Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and Structure–Activity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of Curcumin (L1) and Dimethoxycurcumin (L2) Ligands
2.2. Synthesis and Characterization of Copper Complexes [Cu(Dn)(L1)]NO3 and [Cu(Dn)(L2)]NO3
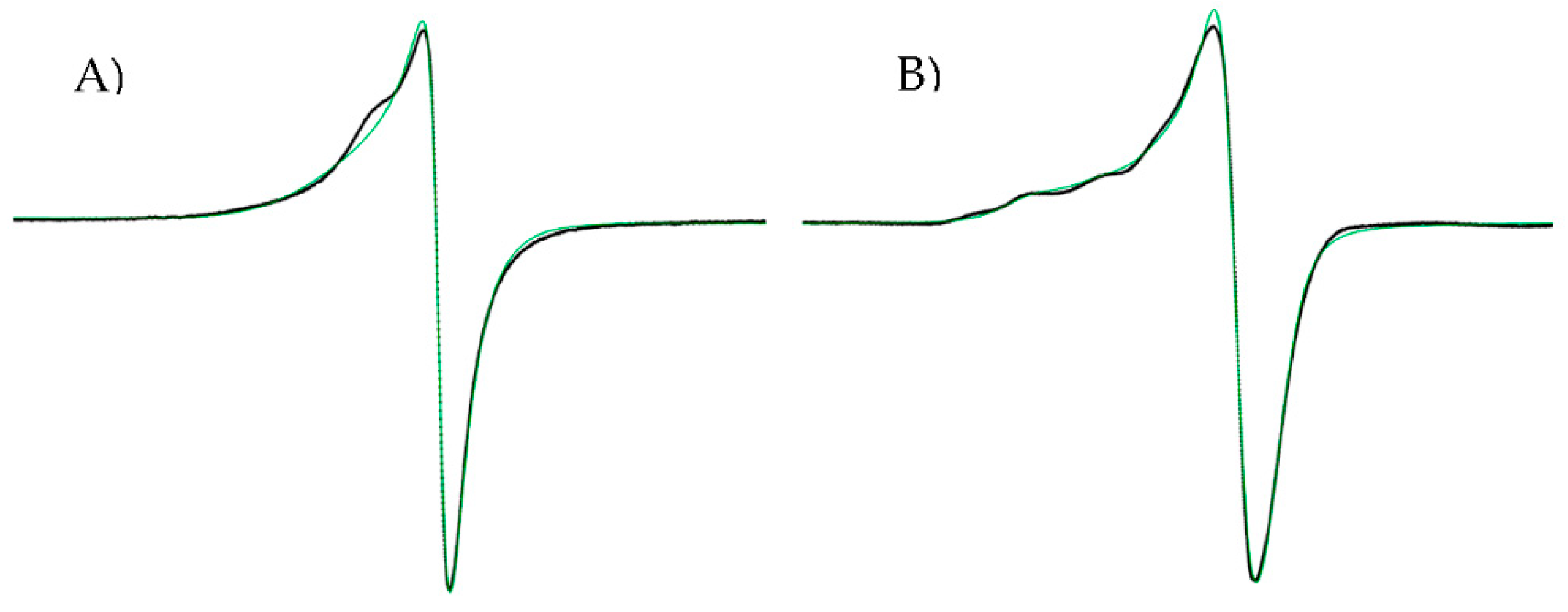
2.2.1. Molar Conductivity, Magnetic Moments and Electron Paramagnetic Resonance (EPR) Spectroscopy
2.2.2. Infrared Spectra
2.2.3. UV-Vis Spectrophotometric Analysis
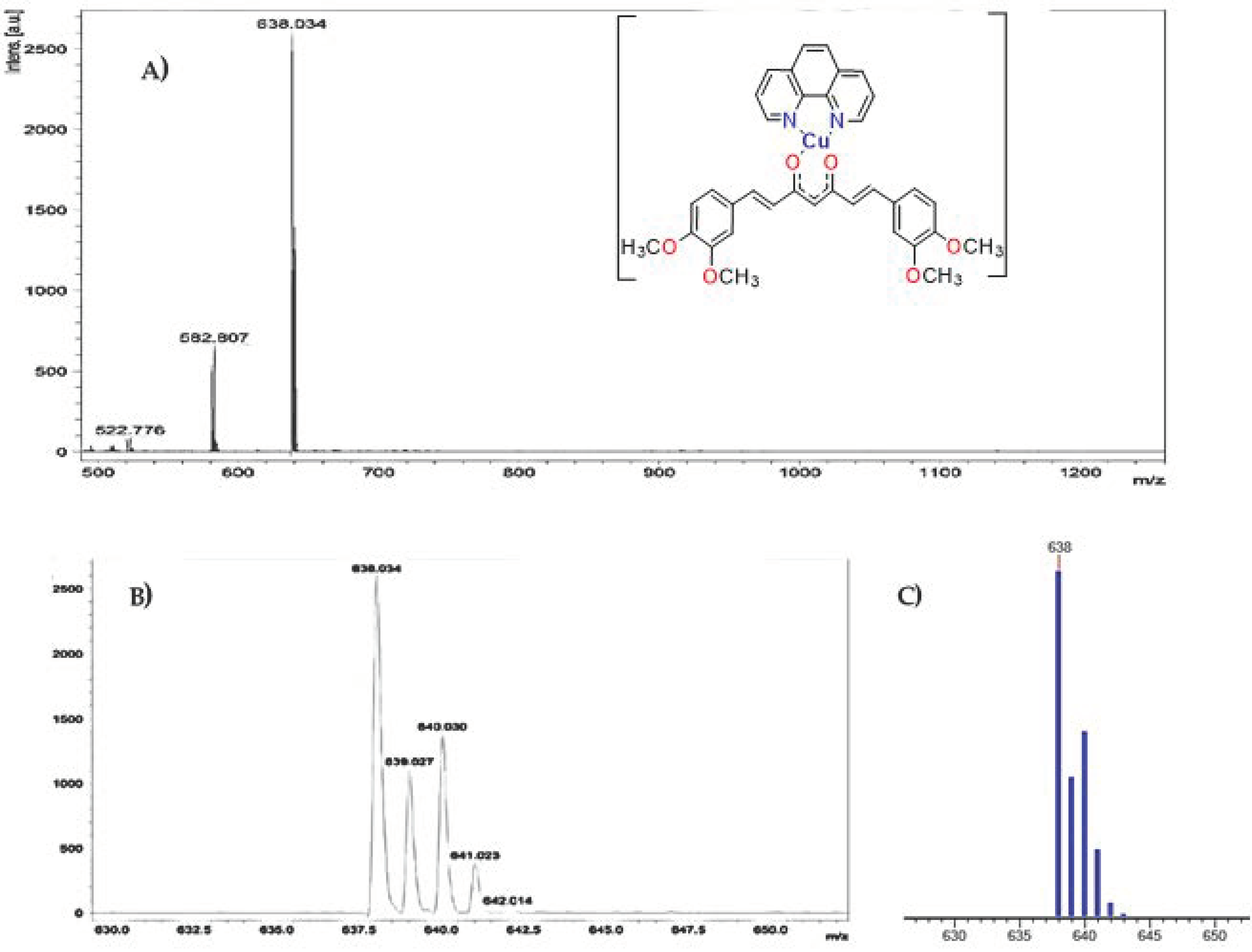
2.2.4. Mass Spectra
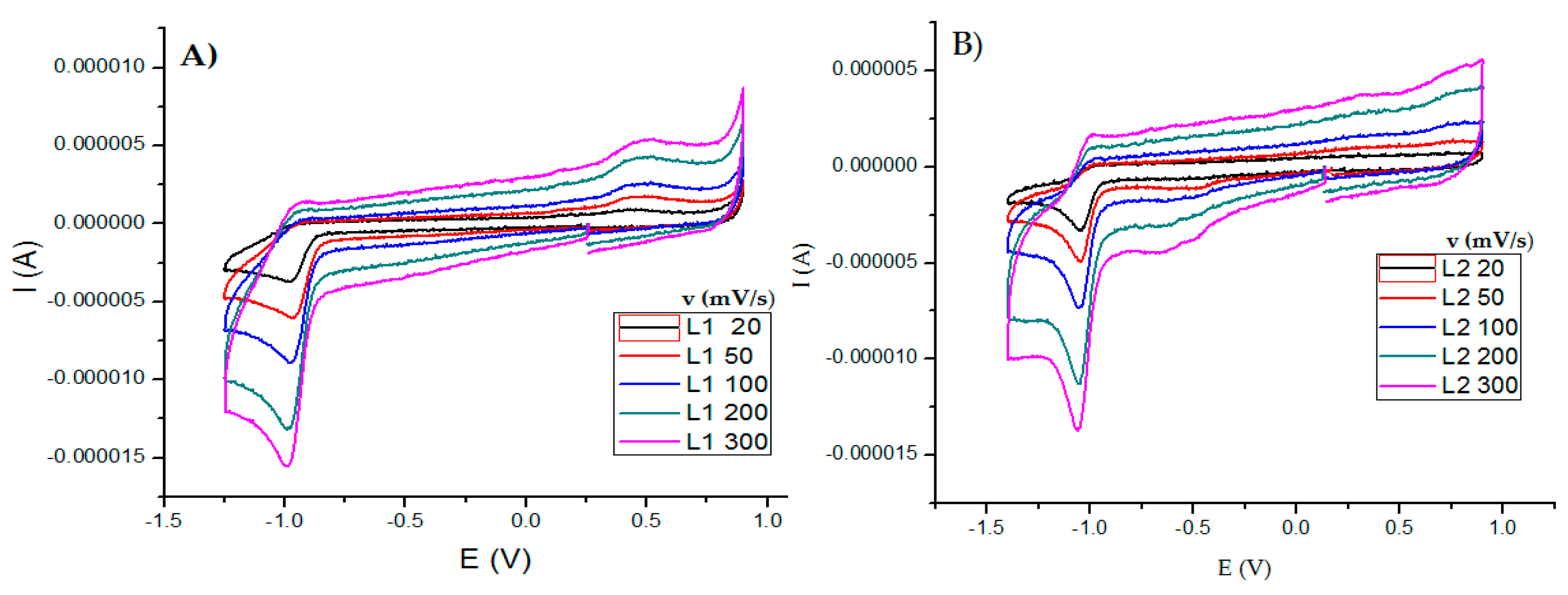
2.2.5. Cyclic Voltammetry Measurements
2.2.6. X-ray
2.3. Biological Assays
Cytotoxicity Activity
| Compound | IC50 SKLU-1 (μM) | IC50 HeLa (μM) | IC50 HeLa (μM) of First-Generation Casiopeinas |
|---|---|---|---|
| L1 | 52.56 ± 2.1 | 50.44 ± 2.3 | - |
| D1CuL1 | 41.53 ± 0.5 | N/A | 42 ± 3.1 [25] |
| D2CuL1 | 14.36 ± 0.8 | 26.4 9 ± 0.7 | 41.7 ± 0.31 [75] |
| D3CuL1 | 8.90 ± 0.2 | 35.78 ± 0.7 | - |
| D4CuL1 | 5.29 ± 0.9 | 7.61 ± 0.6 | 10.7 ± 0.9 [25] |
| D5CuL1 | 2.68 ± 0.2 | 1.91 ± 0.8 | 3.2 ± 0.03 [75] |
| D6CuL1 | 2.60 ± 0.5 | 2.21 ± 0.5 | 2.83 ± 0.09 [75] |
| D7CuL1 | 4.48 ± 1.5 | 4.48 ± 0.5 | 2.37 ± 0.4 [75] |
| D8CuL1 | 6.25 ± 0.9 | 6.74 ± 0.5 | 4.2 ± 0.6 [25] |
| L2 | 56.58 ± 1.8 | 109.05 ± 0.5 | - |
| D1CuL2 | 21.82 ± 0.4 | 44.62 ± 1.1 | 42 ± 3.1 [25] |
| D2CuL2 | 9.49 ± 1.3 | 37.6 ± 1.9 | 41.7 ± 0.31 [75] |
| D3CuL2 | 6.84 ± 1.1 | 30.56 ± 2.5 | - |
| D4CuL2 | 4.56 ± 0.6 | 13.26 ± 1.5 | 10.7 ± 0.9 [25] |
| D5CuL2 | 1.22 ± 0.1 | 2.01 ± 0.0005 | 3.2 ± 0.03 [75] |
| D6CuL2 | 1.19 ± 0.007 | 1.78 ± 0.1 | 2.83 ± 0.09 [75] |
| D7CuL2 | 2.68 ± 0.1 | 1.2 ± 0.05 | 2.37 ± 0.4 [75] |
