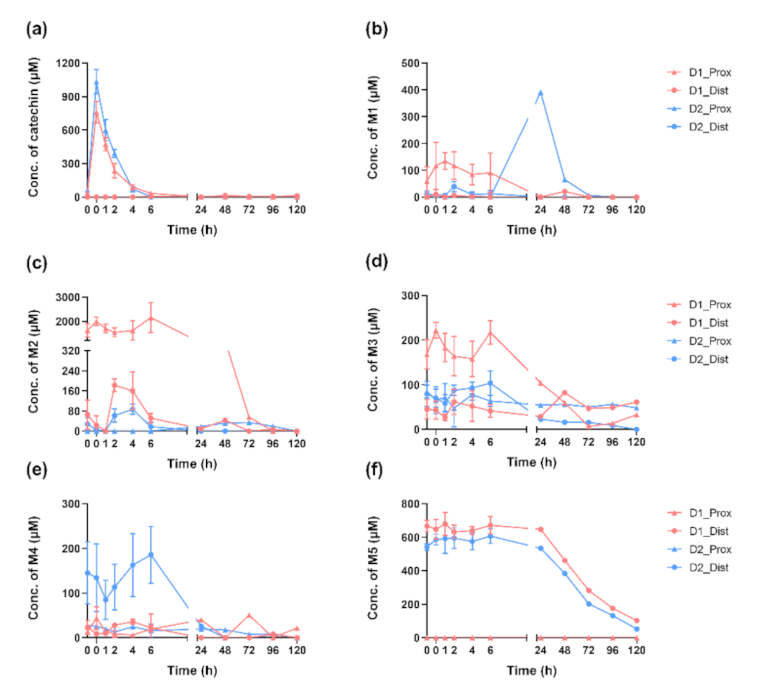
2.3. Dynamics of (+)-Catechin Metabolism in SHIME Is Colon-Region- and Donor-Dependent
3. Materials and Methods
3.1. Chemicals
3.2. Batch Incubation
3.3. Experimental Design of SHIME
3.4. UHPLC-MS Analysis
3.5. UPLC-IMS-Q-TOF-MS Analysis
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zanwar, A.A.; Badole, S.L.; Shende, P.S.; Hegde, M.V.; Bodhankar, S.L. Antioxidant Role of Catechin in Health and Disease. In Polyphenols in Human Health and Disease; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Academic Press: San Diego, CA, USA, 2014; Chapter 21; pp. 267–271. [Google Scholar]
- Wu, L.; Zhang, Q.L.; Zhang, X.Y.; Lv, C.; Li, J.; Yuan, Y.; Yin, F.X. Pharmacokinetics and blood-brain barrier penetration of (+)-catechin and (−)-epicatechin in rats by microdialysis sampling coupled to high-performance liquid chromatography with chemiluminescence detection. J. Agric. Food Chem. 2012, 60, 9377–9383. [Google Scholar] [CrossRef]
- Shabbir, U.; Rubab, M.; Daliri, E.B.; Chelliah, R.; Javed, A.; Oh, D.-H. Curcumin, quercetin, catechins and metabolic diseases: The role of gut microbiota. Nutrients 2021, 13, 206. [Google Scholar] [CrossRef]
- Manach, C.; Milenkovic, D.; Van de Wiele, T.; Rodriguez-Mateos, A.; de Roos, B.; Garcia-Conesa, M.T.; Landberg, R.; Gibney, E.R.; Heinonen, M.; Tomás-Barberán, F. Addressing the inter-individual variation in response to consumption of plant food bioactives: Towards a better understanding of their role in healthy aging and cardiometabolic risk reduction. Mol. Nutr. Food Res. 2017, 61, 1600557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezzat, H.M.; Elnaggar, Y.S.R.; Abdallah, O.Y. Improved oral bioavailability of the anticancer drug catechin using chitosomes: Design, in-vitro appraisal and in-vivo studies. Int. J. Pharm. 2019, 565, 488–498. [Google Scholar] [CrossRef]
- Lambert, J.D.; Sang, S.; Yang, C.S. Biotransformation of green tea polyphenols and the biological activities of those metabolites. Mol. Pharm. 2007, 4, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, R.; Silvestrini, A.; Mancuso, C. Ginsenosides, catechins, quercetin and gut microbiota: Current evidence of challenging interactions. Food Chem. Toxicol. 2019, 123, 42–49. [Google Scholar] [CrossRef]
- Li, Q.; Van de Wiele, T. Gut microbiota as a driver of the interindividual variability of cardiometabolic effects from tea polyphenols. Crit. Rev. Food Sci. Nutr. 2021, 61, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [Green Version]
- Van Duynhoven, J.; Vaughan, E.E.; Jacobs, D.M.; Kemperman, R.A.; van Velzen, E.J.; Gross, G.; Roger, L.C.; Possemiers, S.; Smilde, A.K.; Doré, J. Metabolic fate of polyphenols in the human superorganism. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), S4531–S4538. [Google Scholar] [CrossRef] [Green Version]
- Luca, S.V.; Macovei, I.; Bujor, A.; Miron, A.; Skalicka-Woźniak, K.; Aprotosoaie, A.C.; Trifan, A. Bioactivity of dietary polyphenols: The role of metabolites. Crit. Rev. Food Sci. Nutr. 2020, 60, 626–659. [Google Scholar] [CrossRef]
