Three-Dimensional Bioprinting of Decellularized Extracellular Matrix-Based Bioinks for Tissue Engineering
Abstract
:1. Introduction
2. Extracellular Matrix (ECM)
2.1. Components
2.2. Biological Roles
3. Decellularization Methods and Evaluation
3.1. Decellularization Methods
3.1.1. Physical Methods
3.1.2. Chemical Methods
3.1.3. Enzymatic Methods
3.2. Evaluating the Prepared dECM
4. Strategies for Preparing dECM-Based Bioinks
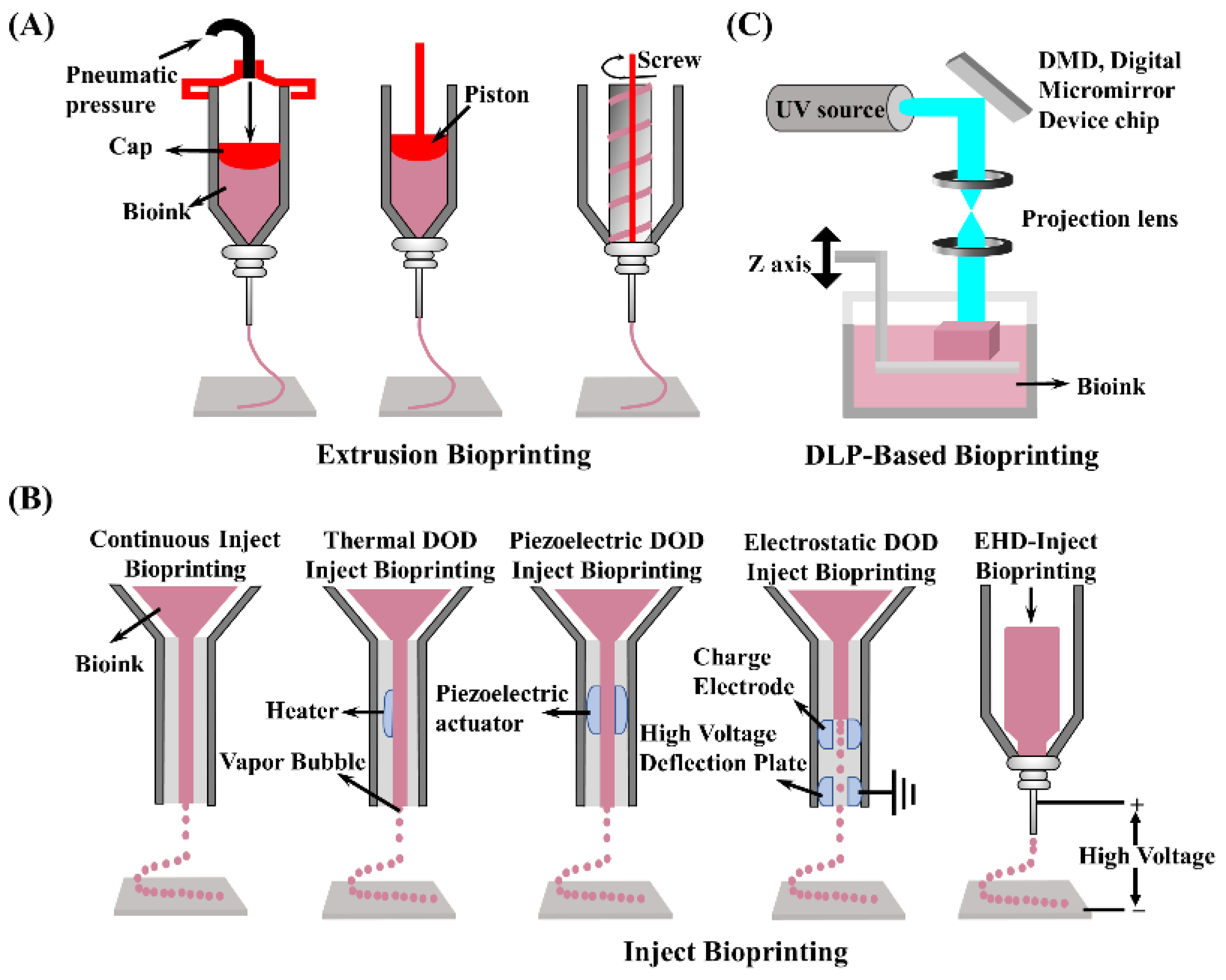
5. 3D Bioprinting Technologies
6. Applications
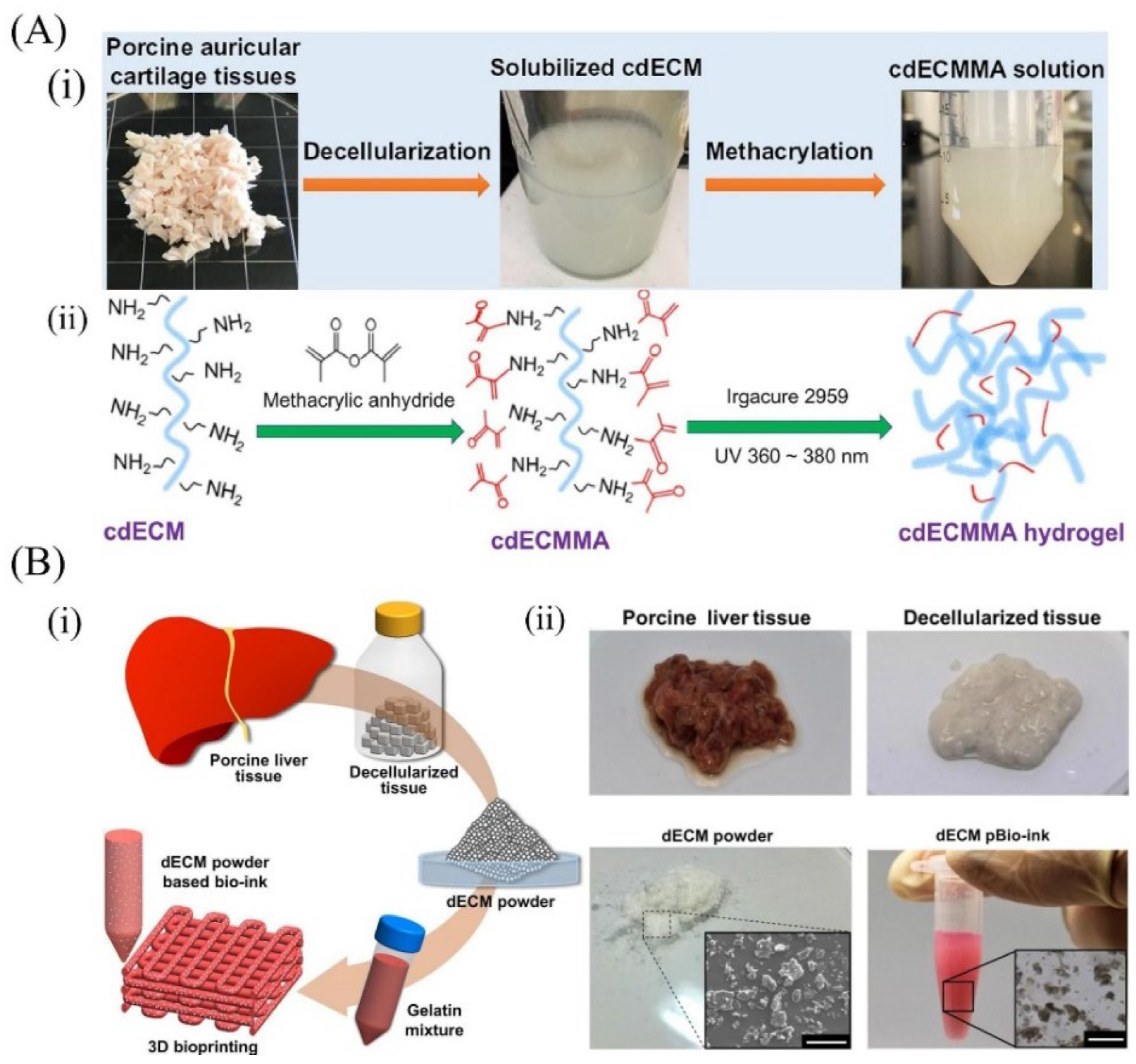
6.1. Cartilage-Derived dECM Bioinks
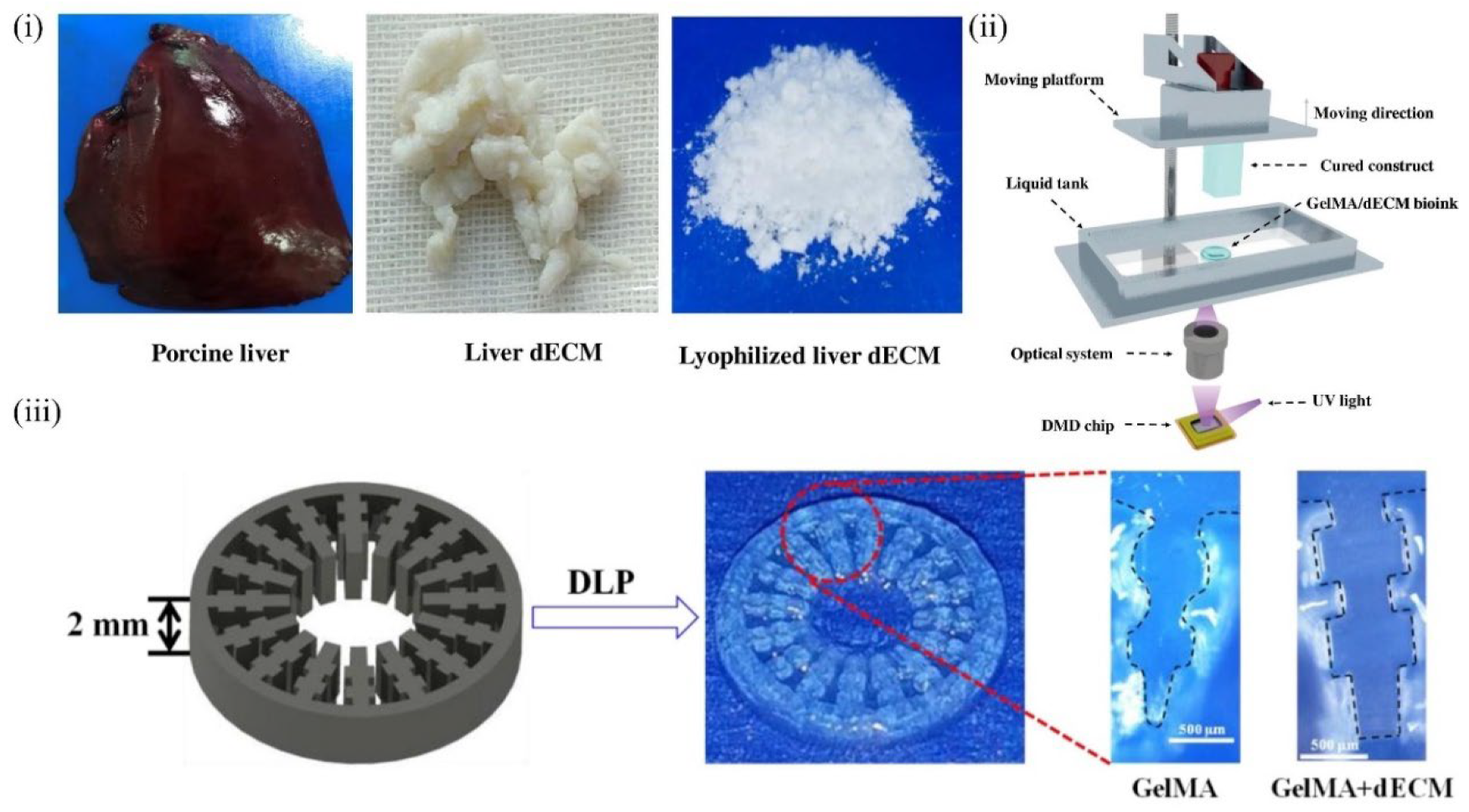
6.2. Liver-Derived dECM Bioinks
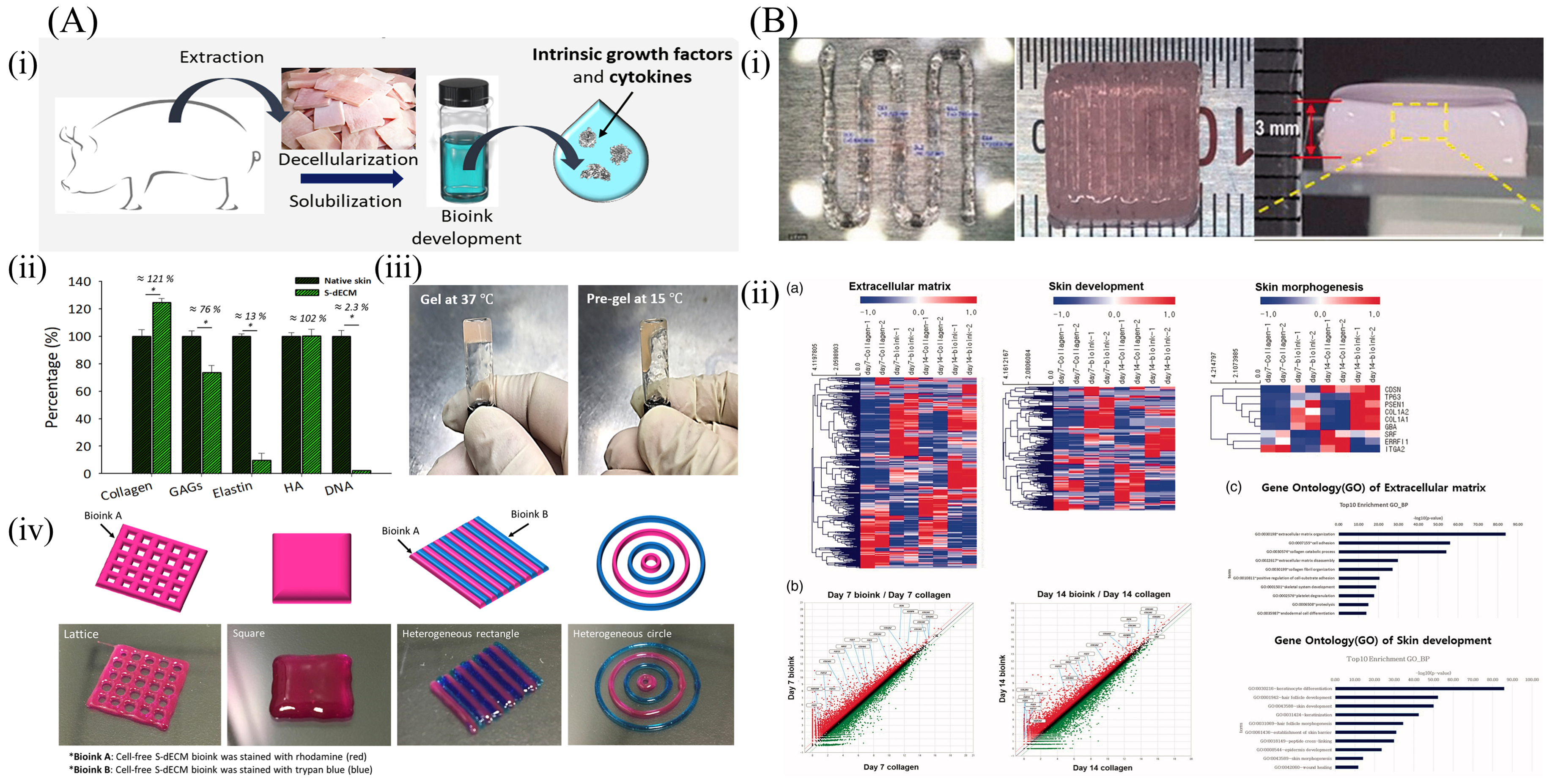
6.3. Skin-Derived dECM Bioinks
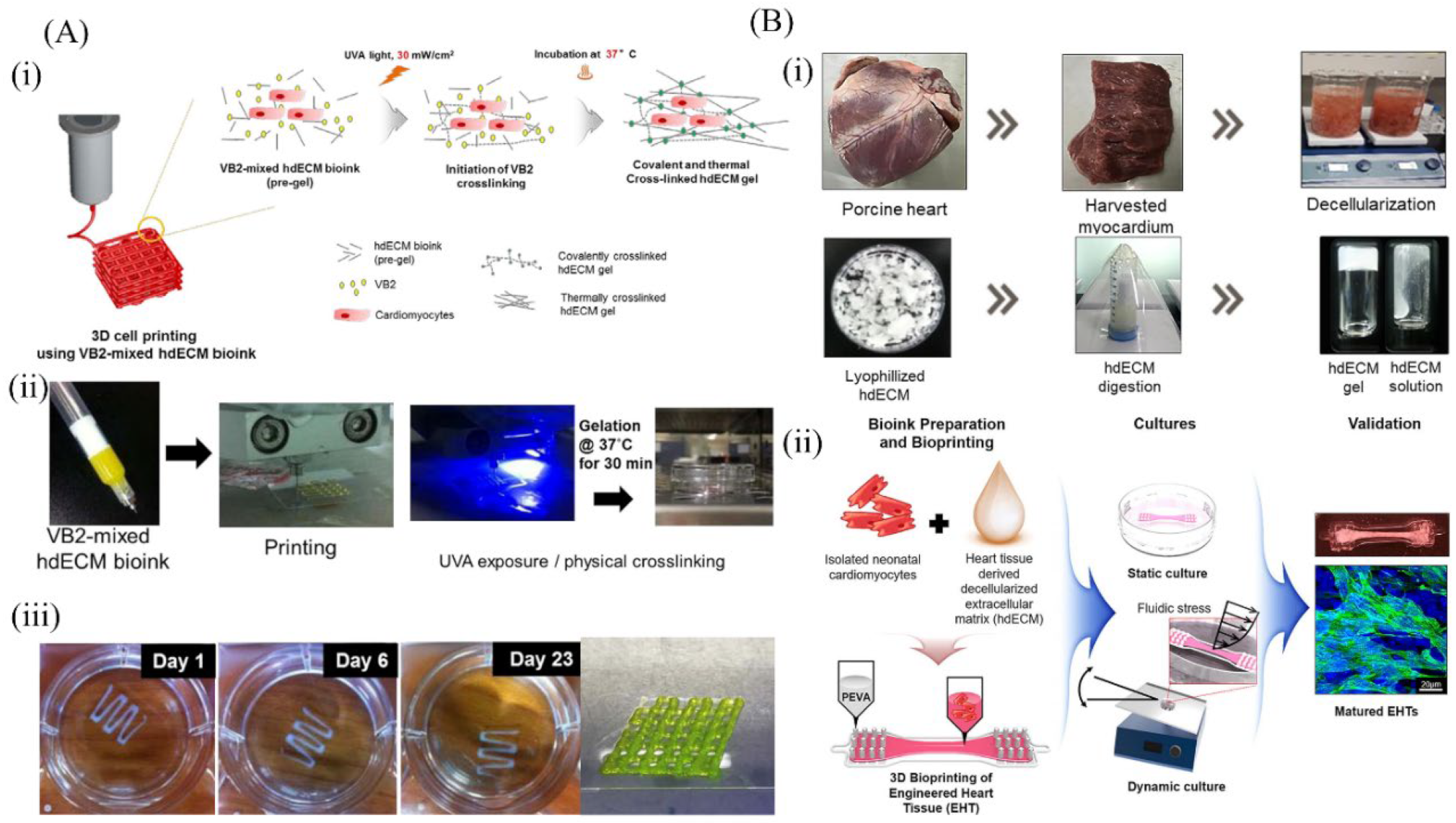
6.4. Cardiac-Derived dECM Bioinks
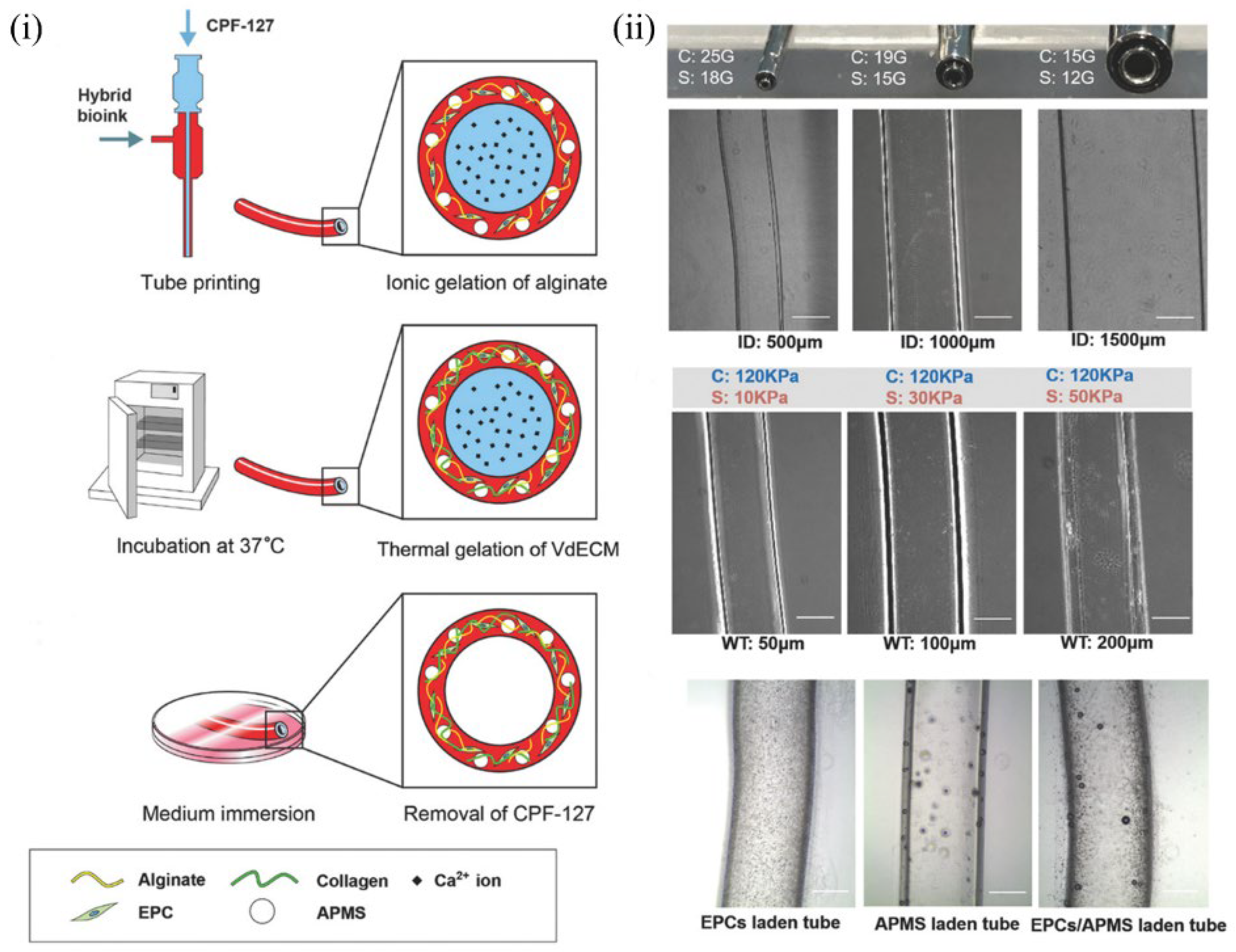
6.5. Blood Vessel–Derived dECM Bioinks
6.6. Kidney-Derived dECM Bioinks
7. Possible Challenges and Solutions
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, W.; Yong, T.; Teo, W.E.; Ma, Z.W.; Ramakrishna, S. Fabrication and endothelialization of collagen-blended biodegradable polymer nanofibers: Potential vascular graft for blood vessel tissue engineering. Tissue Eng. 2005, 11, 1574–1588. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Lu, B.; Zhao, Y.; Jiang, X. Fabrication of aligned fibirous arrays by magnetic electrospinning. Adv. Mater. 2007, 19, 3702. [Google Scholar] [CrossRef]
- Badami, A.S.; Kreke, M.R.; Thompson, M.S.; Riffle, J.S.; Goldstein, A.S. Effect of fiber diameter on spreading, proliferation, and differentiation of osteoblastic cells on electrospun poly (lactic acid) substrates. Biomaterials 2006, 27, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Jeong, G.S.; Choi, Y.Y.; Lee, K.H.; Khademhosseini, A.; Lee, S. Digitally tunable physicochemical coding of material composition and topography in continuous microfibres. Nat. Mater. 2011, 10, 877–883. [Google Scholar] [CrossRef]
- Yamada, M.; Utoh, R.; Ohashi, K.; Tatsumi, K.; Yamato, M.; Okano, T.; Seki, M. Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions. Biomaterials 2012, 33, 8304–8315. [Google Scholar] [CrossRef] [PubMed]
- Onoe, H.; Okitsu, T.; Itou, A.; Kato-Negishi, M.; Gojo, R.; Kiriya, D.; Sato, K.; Miura, S.; Iwanaga, S.; Kuribayashi-Shigetomi, K.; et al. Metre-long cell-laden microfibres exhibit tissue morphologies and functions. Nat. Mater. 2013, 12, 584–590. [Google Scholar] [CrossRef]
- Shao, L.; Gao, Q.; Xie, C.; Fu, J.; Xiang, M.; Liu, Z.; Xiang, L.; He, Y. Sacrificial microgel-laden bioink-enabled 3D bioprinting of mesoscale pore networks. Bio-Des. Manuf. 2020, 3, 30–39. [Google Scholar] [CrossRef]
- Gao, Q.; Niu, X.; Shao, L.; Zhou, L.; Lin, Z.; Sun, A.; Fu, J.; Chen, Z.; Hu, J.; Liu, Y.; et al. 3D printing of complex GelMA-based scaffolds with nanoclay. Biofabrication 2019, 11, 035006. [Google Scholar] [CrossRef]
- Xie, C.; Gao, Q.; Wang, P.; Shao, L.; Yuan, H.; Fu, J.; Chen, W.; He, Y. Structure-induced cell growth by 3D printing of heterogeneous scaffolds with ultrafine fibers. Mater. Des. 2019, 181, 108092. [Google Scholar] [CrossRef]
- Suntornnond, R.; An, J.; Chua, C.K. Bioprinting of Thermoresponsive Hydrogels for next generation tissue engineering: A review. Macromol. Mater. Eng. 2017, 302, 1600266. [Google Scholar] [CrossRef]
- Li, X.; Su, Y.; Liu, S.; Tan, L.; Mo, X.; Ramakrishna, S. Encapsulation of proteins in poly(L-lactide-co-caprolactone) fibers by emulsion electrospinning. Colloids Surf. B 2010, 75, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Wang, D. Decellularized orthopaedic tissue-engineered grafts: Biomaterial scaffolds synthesised by therapeutic cells. Biomater. Sci. 2018, 6, 2798–2811. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Yang, R.; Ma, X.; Chen, W.; Liu, S.; Liu, X.; Cai, X.; Xu, H.; Chi, B. Bionic Poly (gamma-Glutamic Acid) electrospun fibrous scaffolds for preventing hypertrophic scars. Adv. Healthc. Mater. 2019, 8, 1900123. [Google Scholar] [CrossRef]
- Patenaude, M.; Smeets, N.M.B.; Hoare, T. Designing Injectable, Covalently cross-linked hydrogels for biomedical applications. Macromol. Rapid Commun. 2014, 35, 598–617. [Google Scholar] [CrossRef] [PubMed]
- Hussey, G.S.; Cramer, M.C.; Badylak, S.F. Extracellular matrix bioscaffolds for building gastrointestinal tissue. Cell. Mol. Gastroenter. 2018, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalcanti-Adam, E.A.; Micoulet, A.; Blummel, J.; Auernheimer, J.; Kessler, H.; Spatz, J.P. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell Biol. 2006, 85, 219–224. [Google Scholar] [CrossRef]
- Kim, D.; Provenzano, P.P.; Smith, C.L.; Levchenko, A. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012, 197, 351–360. [Google Scholar] [CrossRef]
- Rana, D.; Zreiqat, H.; Benkirane-Jessel, N.; Ramakrishna, S.; Ramalingam, M. Development of decellularized scaffolds for stem cell-driven tissue engineering. J. Tissue Eng. Regen. Med. 2017, 11, 942–965. [Google Scholar] [CrossRef]
- Zhang, B.; Luo, Y.; Ma, L.; Gao, L.; Li, Y.; Xue, Q.; Yang, H.; Cui, Z. 3D bioprinting: An emerging technology full of opportunities and challenges. Bio-Des. Manuf. 2018, 1, 2–13. [Google Scholar] [CrossRef]
- Ahadian, S.; Khademhosseini, A. A perspective on 3D bioprinting in tissue regeneration. Bio-Des. Manuf. 2018, 1, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Groll, J.; Burdick, J.A.; Cho, D.; Derby, B.; Gelinsky, M.; Heilshorn, S.C.; Juengst, T.; Malda, J.; Mironov, V.A.; Nakayama, K.; et al. A definition of bioinks and their distinction from biomaterial inks. Biofabrication 2019, 11, 013001. [Google Scholar] [CrossRef] [PubMed]