| D8CuL2 | 2.38 ± 0.3 | 2.07 ± 0.3 | 4.2 ± 0.6 [25] |
| Cisplatin | 9.56 | 5.1 ± 0.4 [25] | 42 ± 3.1 [25] |
2.4. Structure–Activity Relationships
3. Materials and Methods
3.1. Instrumentation
3.2. Syntheses of Ligands and Coordination Compounds
3.2.1. Syntheses of Ligands Curcumin (L1) and Dimethoxycurcumin (L2)
3.2.2. General Synthesis Procedure of Ternary Complexes of [Cu(Dn)(L1)]NO3 and [Cu(Dn)(L2)]NO3
3.3. Cyclic Voltammetry Studies
3.4. X-ray Crystallography
3.5. Biological Assays
Cytotoxicity Activity
3.6. Structure–ActivityRrelationships
3.6.1. Computational Methods
3.6.2. Computational Details
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guzmán-Méndez, O.; González, F.; Bernès, S.; Flores-Álamo, M.; Ordóñez-Hernández, J.; García-Ortega, H.; Guerrero, J.; Qian, W.; Aliaga-Alcalde, N.; Gasque, L. Coumarin Derivative Directly Coordinated to Lanthanides Acts as an Excellent Antenna for UV–Vis and Near-IR Emission. Inorg. Chem. 2018, 57, 908–911. [Google Scholar] [CrossRef] [PubMed]
- Farrell, N. Transition Metal Complexes as Drugs and Chemotherapeutic Agents, Met. Complexes as Drugs Chemother. Agents 1989, 11, 809–840. [Google Scholar] [CrossRef]
- Annaraj, J.; Srinivasan, S.; Ponvel, K.; Athappan, P. Mixed ligand copper(II) complexes of phenanthroline/bipyridyl and curcumin diketimines as DNA intercalators and their electrochemical behavior under Nafion® and clay modified electrodes. J. Inorg. Biochem. 2005, 99, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Akimenko, N.; Cheltsov, P.; Balcarová, Z.; Kleinwächter, V.; Yevdokimov, Y.U. A study of interactions of platinum (II) compounds with DNA by means of CD spectra of solutions and liquid crystalline microphases of DNA. Gen. Physiol. Biophys. 1985, 4, 597–608. [Google Scholar]