- Mena, P.; González de Llano, D.; Brindani, N.; Esteban-Fernández, A.; Curti, C.; Moreno-Arribas, M.V.; Del Rio, D.; Bartolomé, B. 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone and its sulphate conjugates, representative circulating metabolites of flavan-3-ols, exhibit anti-adhesive activity against uropathogenic Escherichia coli in bladder epithelial cells. J. Funct. Foods 2017, 29, 275–280. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, X.; Lu, Q.; Zhang, L.; Wang, X.; Liu, R. C-ring cleavage metabolites of catechin and epicatechin enhanced antioxidant activities through intestinal microbiota. Food Res. Int. 2020, 135, 109271. [Google Scholar] [CrossRef] [PubMed]
- Mena, P.; Ludwig, I.A.; Tomatis, V.B.; Acharjee, A.; Calani, L.; Rosi, A.; Brighenti, F.; Ray, S.; Griffin, J.L.; Bluck, L.J.; et al. Inter-individual variability in the production of flavan-3-ol colonic metabolites: Preliminary elucidation of urinary metabotypes. Eur. J. Nutr. 2019, 58, 1529–1543. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, K.M.; Neilson, A.P. Simultaneous UPLC-MS/MS analysis of native catechins and procyanidins and their microbial metabolites in intestinal contents and tissues of male Wistar Furth inbred rats. J. Chromatography. B Anal. Technol. Biomed. Life Sci. 2014, 958, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; de Bruijn, W.J.C.; Bruins, M.E.; Vincken, J.P. Reciprocal interactions between epigallocatechin-3-gallate (EGCG) and human gut microbiota in vitro. J. Agric. Food Chem. 2020, 68, 9804–9815. [Google Scholar] [CrossRef]
- Ottaviani, J.I.; Borges, G.; Momma, T.Y.; Spencer, J.P.E.; Keen, C.L.; Crozier, A.; Schroeter, H. The metabolome of [2-14C](−)-epicatechin in humans: Implications for the assessment of efficacy, safety and mechanisms of action of polyphenolic bioactives. Sci. Rep. 2016, 6, 29034. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Vervoort, J.; Beekmann, K.; Baccaro, M.; Kamelia, L.; Wesseling, S.; Rietjens, I. Interindividual differences in human intestinal microbial conversion of (−)-epicatechin to bioactive phenolic compounds. J. Agric. Food Chem. 2020, 68, 14168–14181. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Vervoort, J.; van den Elzen, J.; Beekmann, K.; Baccaro, M.; de Haan, L.; Rietjens, I.M. Interindividual differences in human in vitro intestinal microbial conversion of green tea (−)-epigallocatechin-3-o-gallate and consequences for activation of Nrf2 mediated gene expression. Mol. Nutr. Food Res. 2021, 65, 2000934. [Google Scholar] [CrossRef]
- Gleńsk, M.; Hurst, W.J.; Glinski, V.B.; Bednarski, M.; Gliński, J.A. Isolation of 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-propan-2-ol from grape seed extract and evaluation of its antioxidant and antispasmodic potential. Molecules 2019, 24, 2466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutschera, M.; Engst, W.; Blaut, M.; Braune, A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011, 111, 165–175. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, N.; Arikawa, A.Y.; Chen, C. Inhibitory effects of green tea polyphenols on microbial metabolism of aromatic amino acids in humans revealed by metabolomic analysis. Metabolites 2019, 9, 96. [Google Scholar] [CrossRef] [Green Version]