- Visscher, D.O.; Lee, H.; Van Zuijlen, P.P.M.; Helder, M.N.; Atala, A.; Yoo, J.J.; Lee, S.J. A photo-crosslinkable cartilage-derived extracellular matrix bioink for auricular cartilage tissue engineering. Acta Biomater. 2021, 121, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, T.; Li, X.; Deng, Q.; Zhao, W.; Lin, B.; Luo, Y.; Zhang, X. A GelMA/DECM/nanoclay composite biomaterial ink for printing 3D scaffolds for primary hepatocytes cultivation. Mater. Lett. 2020, 274, 128034. [Google Scholar] [CrossRef]
- Kim, M.K.; Jeong, W.; Lee, S.M.; Kim, J.B.; Jin, S.; Kang, H. Decellularized extracellular matrix-based bio-ink with enhanced 3D printability and mechanical properties. Biofabrication 2020, 12, 025003. [Google Scholar] [CrossRef]
- Brassard, J.A.; Nikolaev, M.; Huebscher, T.; Hofer, M.; Lutolf, M.P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021, 20, 22–29. [Google Scholar] [CrossRef]
- Ha, D.; Chae, S.; Lee, J.Y.; Kim, J.Y.; Yoon, J.; Sen, T.; Lee, S.; Kim, H.J.; Cho, J.H.; Cho, D. Therapeutic effect of decellularized extracellular matrix-based hydrogel for radiation esophagitis by 3D printed esophageal stent. Biomaterials 2021, 266, 120477. [Google Scholar] [CrossRef]
- Aamodt, J.M.; Grainger, D.W. Extracellular matrix-based biomaterial scaffolds and the host response. Biomaterials 2016, 86, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, K.; Kumar, P.; Choonara, Y.E.; Du Toit, L.C.; Pillay, V. Three-dimensional printing of extracellular matrix (ECM)-mimicking scaffolds: A critical review of the current ECM materials. J. Biomed. Mater. Res. A 2020, 108, 2324–2350. [Google Scholar] [CrossRef]
- Patil, V.A.; Masters, K.S. Engineered collagen matrices. Bioengineering 2020, 7, 163. [Google Scholar] [CrossRef]
- Reddel, C.J.; Weiss, A.S.; Burgess, J.K. Elastin in asthma. Pulm. Pharmacol. Ther. 2012, 25, 144–153. [Google Scholar] [CrossRef]
- Vakonakis, I.; Staunton, D.; Ellis, I.R.; Sarkies, P.; Flanagan, A.; Schor, A.M.; Schor, S.L.; Campbell, I.D. Motogenic sites in human fibronectin are masked by long range interactions. J. Biol. Chem. 2009, 284, 15668–15675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belyanina, I.V.; Zamay, T.N.; Zamay, G.S.; Zamay, S.S.; Kolovskaya, O.S.; Ivanchenko, T.I.; Denisenko, V.V.; Kirichenko, A.K.; Glazyrin, Y.E.; Garanzha, I.V.; et al. In vivo cancer cells elimination guided by aptamer-functionalized gold-coated magnetic nanoparticles and controlled with low frequency alternating magnetic field. Theranostics 2017, 7, 3326–3337. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.P.; Nandini, C.D.; Salimath, P.V. Structural characterization of N-linked oligosaccharides of laminin from rat kidney: Changes during diabetes and modulation by dietary fiber and butyric acid. FEBS J. 2011, 278, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lin, L.; Huang, H.; Linhardt, R.J. Chemoenzymatic synthesis of glycosaminoglycans. Acc. Chem. Res. 2020, 53, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Farage, M.A.; Miller, K.W.; Elsner, P.; Maibach, H.I. Intrinsic and extrinsic factors in skin ageing: A review. Int. J. Cosmet. Sci. 2008, 30, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Pastwinska, J.; Zelechowska, P.; Walczak-Drzewiecka, A.; Brzezinska-Blaszczyk, E.; Dastych, J. The art of mast cell adhesion. Cells 2020, 9, 2664. [Google Scholar] [CrossRef]
- Green, H.J.; Brown, N.H. Integrin intracellular machinery in action. Exp. Cell Res. 2019, 378, 226–231. [Google Scholar] [CrossRef]
- Bianconi, D.; Unseld, M.; Prager, G.W. Integrins in the spotlight of cancer. Int. J. Mol. Sci. 2016, 17, 2037. [Google Scholar] [CrossRef] [Green Version]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—A model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Vasquez, C.G.; Martin, A.C. Force transmission in epithelial tissues. Dev. Dynam. 2016, 245, 361–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, J.J.; Adair-Kirk, T.L.; Kelley, D.G.; DeMello, D.; Senior, R.M. Clara cell adhesion and migration to extracellular matrix. Resp. Res. 2008, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.; Han, S.; Kim, D. Cell-ECM contact-guided intracellular polarization is mediated via lamin A/C dependent nucleus-cytoskeletal connection. Biomaterials 2021, 268, 120548. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.; Lin, H.; Lin, S. In Situ Altering of the extracellular matrix to direct the programming of endogenous stem cells. Stem Cells 2014, 32, 1989–1990. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G. Recent Developments in Breast Muscle Myopathies Associated with Growth in Poultry. Annu. Rev. Anim. Biosci. 2019, 7, 289–308. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Y.; Qian, W.X.; Xu, Y.H.; Gopalakrishnan, S.; Wang, J.Q.; Lam, Y.W.; Pang, S.W. Control of cell migration direction by inducing cell shape asymmetry with patterned topography. J. Biomed. Mater. Res. A 2015, 103, 2383–2393. [Google Scholar] [CrossRef]
- Yeung, V.; Zhang, T.C.; Yuan, L.; Parekh, M.; Cortinas, J.A.; Delavogia, E.; Hutcheon, A.E.K.; Guo, X.; Ciolino, J.B. Extracellular vesicles secreted by corneal myofibroblasts promote corneal epithelial cell migration. Int. J. Mol. Sci. 2022, 23, 3136. [Google Scholar] [CrossRef] [PubMed]
- Buckenmeyer, M.J.; Meder, T.J.; Prest, T.A.; Brown, B.N. Decellularization techniques and their applications for the repair and regeneration of the nervous system. Methods 2020, 171, 41–61. [Google Scholar] [CrossRef]