- Sharma, K.; Chandra, S.; Basu, D. Synthesis and antiarthritic study of a new orally active diferuloyl methane (curcumin) gold complex. Inorg. Chim. Acta 1987, 135, 47–48. [Google Scholar] [CrossRef]
- Manikandan, P.; Anandan, R.; Nagini, S. Evaluation of Azadirachta indica Leaf Fractions for in Vitro Antioxidant Potential and Protective Effects against H2O2-Induced Oxidative Damage to pBR322 DNA and Red Blood Cells. J. Agric. Food Chem. 2009, 57, 6990–6996. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Shen, L.; Mohan, H.; Kadam, R.; Dutta, S.; Zhang, H.-Y.; Priyadarsini, K.I. Evaluation of a new copper(II)–curcumin complex as superoxide dismutase mimic and its free radical reactions. Free Radic. Biol. Med. 2005, 39, 811–822. [Google Scholar] [CrossRef]
- Huang, W.; Wei, W.; Shen, Z. Drug-like chelating agents: A potential lead for Alzheimer’s disease. RSC Adv. 2014, 4, 52088–52099. [Google Scholar] [CrossRef]
- Binolfi, A.; Lamberto, G.R.; Duran, R.; Quintanar, L.; Bertoncini, C.W.; Souza, J.M.; Cerveñansky, C.; Zweckstetter, M.; Griesinger, C.; Fernández, C.O. Site-Specific Interactions of Cu(II) with α and β-Synuclein: Bridging the Molecular Gap between Metal Binding and Aggregation. J. Am. Chem. Soc. 2008, 130, 11801–11812. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2013, 114, 815–862. [Google Scholar] [CrossRef]
- WHO. Cancer Today. 2020. Available online: https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=1&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=1&include_nmsc_other=1 (accessed on 22 February 2022).