- Takagaki, A.; Nanjo, F. Catabolism of (+)-catechin and (−)-epicatechin by rat intestinal microbiota. J. Agric. Food Chem. 2013, 61, 4927–4935. [Google Scholar] [CrossRef]
- Ou, K.; Gu, L. Absorption and metabolism of proanthocyanidins. J. Funct. Foods 2014, 7, 43–53. [Google Scholar] [CrossRef]
- Chen, H.; Sang, S. Biotransformation of tea polyphenols by gut microbiota. J. Funct. Foods 2014, 7, 26–42. [Google Scholar] [CrossRef]
- Winter, J.; Moore, L.H.; Dowell, V.R., Jr.; Bokkenheuser, V.D. C-ring cleavage of flavonoids by human intestinal bacteria. Appl. Environ. Microbiol. 1989, 55, 1203–1208. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.-S.; Hattori, M. Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT-1 capable of cleaving the C-ring of (+)-catechin and (−)-epicatechin, followed by p-dehydroxylation of the B-ring. Biol. Pharm. Bull. 2012, 35, 2252–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104 (Suppl. S2), S1–S63. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Patán, F.; Cueva, C.; Monagas, M.; Walton, G.E.; Gibson, G.R.; Martín-Álvarez, P.J.; Victoria Moreno-Arribas, M.; Bartolomé, B. Gut microbial catabolism of grape seed flavan-3-ols by human faecal microbiota. Targetted analysis of precursor compounds, intermediate metabolites and end-products. Food Chem. 2012, 131, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Calani, L.; Del Rio, D.; Luisa Callegari, M.; Morelli, L.; Brighenti, F. Updated bioavailability and 48 h excretion profile of flavan-3-ols from green tea in humans. Int. J. Food Sci. Nutr. 2012, 63, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Borges, G.; Ottaviani, J.I.; van der Hooft, J.J.; Schroeter, H.; Crozier, A. Absorption, metabolism, distribution and excretion of (−)-epicatechin: A review of recent findings. Mol. Aspects Med. 2018, 61, 18–30. [Google Scholar] [CrossRef]
- Castello, F.; Costabile, G.; Bresciani, L.; Tassotti, M.; Naviglio, D.; Luongo, D.; Ciciola, P.; Vitale, M.; Vetrani, C.; Galaverna, G.; et al. Bioavailability and pharmacokinetic profile of grape pomace phenolic compounds in humans. Arch. Biochem. Biophys. 2018, 646, 1–9. [Google Scholar] [CrossRef]
- Mena, P.; Bresciani, L.; Brindani, N.; Ludwig, I.A.; Pereira-Caro, G.; Angelino, D.; Llorach, R.; Calani, L.; Brighenti, F.; Clifford, M.N.; et al. Phenyl-gamma-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: Synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019, 36, 714–752. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Meng, X.; Winnik, B.; Lee, M.J.; Lu, H.; Sheng, S.; Buckley, B.; Yang, C.S. Analysis of urinary metabolites of tea catechins by liquid chromatography/electrospray ionization mass spectrometry. Chem. Res. Toxicol. 2001, 14, 702–707. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, K.; Kerckhof, F.M.; Verspreet, J.; Courtin, C.M.; Van de Wiele, T. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environ. Microbiol. 2017, 19, 3251–3267. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 305–317. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Donor No. | Conversion Percentage (%) |