- Cebotari, S.; Tudorache, I.; Jaekel, T.; Hilfiker, A.; Dorfman, S.; Ternes, W.; Haverich, A.; Lichtenberg, A. Detergent decellularization of heart valves for tissue engineering: Toxicological effects of residual detergents on human endothelial cells. Artif. Organs 2010, 34, 206–209. [Google Scholar] [CrossRef]
- Nonaka, P.N.; Campillo, N.; Uriarte, J.J.; Garreta, E.; Melo, E.; De Oliveira, L.V.F.; Navajas, D.; Farre, R. Effects of freezing/thawing on the mechanical properties of decellularized lungs. J. Biomed. Mater. Res. A 2014, 102, 413–419. [Google Scholar] [CrossRef]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef] [Green Version]
- Seo, Y.; Jung, Y.; Kim, S.H. Decellularized heart ECM hydrogel using supercritical carbon dioxide for improved angiogenesis. Acta Biomater. 2018, 67, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.M.; Ribeiro, N.; Silva, I.V.; Dias, J.R.; Alves, N.M.; Oliveira, A.L. Fast decellularization process using supercritical carbon dioxide for trabecular bone. J. Supercrit. Fluid. 2021, 172, 105194. [Google Scholar] [CrossRef]
- Wang, J.K.; Luo, B.; Guneta, V.; Li, L.; Foo, S.E.M.; Dai, Y.; Tan, T.T.Y.; Tan, N.S.; Choong, C.; Wong, M.T.C. Supercritical carbon dioxide extracted extracellular matrix material from adipose tissue. Mater. Sci. Eng. C 2017, 75, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Jungebluth, P.; Go, T.; Asnaghi, A.; Bellini, S.; Martorell, J.; Calore, C.; Urbani, L.; Ostertag, H.; Mantero, S.; Conconi, M.T.; et al. Structural and morphologic evaluation of a novel detergent-enzymatic tissue-engineered tracheal tubular matrix. J. Thorac. Cardiovasc. Surg. 2009, 138, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wei, X.; Yi, W.; Gu, C.; Kang, X.; Liu, Y.; Li, Q.; Yi, D. RGD-modified acellular bovine pericardium as a bioprosthetic scaffold for tissue engineering. J. Mater. Sci. Mater. Med. 2009, 20, 2327–2336. [Google Scholar] [CrossRef]
- Reing, J.E.; Brown, B.N.; Daly, K.A.; Freund, J.M.; Gilbert, T.W.; Hsiong, S.X.; Huber, A.; Kullas, K.E.; Tottey, S.; Wolf, M.T.; et al. The effects of processing methods upon mechanical and biologic properties of porcine dermal extracellular matrix scaffolds. Biomaterials 2010, 31, 8626–8633. [Google Scholar] [CrossRef] [Green Version]
- Prasertsung, I.; Kanokpanont, S.; Bunaprasert, T.; Thanakit, V.; Damrongsakkul, S. Development of acellular dermis from porcine skin using periodic pressurized technique. J. Biomed. Mater. Res. B 2008, 85B, 210–219. [Google Scholar] [CrossRef]
- Chakraborty, J.; Roy, S.; Ghosh, S. Regulation of decellularized matrix mediated immune response. Biomater. Sci. 2020, 8, 1194–1215. [Google Scholar] [CrossRef]
- Nakayama, K.H.; Batchelder, C.A.; Lee, C.I.; Tarantal, A.F. Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering. Tissue Eng. Part A 2010, 16, 2207–2216. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Williams, J.K.; Greven, M.; Walter, K.A.; Laber, P.W.; Khang, G.; Soker, S. Bioengineering endothelialized neo-corneas using donor-derived corneal endothelial cells and decellularized corneal stroma. Biomaterials 2010, 31, 6738–6745. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Kim, D.-H.; Bansal, S.; Bae, Y.; Mauck, R.L.; Heo, S.-J. Development of a decellularized meniscus matrix-based nanofibrous scaffold for meniscus tissue engineering. Acta Biomater. 2021, 128, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Chen, P.; Chen, Y.; Li, M.; Chen, C.; Lu, H. 3D-printed extracellular matrix/polyethylene glycol diacrylate hydrogel incorporating the anti-inflammatory phytomolecule honokiol for regeneration of osteochondral defects. Am. J. Sport. Med. 2020, 48, 2808–2818. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Luo, C.; Zhai, C.; Li, Z.; Zhang, Y.; Yuan, T.; Dong, S.; Zhang, J.; Fan, W. Crosslinker-free silk/decellularized extracellular matrix porous bioink for 3D bioprinting-based cartilage tissue engineering. Mater. Sci. Eng. C 2021, 118, 111388. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Asawa, Y.; Watanabe, M.; Okubo, R.; Nio, M.; Takato, T.; Hoshi, K.; Hikita, A. The usefulness of the decellularized matrix from three-dimensional regenerative cartilage as a scaffold material. Regen. Ther. 2020, 15, 312–322. [Google Scholar] [CrossRef]
- Zhao, F.; Cheng, J.; Sun, M.; Yu, H.; Wu, N.; Li, Z.; Zhang, J.; Li, Q.; Yang, P.; Liu, Q.; et al. Digestion degree is a key factor to regulate the printability of pure tendon decellularized extracellular matrix bio-ink in extrusion-based 3D cell printing. Biofabrication 2020, 12, 045011. [Google Scholar] [CrossRef]
- Shin, S.; Park, H.Y.; Shin, N.; Jung, D.; Kwon, H.; Kim, J.M.; Wang, S.; Lee, J.; Sung, E.; Park, G.C.; et al. Evaluation of decellularized xenogenic porcine auricular cartilage as a novel biocompatible filler. J. Biomed. Mater. Res. B. 2018, 106, 2708–2715. [Google Scholar] [CrossRef]
- Mao, Q.; Wang, Y.; Li, Y.; Juengpanich, S.; Li, W.; Chen, M.; Yin, J.; Fu, J.; Cai, X. Fabrication of liver microtissue with liver decellularized extracellular matrix (dECM) bioink by digital light processing (DLP) bioprinting. Mater. Sci. Eng. C 2020, 109, 110625. [Google Scholar] [CrossRef]
- Lee, H.; Han, W.; Kim, H.; Ha, D.; Jang, J.; Kim, B.S.; Cho, D. Development of liver decellularized extracellular matrix bioink for three-dimensional cell printing-based liver tissue engineering. Biomacromolecules 2017, 18, 1229–1237. [Google Scholar] [CrossRef]
- Meng, F.; Almohanna, F.; Altuhami, A.; Assiri, A.M.; Broering, D. Vasculature reconstruction of decellularized liver scaffolds via gelatin-based re-endothelialization. J. Biomed. Mater. Res. A 2019, 107, 392–402. [Google Scholar] [CrossRef]