- Alderden, R.A.; Hall, M.D.; Hambley, T. The Discovery and Development of Cisplatin. J. Chem. Educ. 2006, 83, 728–734. [Google Scholar] [CrossRef]
- Guo, Z.; Sadler, P.J. Metals in Medicine. Angew. Chem. Int. Ed. 1999, 38, 1512–1531. [Google Scholar] [CrossRef]
- Solans, X.; Ruiz-Ramírez, L.; Gasque, L.; Briansó, J.L. Structure of (1,10-phenanthroline)(salicylaldehydato)copper(II) nitrate. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1987, 43, 428–430. [Google Scholar] [CrossRef]
- Solans, X.; Ruiz-Ramírez, L.; Martinez, A.; Gasque, L.; Briansó, J.L. Structures of chloro(glycinato)(1,10-phenanthroline)copper(II) monohydrate (I) and aqua(1,10-phenanthroline)(L-phenylalaninato)copper(II) nitrate monohydrate (II). Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1988, 44, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Carvallo-Chaigneau, F.; Trejo-Solís, C.; Gómez-Ruiz, C.; Rodríguez-Aguilera, E.; Macías-Rosales, L.; Cortés-Barberena, E.; Cedillo-Peláez, C.; Gracia-Mora, I.; Ruiz-Azuara, L.; Madrid-Marina, V.; et al. Constantino-Casas, Casiopeina III-ia induces apoptosis in HCT-15 cells in vitro through caspase-dependent mechanisms and has antitumor effect in vivo. BioMetals 2007, 21, 17–28. [Google Scholar] [CrossRef]
- Trejo-Solís, C.; Palencia, G.; Zuñiga, S.; Rodríguez-Ropon, A.; Osorio-Rico, L.; Luvia, S.T.; Gracia-Mora, I.; Marquez-Rosado, L.; Sánchez, A.; Moreno-García, M.E.; et al. Sotelo, Cas Ilgly Induces Apoptosis in Glioma C6 Cells In Vitro and In Vivo through Caspase-Dependent and Caspase-Independent Mechanisms. Neoplasia 2005, 7, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Serment-Guerrero, J.; Cano-Sanchez, P.; Reyes-Perez, E.; Velazquez-Garcia, F.; Bravo-Gomez, M.; Ruiz-Azuara, L. Genotoxicity of the copper antineoplastic coordination complexes casiopeinas®. Toxicol. Vitr. 2011, 25, 1376–1384. [Google Scholar] [CrossRef]
- Figueroa-DePaz, Y.; Resendiz-Acevedo, K.; Dávila-Manzanilla, S.G.; García-Ramos, J.C.; Ortiz-Frade, L.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA, a target of mixed chelate copper(II) compounds (Casiopeinas®) studied by electrophoresis, UV–vis and circular dichroism techniques. J. Inorg. Biochem. 2022, 231, 111772. [Google Scholar] [CrossRef]
- Gutiérrez, A.G.; Vázquez-Aguirre, A.; García-Ramos, J.C.; Flores-Alamo, M.; Hernández-Lemus, E.; Ruiz-Azuara, L.; Mejía, C. Copper(II) mixed chelate compounds induce apoptosis through reactive oxygen species in neuroblastoma cell line CHP-212. J. Inorg. Biochem. 2013, 126, 17–25. [Google Scholar] [CrossRef]
- Hernández-Esquivel, L.; Marín-Hernández, A.; Pavón, N.; Carvajal, K.; Moreno-Sánchez, R. Cardiotoxicity of copper-based antineoplastic drugs casiopeinas is related to inhibition of energy metabolism. Toxicol. Appl. Pharmacol. 2006, 212, 79–88. [Google Scholar] [CrossRef]
- Ramirez-Palma, L.G.; Espinoza-Guillen, A.; Nieto-camacho, F.; López-Guerra, A.; Gómez-Vidales, V.; Cortés-Guzmán, F.; Ruiz-Azuara, L. Intermediate Detection in the Casiopeina—Cysteine Interaction Ending in the Disulfide Bond Formation and Copper Reduction. Molecules 2021, 26, 5729. [Google Scholar] [CrossRef] [PubMed]
- Marín-Hernández, A.; Gracia-Mora, I.; Ruiz-Ramírez, L.; Moreno-Sánchez, R. Toxic effects of copper-based antineoplastic drugs (Casiopeinas®) on mitochondrial functions. Biochem. Pharmacol. 2003, 65, 1979–1989. [Google Scholar] [CrossRef]
- Kachadourian, R.; Brechbuhl, H.M.; Ruiz-Azuara, L.; Gracia-Mora, I.; Day, B.J. Casiopeína IIgly-induced oxidative stress and mitochondrial dysfunction in human lung cancer A549 and H157 cells. Toxicology 2010, 268, 176–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo-Gómez, M.E.; García-Ramos, J.C.; Gracia-Mora, I.; Ruiz-Azuara, L. Antiproliferative activity and QSAR study of copper(II) mixed chelate [Cu(N–N)(acetylacetonato)]NO3 and [Cu(N–N)(glycinato)]NO3 complexes, (Casiopeínas®). J. Inorg. Biochem. 2009, 103, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Stanić, Z. Curcumin, a Compound from Natural Sources, a True Scientific Challenge—A Review. Plant Foods Hum. Nut. 2016, 72, 1–12. [Google Scholar] [CrossRef]