|---|---|
| D1 | 44.71% (fast) |
| D2 | −1.13% (slow) 1 |
| D3 | 35.16% |
| D4 | 32.36% |
| D5 | 30.15% |
| D6 | 3.03% |
| D7 | 49.16% |
| D8 | −3.84% |
| D9 | −1.54% |
| D10 | 19.66% |
| D11 | 36.65% |
| D12 | 54.99% |
| Chemical | MW (g/mol) | RT (min) | [M-H]− (m/z) | [MS/MS] (m/z) Profiles | |
|---|---|---|---|---|---|
| M1 | 1-(3′,4′-dihydroxyphenyl)-3-(2′′,4′′,6′′-trihydroxyphenyl) propan-2-ol | 292.28 | 8.28 | 291.0877 | 289.07, 123.04, 247.10, 135.04, 167.03, 205.05, 161.06, 151.04, 189.06 |
| M2 | 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone | 208.21 | 8.35 | 207.0664 | 163.08, 123.04, 122.03, 161.06, 164.08, 147.09, 109.02, 153.00, 123.04 |
| M3 | 4-hydroxy-(3′,4′-dihydroxyphenyl)-valeric acid | 226.23 | 5.98 | 225.0744 | 123.04, 163.07, 207.06, 101.02, 223.05, 161.06, 179.07, 122.03, 109.03 |
| M4 | 5-(3′,4′-dihydroxyphenyl)-valeric acid | 210.22 | 11.84 | 209.0812 | 135.05, 191.07, 165.09, 147.08, 163.07, 109.03, 122.04 |
| M5 | 5-(3′-hydroxyphenyl)-valeric acid | 194.23 | 15.23 | 193.0888 | 175.07, 119.05, 147.08, 149.10, 106.04, 99.92, 157,06, 176.08 |
| M6 | 5-(3′-hydroxyphenyl)-γ-valerolactone | 192.21 | 11.6 | 191.0699 | 147.08, 99.92, 106.04, 116.92 |
| M7 | 5-(3′,4′-dihydroxyphenyl)-4-oxo-valeric acid | 224.21 | 6.63 | 223.0602 | 123.04, 179.07, 177.97, 122.03, 161.06, 201.05, 121.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Van Herreweghen, F.; De Mey, M.; Goeminne, G.; Van de Wiele, T. The Donor-Dependent and Colon-Region-Dependent Metabolism of (+)-Catechin by Colonic Microbiota in the Simulator of the Human Intestinal Microbial Ecosystem. Molecules 2022, 27, 73. https://doi.org/10.3390/molecules27010073
Li Q, Van Herreweghen F, De Mey M, Goeminne G, Van de Wiele T. The Donor-Dependent and Colon-Region-Dependent Metabolism of (+)-Catechin by Colonic Microbiota in the Simulator of the Human Intestinal Microbial Ecosystem. Molecules. 2022; 27(1):73. https://doi.org/10.3390/molecules27010073
Chicago/Turabian StyleLi, Qiqiong, Florence Van Herreweghen, Marjan De Mey, Geert Goeminne, and Tom Van de Wiele. 2022. "The Donor-Dependent and Colon-Region-Dependent Metabolism of (+)-Catechin by Colonic Microbiota in the Simulator of the Human Intestinal Microbial Ecosystem" Molecules 27, no. 1: 73. https://doi.org/10.3390/molecules27010073
APA StyleLi, Q., Van Herreweghen, F., De Mey, M., Goeminne, G., & Van de Wiele, T. (2022). The Donor-Dependent and Colon-Region-Dependent Metabolism of (+)-Catechin by Colonic Microbiota in the Simulator of the Human Intestinal Microbial Ecosystem. Molecules, 27(1), 73. https://doi.org/10.3390/molecules27010073