- Loneker, A.E.; Faulk, D.M.; Hussey, G.S.; D’Amore, A.; Badylak, S.F. Solubilized liver extracellular matrix maintains primary rat hepatocyte phenotype in-vitro (vol 104A, pg 957, 2016). J. Biomed. Mater. Res. A 2016, 104, 1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, P.L.; Su, J.; Yan, M.; Meng, F.; Glaser, S.S.; Alpini, G.D.; Green, R.M.; Sosa-Pineda, B.; Shah, R.N. Complex bile duct network formation within liver decellularized extracellular matrix hydrogels. Sci. Rep. 2018, 8, 12220. [Google Scholar] [CrossRef] [PubMed]
- Faulk, D.M.; Londono, R.; Wolf, M.T.; Ranallo, C.A.; Carruthers, C.A.; Wildemann, J.D.; Dearth, C.L.; Badylak, S.F. ECM hydrogel coating mitigates the chronic inflammatory response to polypropylene mesh. Biomaterials 2014, 35, 8585–8595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Fang, H.; Zheng, S.; Li, L.; Jiao, Z.; Wang, H.; Nie, Y.; Liu, T.; Song, K. A biological functional hybrid scaffold based on decellularized extracellular matrix/gelatin/chitosan with high biocompatibility and antibacterial activity for skin tissue engineering. Int. J. Biol. Macromol. 2021, 187, 840–849. [Google Scholar] [CrossRef]
- Lau, C.S.; Hassanbhai, A.; Wen, F.; Wang, D.; Chanchareonsook, N.; Goh, B.T.; Yu, N.; Teoh, S. Evaluation of decellularized tilapia skin as a tissue engineering scaffold. J. Tissue Eng. Regen. Med. 2019, 13, 1779–1791. [Google Scholar] [CrossRef]
- Zhang, Q.; Johnson, J.A.; Dunne, L.W.; Chen, Y.; Iyyanki, T.; Wu, Y.; Chang, E.I.; Branch-Brooks, C.D.; Robb, G.L.; Butler, C.E. Decellularized skin/adipose tissue flap matrix for engineering vascularized composite soft tissue flaps. Acta Biomater. 2016, 35, 166–184. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Ma, H.; Wu, Z.; Zeng, H.; Li, Z.; Wang, Y.; Liu, G.; Xu, B.; Lin, Y.; Zhang, P.; et al. Enhancement of skin wound healing with decellularized scaffolds loaded with hyaluronic acid and epidermal growth factor. Mater. Sci. Eng. C 2014, 44, 440–448. [Google Scholar] [CrossRef]
- Glynn, J.J.; Polsin, E.G.; Hinds, M.T. Crosslinking decreases the hemocompatibility of decellularized, porcine small intestinal submucosa. Acta Biomater. 2015, 14, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Sun, L.; Pang, Y.; Hu, D.; Xu, H.; Mao, S.; Peng, W.; Wang, Y.; Xu, Y.; Zheng, Y.; et al. Three-dimensional bioprinted hepatorganoids prolong survival of mice with liver failure. Gut 2021, 70, 567–574. [Google Scholar] [CrossRef]
- Won, J.; Lee, M.; Kim, M.; Min, K.; Ahn, G.; Han, J.; Jin, S.; Yun, W.; Shim, J. A potential dermal substitute using decellularized dermis extracellular matrix derived bio-ink. Artif. Cells Nanomed. Biotechnol. 2019, 47, 644–649. [Google Scholar] [CrossRef]
- Tong, C.; Li, C.; Xie, B.; Li, M.; Li, X.; Qi, Z.; Xia, J. Generation of bioartificial hearts using decellularized scaffolds and mixed cells. Biomed. Eng. Online 2019, 18, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roderjan, J.G.; De Noronha, L.; Stimamiglio, M.A.; Correa, A.; Leitolis, A.; Bueno, R.R.L.; Da Costa, F.D.A. Structural assessments in decellularized extracellular matrix of porcine semilunar heart valves: Evaluation of cell niches. Xenotransplantation 2019, 26, e12503. [Google Scholar] [CrossRef] [PubMed]
- Roosens, A.; Somers, P.; De Somer, F.; Carriel, V.; Van Nooten, G.; Cornelissen, R. Impact of detergent-based decellularization methods on porcine tissues for heart valve engineering. Ann. Biomed. Eng. 2016, 44, 2827–2839. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, H.; Gong, W.; Li, S.; Li, H.; Wang, Z.; Zhao, Q. Impact of decellularization on porcine myocardium as scaffold for tissue engineered heart tissue. J. Mater. Sci. Mater. Med. 2016, 27, 70. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.C.W.; Wang, Z.; Missinato, M.A.; Park, D.W.; Long, D.W.; Liu, H.; Zeng, X.; Yates, N.A.; Kim, K.; Wang, Y. Decellularized zebrafish cardiac extracellular matrix induces mammalian heart regeneration. Sci. Adv. 2016, 2, e1600844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simsa, R.; Padma, A.M.; Heher, P.; Hellstrom, M.; Teuschl, A.; Jenndahl, L.; Bergh, N.; Fogelstrand, P. Systematic in vitro comparison of decellularization protocols for blood vessels. PLoS ONE 2018, 13, e0209269. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Zhang, H.; Wu, C.; Zhang, J.; Sun, X.; Zhang, H.; Zhu, Z.; Chang, J. Crosslinking of saphenous vein ECM by procyanidins for small diameter blood vessel replacement. J. Biomed. Mater. Res. B 2014, 102, 1190–1198. [Google Scholar] [CrossRef]
- He, M.; Callanan, A.; Lagaras, K.; Steele, J.A.M.; Stevens, M.M. Optimization of SDS exposure on preservation of ECM characteristics in whole organ decellularization of rat kidneys. J. Biomed. Mater. Res. B 2017, 105, 1352–1360. [Google Scholar] [CrossRef]
- Poornejad, N.; Momtahan, N.; Salehi, A.S.M.; Scott, D.R.; Fronk, C.A.; Roeder, B.L.; Reynolds, P.R.; Bundy, B.C.; Cook, A.D. Efficient decellularization of whole porcine kidneys improves reseeded cell behavior. Biomed. Mater. 2016, 11, 025003. [Google Scholar] [CrossRef]
- Keshvari, M.A.; Afshar, A.; Daneshi, S.; Khoradmehr, A.; Baghban, M.; Muhaddesi, M.; Behrouzi, P.; Miri, M.R.; Azari, H.; Nabipour, I.; et al. Decellularization of kidney tissue: Comparison of sodium lauryl ether sulfate and sodium dodecyl sulfate for allotransplantation in rat. Cell Tissue Res. 2021, 386, 365–378. [Google Scholar] [CrossRef]