- Epstein, J.; Sanderson, I.R.; MacDonald, T.T. Curcumin as a Therapeutic Agent: The Evidence from in vitro, Animal and Human Studies. Br. J. Nutr. 2010, 103, 1545–1557. [Google Scholar] [CrossRef] [Green Version]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The essential medicinal chemistry of curcumin. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Surh, Y.J.; Shishodia, S. (Eds.) The Molecular Targets and Therapeutic Uses of Curcumin in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2007; Volume 595. [Google Scholar]
- Kunnumakkara, A.B.; Bordoloi, D.; Padmavathi, G.; Monisha, J.; Roy, N.K.; Prasad, S.; Aggarwal, B.B. Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br. J. Pharmacol. 2017, 174, 1325–1348. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.L.; Hsu, C.-H.; Lin, J.K.; Hsu, M.M.; Ho, Y.-F.; Shen, T.S.; Ko, J.Y.; Lin, J.T.; Lin, B.-R.; Ming-Shiang, W.; et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001, 21, 2895–2900. [Google Scholar]
- Pabon, H.J.J. Synthesis of curcumin and related compounds. Recueil 1964, 83, 379–386. [Google Scholar] [CrossRef]
- Leung, M.H.M.; Harada, T.; Kee, T.W. Delivery of Curcumin and Medicinal Effects of the Copper(II)-Curcumin Complexes. Curr. Pharm. Des. 2013, 19, 2070–2083. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.A.; Steward, W.P.; Gescher, A.J. Pharmacokinetics and Pharmacodynamics of Curcumin. Mol. Targets Ther. Uses Curcumin Health Dis. 2007, 595, 453–470. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Newman, Bioavailability of Curcumin: Problems and Promises reviews Bioavailability of Curcumin: Problems and Promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef]
- Schneider, C.; Gordon, O.N.; Edwards, R.L.; Luis, P.B. Degradation of Curcumin: From Mechanism to Biological Implications. J. Agric. Food Chem. 2015, 63, 7606–7614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunwar, A.; Simon, E.; Singh, U.; Chittela, R.K.; Sharma, D.; Sandur, S.K.; Priyadarsini, I.K. Interaction of a curcumin analogue dimethoxycurcumin with DNA. Chem. Biol. Drug Des. 2011, 77, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Meza-Morales, W.; Estévez-Carmona, M.M.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Cassani, J.; Ramírez-Apan, M.T.; Escobedo-Martínez, C.; Soriano-García, M.; Reynolds, W.F.; Enríquez, R.G. Full Structural Characterization of Homoleptic Complexes of Diacetylcurcumin with Mg, Zn, Cu, and Mn: Cisplatin-level Cytotoxicity in Vitro with Minimal Acute Toxicity in Vivo. Molecules 2019, 24, 1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahabadi, N.; Falsafi, M.; Moghadam, N.H. DNA interaction studies of a novel Cu(II) complex as an intercalator containing curcumin and bathophenanthroline ligands. J. Photochem. Photobiol. B Biol. 2013, 122, 45–51. [Google Scholar] [CrossRef]
- Banerjee, R. Inhibitory Effect of Curcumin-Cu(II) and Curcumin-Zn(II) Complexes on Amyloid-Beta Peptide Fibrillation. Bioinorg. Chem. Appl. 2014, 2014, 325873. [Google Scholar] [CrossRef]
- Mary, C.P.V.; Vijayakumar, S.; Shankar, R. Metal chelating ability and antioxidant properties of Curcumin-metal complexes—A DFT approach. J. Mol. Graph. Model. 2018, 79, 1–14. [Google Scholar] [CrossRef]
- Correa-Ascencio, M.; Galván-Miranda, E.K.; Rascón-Cruz, F.; Jiménez-Sandoval, O.; Jiménez-Sandoval, S.J.; Cea-Olivares, R.; Jancik, V.; Toscano, R.A.; García-Montalvo, V. Lanthanide(III) Complexes with 4,5-Bis(diphenylphosphinoyl)-1,2,3-triazolate and the Use of 1,10-Phenanthroline As Auxiliary Ligand. Inorg. Chem. 2010, 49, 4109–4116. [Google Scholar] [CrossRef]
- Refat, M.S. Synthesis and characterization of ligational behavior of curcumin drug towards some transition metal ions: Chelation effect on their thermal stability and biological activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 105, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Pröhl, M.; Schubert, U.; Weigand, W.; Gottschaldt, M. Metal complexes of curcumin and curcumin derivatives for molecular imaging and anticancer therapy. Coord. Chem. Rev. 2015, 307, 32–41. [Google Scholar] [CrossRef]
- Rigamonti, L.; Orteca, G.; Asti, M.; Basile, V.; Imbriano, C.; Saladini, M.; Ferrari, E. New curcumin-derived ligands and their affinity towards Ga3+, Fe3+ and Cu2+: Spectroscopic studies on complex formation and stability in solution. N. J. Chem. 2018, 42, 7680–7690. [Google Scholar] [CrossRef]
- Priyadharshini, N.; Iyyam, P.S.; Subramanian, S.; Venkatesh, P. Venkatesh, Synthesis, spectroscopic characterization and DNA interaction of schiff base curcumin Cu(II), Ni(II) and Zn(II) complexes. Pharma Chem. 2015, 7, 186–201. Available online: http://derpharmachemica.com/archive.html (accessed on 28 April 2022).
- Pi, Z.; Wang, J.; Jiang, B.; Cheng, G.; Zhou, S. A curcumin-based TPA four-branched copper(II) complex probe for in vivo early tumor detection. Mater. Sci. Eng. C 2015, 46, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xue, X.; Jiang, B.; Tian, Y. Metal complexes of a novel bis-β-diketone-type ligand and its copper(II) complexes of two-photon biological imaging. Sci. China Chem. 2011, 55, 334–340. [Google Scholar] [CrossRef]
- Xue, X.; Wang, J.; Si, G.; Wang, C.; Zhou, S. Synthesis, DNA-binding properties and cytotoxicity evaluation of two copper(II) complexes based on curcumin. Transit. Met. Chem. 2016, 41, 331–337. [Google Scholar] [CrossRef]
- Wang, J.; Wei, D.; Jiang, B.; Liu, T.; Ni, J.; Zhou, S. Two copper(II) complexes of curcumin derivatives: Synthesis, crystal structure and in vitro antitumor activity. Transit. Met. Chem. 2014, 39, 553–558. [Google Scholar] [CrossRef]
- JPorkodi, J.; Raman, N. Synthesis, characterization and biological screening studies of mixed ligand complexes using flavonoids as precursors. Appl. Organomet. Chem. 2017, 32, e4030. [Google Scholar] [CrossRef]
- Boorman, P.M.; Greenwood, N.N. and Hildon, M.A. Some diphosohine complexes of tungsten-(III) and –(IV). J. Chem. Soc. A 1968, 2466. [Google Scholar] [CrossRef]
- Geary, W. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Co-ord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Ismail, E.H.; Sabry, D.Y.; Mahdy, H.; Khalil, M.M.H. Synthesis and Characterization of some Ternary Metal Complexes of Curcumin with 1,10-phenanthroline and their Anticancer Applications. J. Sci. Res. 2014, 6, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Goswami, T.K.; Gadadhar, S.; Gole, B.; Karande, A.A.; Chakravarty, A.R. Photocytotoxicity of copper(II) complexes of curcumin and N-ferrocenylmethyl-l-amino acids. Eur. J. Med. Chem. 2013, 63, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Deepthi, T.V.; Venugopalan, P. Synthesis, DNA-binding, and cytotoxic studies on three copper(II) complexes of unsymmetrical synthetic analogues of curcumin. J. Coord. Chem. 2016, 69, 3403–3416. [Google Scholar] [CrossRef]
- Banerjee, S.; Chakravarty, A.R. Metal Complexes of Curcumin for Cellular Imaging, Targeting, and Photoinduced Anticancer Activity. Acc. Chem. Res. 2015, 48, 2075–2083. [Google Scholar] [CrossRef]
- Joseph, J.; Suman, A.; Nagashri, K.; Joseyphus, R.S.; Balakrishnan, N. Synthesis, characterization and biological studies of copper(II) complexes with 2-aminobenzimidazole derivatives. J. Mol. Struct. 2017, 1137, 17–26. [Google Scholar] [CrossRef]
- Bain, G.A.; Berry, J.F. Diamagnetic Corrections and Pascal’s Constants. J. Chem. Educ. 2008, 85, 532–536. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G. The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment. J. Chem. Educ. 2006, 83, 1229–1232. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; González-Álvarez, M.; Liu-González, M.; Macías, B.; Borrás, J.; Alzuet, G. The development of metal-based synthetic nucleases: DNA binding and oxidative DNA cleavage of a mixed copper(II) complex with N-(9H-purin-6-yl)benzenesulfonamide and 1,10-phenantroline. Antitumor activity in human Caco-2 cells and Jurkat T lymphocy. J. Inorg. Biochem. 2009, 103, 923–934. [Google Scholar] [CrossRef]
- Raman, N.; Sobha, S. Synthesis, characterization, DNA interaction and antimicrobial screening of isatin-based polypyridyl mixed ligand Cu(II) and Zn(II) complexes. J. Serbian Chem. Soc. 2010, 75, 773–788. [Google Scholar] [CrossRef]
- Masek, A.; Chrzescijanska, E.; Zaborski, M. Characteristics of curcumin using cyclic voltammetry, UV–vis, fluorescence and thermogravimetric analysis. Electrochim. Acta 2013, 107, 441–447. [Google Scholar] [CrossRef]