- Du, C.; Narayanan, K.; Leong, M.F.; Ibrahim, M.S.; Chua, Y.P.; Khoo, V.M.H.; Wan, A.C.A. Functional kidney bioengineering with pluripotent stem-cell-derived renal progenitor cells and decellularized kidney scaffolds. Adv. Healthc. Mater. 2016, 5, 2080–2091. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, F.; Ekhtiari, M.; Moeini Chaghervand, B.; Moradi, L.; Mohammadi, B.; Kajbafzadeh, A. Detection of the residual concentration of sodium dodecyl sulfate in the decellularized whole rabbit kidney extracellular matrix. Cell Tissue Bank. 2022, 23, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Chani, B.; Puri, V.; Sobti, R.C.; Jha, V.; Puri, S. Decellularized scaffold of cryopreserved rat kidney retains its recellularization potential. PLoS ONE 2017, 12, e0173040. [Google Scholar] [CrossRef] [PubMed]
- McKee, R.A.; Wingert, R.A. Repopulating decellularized kidney scaffolds: An avenue for ex vivo organ generation. Materials 2016, 9, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Y.; Liu, S.; Liu, Y.; Sun, C.; Cheng, G.; Luan, Y.; Li, K.; Wang, J.; Xie, X.; Zhao, S. Porcine kidneys as a source of ECM scaffold for kidney regeneration. Mater. Sci. Eng. C 2015, 56, 451–456. [Google Scholar] [CrossRef]
- Ventura, R.D.; Padalhin, A.R.; Park, C.M.; Lee, B.T. Enhanced decellularization technique of porcine dermal ECM for tissue engineering applications. Mater. Sci. Eng. C 2019, 104, 109841. [Google Scholar] [CrossRef]
- Granato, A.E.C.; Da Cruz, E.F.; Rodrigues-Junior, D.M.; Mosini, A.C.; Ulrich, H.; Rodrigues, B.V.M.; Cheffer, A.; Porcionatto, M. A novel decellularization method to produce brain scaffolds. Tissue Cell 2020, 67, 101412. [Google Scholar] [CrossRef]
- Ahmed, E.; Saleh, T.; Yu, L.; Kwak, H.; Kim, B.; Park, K.; Lee, Y.; Kang, B.; Choi, K.; Kang, K.; et al. Micro and ultrastructural changes monitoring during decellularization for the generation of a biocompatible liver. J. Biosci. Bioeng. 2019, 128, 218–225. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Li, L.; Chen, F.; Bao, J.; Li, W. Decellularization of porcine whole lung to obtain a clinical-scale bioengineered scaffold. J. Biomed. Mater. Res. A 2021, 109, 1623–1632. [Google Scholar] [CrossRef]
- Philips, C.; Campos, F.; Roosens, A.; Del Carmen Sanchez-Quevedo, M.; Declercq, H.; Carriel, V. Qualitative and quantitative evaluation of a novel detergent-based method for decellularization of peripheral nerves. Ann. Biomed. Eng. 2018, 46, 1921–1937. [Google Scholar] [CrossRef]
- Kamalvand, M.; Biazar, E.; Daliri-Joupari, M.; Montazer, F.; Rezaei-Tavirani, M.; Heidari-Keshel, S. Design of a decellularized fish skin as a biological scaffold for skin tissue regeneration. Tissue Cell 2021, 71, 101509. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.L.; Castells-Sala, C.; Lopez-Chicon, P.; Nieto-Nicolau, N.; Aiti, A.; Farinas, O.; Casaroli-Marano, R.P.; Porta, O.; Vilarrodona, A. Fast protocol for the processing of split-thickness skin into decellularized human dermal matrix. Tissue Cell 2021, 72, 101572. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tian, Z.; Tian, Q.; Peng, L.; Li, K.; Luo, X.; Wang, D.; Yang, Z.; Jiang, S.; Sui, X.; et al. 3D bioprinting of a biomimetic meniscal scaffold for application in tissue engineering. Bioact. Mater. 2021, 6, 1711–1726. [Google Scholar] [CrossRef]
- Bishop, E.S.; Mostafa, S.; Pakvasa, M.; Luu, H.H.; Lee, M.J.; Wolf, J.M.; Ameer, G.A.; He, T.; Reid, R.R. 3-D bioprinting technologies in tissue engineering and regenerative medicine: Current and future trends. Genes Dis. 2017, 4, 185–195. [Google Scholar] [CrossRef]
- Min, D.; Lee, W.; Bae, I.; Lee, T.R.; Croce, P.; Yoo, S. Bioprinting of biomimetic skin containing melanocytes. Exp. Dermatol. 2018, 27, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Antich, C.; De Vicente, J.; Jimenez, G.; Chocarro, C.; Carrillo, E.; Montanez, E.; Galvez-Martin, P.; Antonio Marchal, J. Bio-inspired hydrogel composed of hyaluronic acid and alginate as a potential bioink for 3D bioprinting of articular cartilage engineering constructs. Acta Biomater. 2020, 106, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.P.; Bandyopadhyay, A.; Mandal, B.B. 3D Bioprinting Using cross-linker-free silk-gelatin bioink for cartilage tissue engineering. ACS Appl. Mater. Interfaces 2019, 11, 33684–33696. [Google Scholar] [CrossRef]
- Kang, D.; Hong, G.; An, S.; Jang, I.; Yun, W.; Shim, J.; Jin, S. Bioprinting of multiscaled hepatic lobules within a highly vascularized construct. Small 2020, 16, e1905505. [Google Scholar] [CrossRef]
- Yang, T.; Lin, S.; Xie, Q.; Ouyang, W.; Tan, T.; Li, J.; Chen, Z.; Yang, J.; Wu, H.; Pan, J.; et al. Impact of 3D printing technology on the comprehension of surgical liver anatomy. Surg. Endosc. 2019, 33, 411–417. [Google Scholar] [CrossRef]
- Xie, F.; Sun, L.; Pang, Y.; Xu, G.; Jin, B.; Xu, H.; Lu, X.; Xu, Y.; Du, S.; Wang, Y.; et al. Three-dimensional bio-printing of primary human hepatocellular carcinoma for personalized medicine. Biomaterials 2021, 265, 120416. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Jeong, Y.H.; Kim, Y.; Choi, Y.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016, 34, 312. [Google Scholar] [CrossRef] [PubMed]
- Gungor-Ozkerim, P.S.; Inci, I.; Zhang, Y.S.; Khademhosseini, A.; Dokmeci, M.R. Bioinks for 3D bioprinting: An overview. Biomater. Sci. 2018, 6, 915–946. [Google Scholar] [CrossRef] [Green Version]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The bioink: A comprehensive review on bioprintable materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Bibb, R.; Harris, R. Engineering design of artificial vascular junctions for 3D printing. Biofabrication 2016, 8, 025018. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Gao, X.; Wang, J.; Chen, H.; Tong, C.; Tan, Y.; Tan, Z. Triple-layer vascular grafts fabricated by combined e-jet 3D printing and electrospinning. Ann. Biomed. Eng. 2018, 46, 1254–1266. [Google Scholar] [CrossRef]