- Călinescu, M.; Fiastru, M.; Bala, D.; Mihailciuc, C.; Negreanu-Pîrjol, T.; Jurca, B. Synthesis, characterization, electrochemical behavior and antioxidant activity of new copper(II) coordination compounds with curcumin derivatives. J. Saudi Chem. Soc. 2019, 23, 817–827. [Google Scholar] [CrossRef]
- Zhu, J.; Sanidad, K.Z.; Sukamtoh, E.; Zhang, G. Potential roles of chemical degradation in the biological activities of curcumin. Food Funct. 2017, 8, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Meza-Morales, W.; Machado-Rodriguez, J.C.; Alvarez-Ricardo, Y.; Obregón-Mendoza, M.A.; Nieto-Camacho, A.; Toscano, R.A.; Soriano-García, M.; Cassani, J.; Enríquez, R.G. A New Family of Homoleptic Copper Complexes of Curcuminoids: Synthesis, Characterization and Biological Properties. Molecules 2019, 24, 910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanninger, S.; Lorenz, V.; Subhan, A.; Edelmann, F.T. Metal complexes of curcumin—Synthetic strategies, structures and medicinal applications. Chem. Soc. Rev. 2015, 44, 4986–5002. [Google Scholar] [CrossRef] [Green Version]
- Pucci, D.; Bellini, T.; Crispini, A.; D’Agnano, I.; Liguori, P.F.; Garcia-Orduña, P.; Pirillo, S.; Valentini, A.; Zanchetta, G. DNA binding and cytotoxicity of fluorescent curcumin-based Zn(ii) complexes. MedChemComm 2012, 3, 462–468. [Google Scholar] [CrossRef]
- Aliaga-Alcalde, N.; Marqués-Gallego, P.; Kraaijkamp, M.; Herranz-Lancho, C.; Dulk, H.D.; Görner, H.; Roubeau, O.; Teat, S.J.; Weyhermüller, T.; Reedijk, J. Copper Curcuminoids Containing Anthracene Groups: Fluorescent Molecules with Cytotoxic Activity. Inorg. Chem. 2010, 49, 9655–9663. [Google Scholar] [CrossRef]
- Chikira, M.; Tomizawa, Y.; Fukita, D.; Sugizaki, T.; Sugawara, N.; Yamazaki, T.; Sasano, A.; Shindo, H.; Palaniandavar, M.; Antholine, W.E. DNA-fiber EPR study of the orientation of Cu(II) complexes of 1,10-phenanthroline and its derivatives bound to DNA: Mono(phenanthroline)-copper(II) and its ternary complexes with amino acids. J. Inorg. Biochem. 2002, 89, 163–173. [Google Scholar] [CrossRef]
- Hirohama, T.; Kuranuki, Y.; Ebina, E.; Sugizaki, T.; Arii, H.; Chikira, M.; Selvi, P.T.; Palaniandavar, M. Copper(II) complexes of 1,10-phenanthroline-derived ligands: Studies on DNA binding properties and nuclease activity. J. Inorg. Biochem. 2005, 99, 1205–1219. [Google Scholar] [CrossRef]
- Bravo-gómez, M.E.; Dávila-manzanilla, S.; Flood-garibay, J.; Muciño, M. Á.; Mendoza, Á.; García-ramos, J.C.; Moreno-esparza, R.; Ruiz-azuara, L. Secondary Ligand Effects on the Cytotoxicity of Several Casiopeína’s Group II Compounds. J. Mex. Chem. Soc. 2012, 56, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, V.T.; Icsel, C.; Suyunova, F.; Aygun, M.; Aztopal, N.; Ulukaya, E. Ni(ii)/Cu(ii)/Zn(ii) 5,5-diethylbarbiturate complexes with 1,10-phenanthroline and 2,2′-dipyridylamine: Synthesis, structures, DNA/BSA binding, nuclease activity, molecular docking, cellular uptake, cytotoxicity and the mode of cell death. Dalton Trans. 2016, 45, 10466–10479. [Google Scholar] [CrossRef] [PubMed]
- Moghassemi, S.; Hadjizadeh, A. Nano-niosomes as nanoscale drug delivery systems: An illustrated review. J. Control. Release 2014, 185, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Davila-Manzanilla, S.G.; Figueroa-De-Paz, Y.; Mejia, C.; Ruiz-Azuara, L. Synergistic effects between a copper-based metal Casiopeína III-ia and cisplatin. Eur. J. Med. Chem. 2017, 129, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Tovar-Tovar, A.; Ruiz-Ramírez, L.; Campero, A.; Romerosa, A.; Moreno-Esparza, R.; Rosales-Hoz, M.J. Structural and reactivity studies on 4,4-dimethyl-2,2-bipyridine acetylacetonate copper(II) nitrate (CASIOPEINA III-ia) with methionine, by UV-visible and EPR techniques. J. Inorg. Biochem. 2004, 98, 1045–1053. [Google Scholar] [CrossRef]
- Reina, M.; Hernández-Ayala, L.F.; Bravo-Gómez, M.E.; Gómez, V.; Ruiz-Azuara, L. Second generation of Casiopeinas®: A joint experimental and theoretical study. Inorganica Chim. Acta 2020, 517, 120201. [Google Scholar] [CrossRef]
- Stoll, S.; Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef]
- Ruiz-Azuara, L. United States Patent 07/628,843: RE 35,458, 1997.