- Jia, L.T.; Hua, Y.J.; Zeng, J.S.; Liu, W.S.; Wang, D.; Zhou, G.D.; Liu, X.; Jiang, H.Y. Bioprinting and regeneration of auricular cartilage using a bioactive bioink based on microporous photocrosslinkable acellular cartilage matrix. Bioact. Mater. 2020, 16, 66–81. [Google Scholar] [CrossRef]
- Pi, Q.; Maharjan, S.; Yan, X.; Liu, X.; Singh, B.; Van Genderen, A.M.; Robledo-Padilla, F.; Parra-Saldivar, R.; Hu, N.; Jia, W.; et al. Digitally tunable microfluidic bioprinting of multilayered cannular tissues. Adv. Mater. 2018, 30, e1706913. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, H.; Huang, X.; Hang, R.; Zhang, X.; Wang, Y.; Yao, X. DLP printing photocurable chitosan to build bio-constructs for tissue engineering. Carbohydr. Polym. 2020, 235, 115970. [Google Scholar] [CrossRef]
- Ozbolat, I.T.; Hospodiuk, M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials 2016, 76, 321–343. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Schuurmans, C.C.L.; Van Genderen, A.M.; Cao, X.; Li, W.; Cheng, F.; He, J.J.; Lopez, A.; Huerta, V.; Manriquez, J.; et al. Complexation-induced resolution enhancement of 3D-printed hydrogel constructs. Nat. Commun. 2020, 11, 1267. [Google Scholar] [CrossRef]
- Klak, M.; Kowalska, P.; Dobrzanski, T.; Tymicki, G.; Cywoniuk, P.; Gomolka, M.; Kosowska, K.; Bryniarski, T.; Berman, A.; Dobrzyn, A.; et al. Bionic organs: Shear forces reduce pancreatic islet and mammalian cell viability during the process of 3D bioprinting. Micromachines 2021, 12, 304. [Google Scholar] [CrossRef]
- Shin, Y.J.; Shafranek, R.T.; Tsui, J.H.; Walcott, J.; Nelson, A.; Kim, D.-H. 3D bioprinting of mechanically tuned bioinks derived from cardiac decellularized extracellular matrix. Acta Biomater. 2020, 119, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Ahn, G.; Kim, C.; Lee, J.-S.; Lee, I.-G.; An, S.-H.; Yun, W.-S.; Kim, S.-Y.; Shim, J.-H. Synergistic Effects of Beta Tri-Calcium Phosphate and Porcine-Derived Decellularized Bone Extracellular Matrix in 3D-Printed Polycaprolactone Scaffold on Bone Regeneration. Macromol. Biosci. 2018, 18, e1800025. [Google Scholar] [CrossRef] [PubMed]
- Rueda-Gensini, L.; Serna, J.A.; Cifuentes, J.; Cruz, J.C.; Muñoz-Camargo, C. Graphene oxide-embedded extracellular matrix-derived hydrogel as a multiresponsive platform for 3D bioprinting applications. Int. J. Bioprint. 2021, 7, 353. [Google Scholar] [CrossRef]
- Thanh, L.C.; Garima, T.; Myeongki, P.; Sang-Ho, B.; Byong-Taek, L. In-vitro and in-vivo biocompatibility of decmalginate as a promising candidate in cell delivery for kidney regeneration. Int. J. Biol. Macromol. 2022, 211, 616–625. [Google Scholar] [CrossRef]
- Kort-Mascort, J.; Bao, G.; Elkashty, O.; Flores-Torres, S.; Munguia-Lopez, J.G.; Jiang, T.; Ehrlicher, A.J.; Mongeau, L.; Tran, S.D.; Kinsella, J.M. Decellularized Extracellular Matrix Composite Hydrogel Bioinks for the Development of 3D Bioprinted Head and Neck in Vitro Tumor Models. ACS Biomater. Sci. Eng. 2021, 7, 5288–5300. [Google Scholar] [CrossRef]
- Elomaa, L.; Keshi, E.; Sauer, I.M.; Weinhart, M. Development of GelMA/PCL and dECM/PCL resins for 3D printing of acellular in vitro tissue scaffolds by stereolithography. Mater. Sci. Eng. C 2020, 112, 110958. [Google Scholar] [CrossRef]
- Basara, G.; Ozcebe, S.; Ellis, B.; Zorlutuna, P. Tunable Human Myocardium Derived Decellularized Extracellular Matrix for 3D Bioprinting and Cardiac Tissue Engineering. Gels 2021, 7, 70. [Google Scholar] [CrossRef]
- Tang-Quan, K.R.; Xi, Y.; Hochman-Mendez, C.; Xiang, Q.; Lee, P.; Sampaio, L.C.; Taylor, D.A. Gelatin promotes cell reten-tion within decellularized heart extracellular matrix vasculature and parenchyma. Cell. Mol. Bioeng. 2020, 13, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Veiga, A.; Silva, I.V.; Duarte, M.M.; Oliveira, A.L. Current Trends on Protein Driven Bioinks for 3D Printing. Pharmaceutics 2021, 13, 1444. [Google Scholar] [CrossRef] [PubMed]
- Gudapati, H.; Dey, M.; Ozbolat, I. A comprehensive review on droplet-based bioprinting: Past, present and future. Biomaterials 2016, 102, 20–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-González, A.C.; Téllez-Jurado, L.; Rodríguez-Lorenzo, L.M. Alginate hydrogels for bone tissue engineering, from injectables to bioprinting: A review. Carbohydr. Polym. 2019, 229, 115514. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Ma, X.; Gou, M.; Mei, D.; Zhang, K.; Chen, S. 3D printing of functional biomaterials for tissue engineering. Curr. Opin. Biotechnol. 2016, 40, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, B.D.; Allen, J.B. Vascular Endothelial Cell Behavior in Complex Mechanical Microenvironments. ACS Biomater. Sci. Eng. 2018, 4, 3818–3842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, J.; Jiang, Q.; Ding, X.; Li, Y.; Chen, C.; Yang, W.; Chen, S. Highly biosafe biomimetic stem cell mem-brane-disguised nanovehicles for cartilage regeneration. J. Mater. Chem. B 2020, 8, 8884–8893. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.; Lee, S.; Choi, Y.; Hong, D.H.; Gao, G.; Wang, J.H.; Cho, D. 3d cell-printing of biocompatible and functional meniscus constructs using meniscus-derived bioink. Biomaterials 2020, 267, 120466. [Google Scholar] [CrossRef]
- Xu, D.; Bartelt, S.M.; Rasoulinejad, S.; Chen, F.; Wegner, S.V. Green light lithography: A general strategy to create active protein and cell micropatterns. Mater. Horiz. 2019, 6, 1222–1229. [Google Scholar] [CrossRef] [Green Version]
- Lang, R.; Stern, M.M.; Smith, L.; Liu, Y.; Bharadwaj, S.; Liu, G.; Baptista, P.M.; Bergman, C.R.; Soker, S.; Yoo, J.J.; et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials 2011, 32, 7042–7052. [Google Scholar] [CrossRef]
- Ahn, G.; Min, K.; Kim, C.; Lee, J.; Kang, D.; Won, J.; Cho, D.; Kim, J.; Jin, S.; Yun, W.; et al. Precise stacking of decellularized extracellular matrix based 3d cell-laden constructs by a 3d cell printing system equipped with heating modules. Sci. Rep. 2017, 7, 8624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.S.; Kwon, Y.W.; Kong, J.-S.; Park, G.T.; Gao, G.; Han, W.; Kim, M.-B.; Lee, H.; Kim, J.H.; Cho, D.-W. 3D cell printing of in vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: A step towards advanced skin tissue engineering. Biomaterials 2018, 168, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Park, H.-J.; Kim, S.-W.; Kim, H.; Park, J.Y.; Na, S.J.; Kim, H.J.; Park, M.N.; Choi, S.H.; Park, S.H.; et al. 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 2017, 112, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Kim, T.G.; Kim, B.S.; Kim, S.-W.; Kwon, S.-M.; Cho, D.-W. Tailoring mechanical properties of decellularized extracellular matrix bioink by vitamin B2-induced photo-crosslinking. Acta Biomater. 2016, 33, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kim, S.-W.; Choi, Y.-J.; Lee, S.; Lee, S.-H.; Kong, J.-S.; Park, H.-J.; Cho, D.-W.; Jang, J. Decellularized extracellular matrix bioinks and the external stimuli to enhance cardiac tissue development in vitro. Acta Biomater. 2019, 95, 188–200. [Google Scholar] [CrossRef]
- Gao, G.; Lee, J.H.; Jang, J.; Lee, D.H.; Kong, J.; Kim, B.S.; Choi, Y.; Jang, W.B.; Hong, Y.J.; Kwon, S.; et al. Tissue engineered bio-blood-vessels constructed using a tissue-specific bioink and 3d coaxial cell printing technique: A novel therapy for ischemic disease. Adv. Funct. Mater. 2017, 27, 1700798. [Google Scholar] [CrossRef]
- Ali, M.; Pr, A.K.; Yoo, J.J.; Zahran, F.; Atala, A.; Lee, S.J. A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation. Adv. Healthc. Mater. 2019, 8, 1800992. [Google Scholar] [CrossRef]
- Meihan, T.; Tianrang, A.; Xiaoyan, M.; Xinzhu, Y.; Rabia, J.; Weijian, H.; Yang, W.; Cong, S.; Shuang, L.; Tianhao, Y.; et al. Sterilization and disinfection methods for decellularized matrix materials: Review, consideration and proposal. Bioact. Mater. 2021, 9, 2927–2945. [Google Scholar] [CrossRef]
- Uriarte, J.J.; Nonaka, P.N.; Campillo, N.; Palma, R.K.; Melo, E.; De Oliveira, L.V.F.; Navajas, D.; Farré, R. Mechanical prop-erties of acellular mouse lungs after sterilization by gamma irradiation. J. Mech. Behav. Biomed. 2014, 40, 168–177. [Google Scholar] [CrossRef]
- Rosario, D.J.; Reilly, G.C.; Salah, E.A.; Glover, M.; Bullock, A.J.; MacNeil, S. Decellularization and sterilization of porcine urinary bladder matrix for tissue engineering in the lower urinary tract. Regen. Med. 2008, 3, 145–156. [Google Scholar] [CrossRef]
- Gilbert, T.W.; Sellaro, T.L.; Badylak, S.F. Decellularization of tissues and organs. Biomaterials 2006, 27, 3675–3683. [Google Scholar] [CrossRef]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.N.; Zheng, Q.; Wu, Z.L. Recent advances in 3D printing of tough hydrogel: A review. Compos. Part B Eng. 2022, 238, 109895. [Google Scholar] [CrossRef]
- Yu, C.; Ma, X.; Zhu, W.; Wang, P.; Miller, K.L.; Stupin, J.; Koroleva-Maharajh, A.; Hairabedian, A.; Chen, S. Scanningless and continuous 3D bioprinting of human tissues with decellularized extracellular matrix. Biomaterials 2019, 194, 1–13. [Google Scholar] [CrossRef]
- Fitzpatrick, V.; Martín-Moldes, Z.; Deck, A.; Torres-Sanchez, R.; Valat, A.; Cairns, D.; Li, C.; Kaplan, D.L. Functionalized 3D-printed silk-hydroxyapatite scaffolds for enhanced bone regeneration with innervation and vascularization. Biomaterials 2021, 276, 120995. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.; Mitra, I.; Bose, S. 3D Printing for Bone Regeneration. Curr. Osteoporos. Rep. 2020, 18, 505–514. [Google Scholar] [CrossRef]




| Tissue or Organ Sources | Decellularized Method | Mode of Digestion | Ref. |
|---|---|---|---|
| Porcine lateral and medial menisci | Frozen for 5 min and thawed at 21 °C for 10 min 6 times; 0.25% trypsin for 8 h; 3% SDS for 72 h; 50 U/mL DNAse in PBS for 48 h | 0.1% peracetic acid | [62] |
| Porcine cartilage tissue | Freeze–thaw cycles 3 times; 1% Triton X-100 for 1 d; immersed in 1% SDS for 24 h; 200 U/mL DNase I for 12 h | 0.5 M acetic acid with 30 mg of pepsin for 48 h | [63] |
| Goat articular cartilage tissue | 0.1% EDTA and 3.5% PMSF for 24 h; 1% Triton X-100 in Tris-HCl (pH = 7.5) with a protease inhibitor cocktail for 24 h; 50 U/mL DNAse and 1 U/mL RNAse for 12 h | 1 mL of 0.1 M HCl containing 1 mg of pepsin for 48 h | [64] |
| Human auricular cartilage | 4% SDS for 3 h; 1000 U/mL DNase for 3 h | - | [65] |
| Porcine tendon tissues | 100% acetone for 30 min; 0.25% trypsin-EDTA; 2% SDS for 96 h | 3 mg mL−1 pepsin in 0.1 M HCl | [66] |
| Porcine auricular cartilage | Immersed in 0.02% Tris/EDTA with protease inhibitor for 48 h; 1% Triton X-100; incubated with DNAse/RNAse (15 μg/mL) for 24 h; retreated with 0.02% Tris/EDTA solution for 48 h | - | [67] |
| Porcine liver | 0.025% trypsin for 30 min 1% Triton solution for 24 h; 2% SDS for 36 h | Digested in 0.5 M acetic acid and pepsin solution for 96 h | [68] |
| Rat liver | 1% Triton X-100 for 2 h; 0.1% SDS for 1 h; 750 U/mL DNAse and 25 U/mL RNAse for 30 min | Digested in 1 mg/mL of HCl (0.1 M) of pepsin for 72 h | [24,69] |
| Porcine liver | 0.5% Triton X-100 for 9 h; 1% SDS for 3 h | - | [69] |
| Rat liver | 1% Triton x-100 with 0.1% NH4OH (15 mL/min, 1 h; 20 mL/min, 2 h); sterile DI water (5 mL/min, 40 min; 15 mL/min, 15 min; 20 mL/min, 45 min); 0.1% peracetic acid (PAA) in 4% alcohol (5 mL/min, 40 min); submerged in PAA (30 min); sterile DI water (5 mL/min, overnight) | - | [70] |
| Porcine, canine, human, rat liver | Exposed the liver tissue to trypsin/EGTA and Triton X-100 | Digested in pepsin solution | [71] |
| Porcine liver | 0.1% SDS wash overnight | Digested at a 10 mg/mL dECM and 1 mg/mL pepsin at 0.01 M HCl for 48 h | [72] |
| Porcine skin | 0.25% trypsin for 6 h; 70% ethanol for 10 h; 3% H2O2 for 15 min; 1% Triton X-100 in 0.26% EDTA/0.69% Tris for 6 h with a solution change for an additional 16 h; 0.1% peracetic acid/4% ethanol for 2 h | Digested in a 1 mg/mL pepsin solution in 0.01 N HCl for 48 h at 10 mg ECM/mL solution | [73] |
| Porcine skin | 0.25% trypsin for 6 h; 1% Triton X-100 for 24 h; 10% isopropanol for 24 h; 30 U/mL DNase for 24 h; 0.1% peracetic acid in 4% ethanol for 2 h | Digested in papain solution (125 μg/mL) for 16 h | [74] |
| Nile tilapia skin | 2.5 U/mL disperse for 3 h; 1% SDS for 6 h; 25 U/mL Pierce Universal Nuclease for 3 h; 1% SDS for 1 h | - | [75] |
| Groin skin | Cycle freeze–thaw 3 times; 0.25% trypsin/EDTA for 2 h; processed with isopropanol overnight; treated with 1% Triton X-100 for 48 h | - | [76] |
| Porcine peritoneum | Treated with a solution (pH 5.6) containing 2% SDS and 0.3% NaCl; ultrasonic treatment for 24 h; | - | [77] |
| Porcine small intestinal submucosa | Treated with mechanical removal of the tunica mucosa, the tunica serosa, and the tunica muscularis externa; treated with peracetic acid to remove remaining cells, RNA, and DNA | - | [78] |
| Porcine skin | 0.25 wt% trypsin and 1 mM EDTA for 6 h; 1 wt% TritonX-100 for 24 h; 30 U/mL DNase for 24 h | 0.5 M acetic acid solution containing 15 mg of pepsin per 100 mg dECM for 120 h | [79] |
| Porcine lateral and medial menisci | Frozen in liquid nitrogen for 5 min and then thawed at 21 °C for 10 min repeated 6 times; 0.25% (w/v) trypsin for 8 h; 3% (w/v) sodium deoxycholate for 3 d; 50 U/mL DNAse for 48 h | Lyophilized and pulverized into fine powder | [80] |
| Rat heart | Perfused through the ascending aorta with 200 mL of PBS containing heparin (20 U/mL) and 10 mM adenosine followed by 0.1% SDS, deionized water, 1% Triton X-100, 100 U/mL penicillin-G (Gibco), 100 U/mL streptomycin, and 100 U/mL amphotericin B | - | [81] |
| Porcine heart | 0.1% SDS containing 7 mmol/L EDTA for 24 h, washed with 70% ethanol | 2.0 mL of 6.0 N HCl for 24 h | [82] |
| porcine aortic valves and pericardia | 5 mM Tris buffer with 1% Triton X-100 for 24 h; HBSS medium supplemented with 100 mg/L DNase, 20 mg/L RNase and 100 mg/L trypsin for 90 min; new 5 mM Tris buffer with 1% Triton X-100 for 24 h | - | [83] |
| Porcine myocardium | PBS solution with 1.0% Triton X-100 for 72 h; 20 mg/mL ribonuclease A and 0.2 mg/mL deoxyribonuclease for 48 h | 0.05% collagenase, type IV, 0.5 mg/mL pancreatin, 1 mg/mL BSA solution | [84] |
| Zebrafish ventricular wall | Repeated freeze–thaw cycles, red blood cells, and DNA/RNA are removed by the erythrolysis buffer and deoxyribonuclease/ribonuclease | Mechanically ground into fine powders in liquid nitrogen | [85] |
| Porcine vena cava | 0.1% SDS for 16 h; 40 U/mL DNase for 2 h | - | [86] |
| Saphenous vein | 0.25% trypsin with 0.02% EDTA for 5 min; 10 mmol/L Tris, 5 mmol/L EDTA for 72 h; frozen at −80 °C for 2 h and thawing at 37 °C for 30 min | 50 mL 10 mM ethylenediaminetetraactic acid | [87] |
| Wistar rat kidney | Perfusated by 1% SDS | 5 mL of papain solution for 24 h | [88] |
| Porcine kidney | Repetitive cycle of: perfused with 0.5 M NaCl solution for 30 min; 0.5% SDS solution for 30 min; deionized (DI) water for 30 min | Lyophilize and mechanically ground into fine powders | [89] |
| Rat kidney | Perfused with 1% SDS for 4 h or 1% SLES for 6 h | - | [90] |
| Rat kidney | perfused with 1% SDS for 3 h and 1% Triton X-100 for 16 h | - | [91] |
| Rabbit kidney | Perfused with 1% SDS for 90 h, 2% Triton X-100 for 12 h | - | [92] |
| Rat kidney | Perfused with 1% SDS for 48 h, 0.2 mg/mL deoxyribonuclease I and 10 mM MgCl2 for 16 h | - | [93] |
| Rhesus monkey kidney | Perfused with 1% SDS and 1% Triton X-100 | - | [94] |
| Porcine kidney | Perfused with 1% SDS for 28 h, 1% Triton X-100 for 2 h | Incubation with papain extraction reagent for 3 h | [95] |
| Porcine skin | 0.25% trypsin for 6 h; 0.1% SDS in 0.26% EDTA with 0.69% Tris for 6 h; 1% Triton X-100 in 0.26% EDTA with 0.69% Tris for 12 h | Lyophilized and dried for 72 h | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.-Y.; Fu, C.-P.; Li, X.-Y.; Lu, X.-C.; Hu, L.-G.; Kankala, R.K.; Wang, S.-B.; Chen, A.-Z. Three-Dimensional Bioprinting of Decellularized Extracellular Matrix-Based Bioinks for Tissue Engineering. Molecules 2022, 27, 3442. https://doi.org/10.3390/molecules27113442
Zhang C-Y, Fu C-P, Li X-Y, Lu X-C, Hu L-G, Kankala RK, Wang S-B, Chen A-Z. Three-Dimensional Bioprinting of Decellularized Extracellular Matrix-Based Bioinks for Tissue Engineering. Molecules. 2022; 27(11):3442. https://doi.org/10.3390/molecules27113442
Chicago/Turabian StyleZhang, Chun-Yang, Chao-Ping Fu, Xiong-Ya Li, Xiao-Chang Lu, Long-Ge Hu, Ranjith Kumar Kankala, Shi-Bin Wang, and Ai-Zheng Chen. 2022. "Three-Dimensional Bioprinting of Decellularized Extracellular Matrix-Based Bioinks for Tissue Engineering" Molecules 27, no. 11: 3442. https://doi.org/10.3390/molecules27113442