- Ruiz-Azuara, L. United States Patent 07/628,628: 5,576,326, 1996.
- Ruiz-Azuara, L. United States Patent 07/628,628: RE 35,458, 1997.


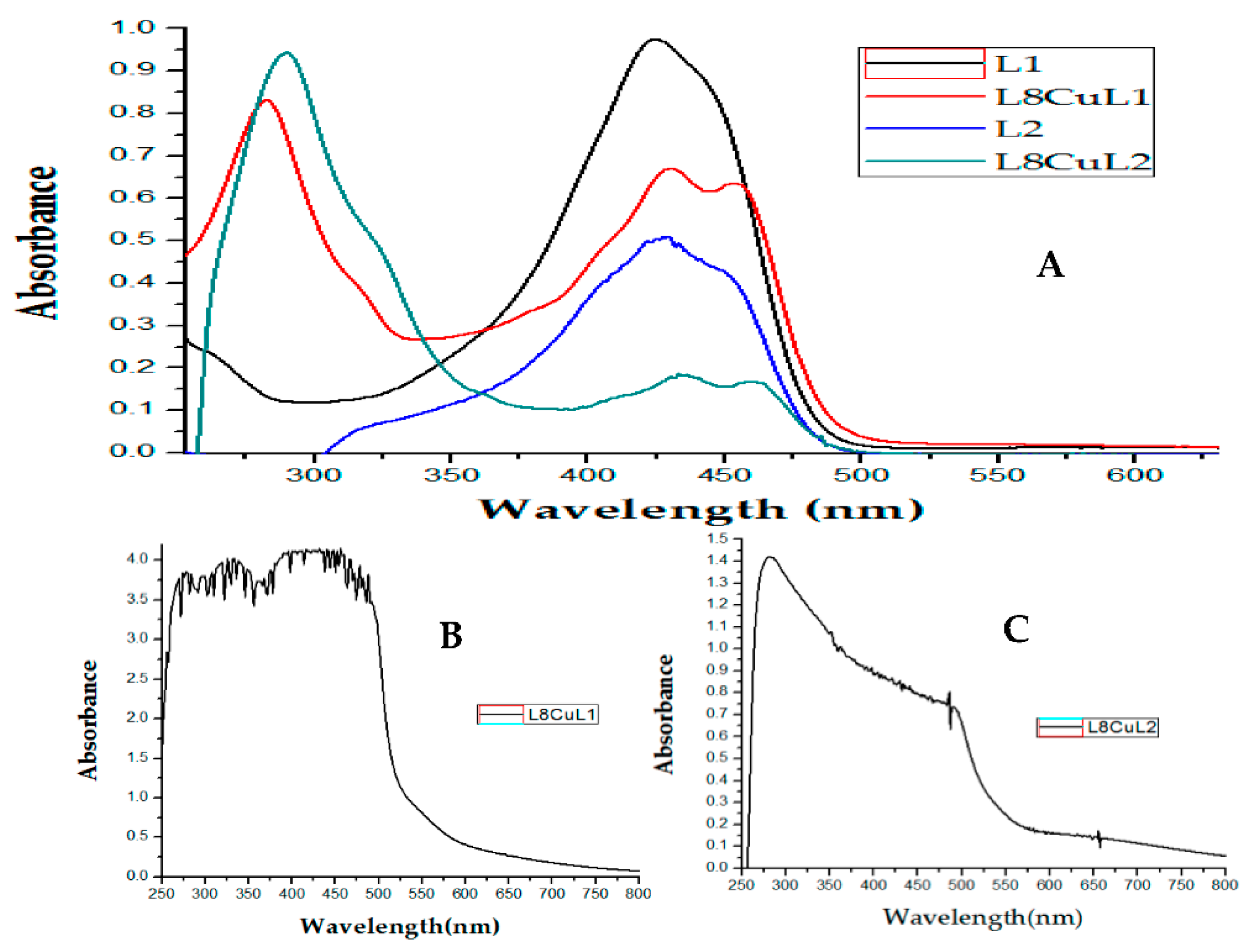
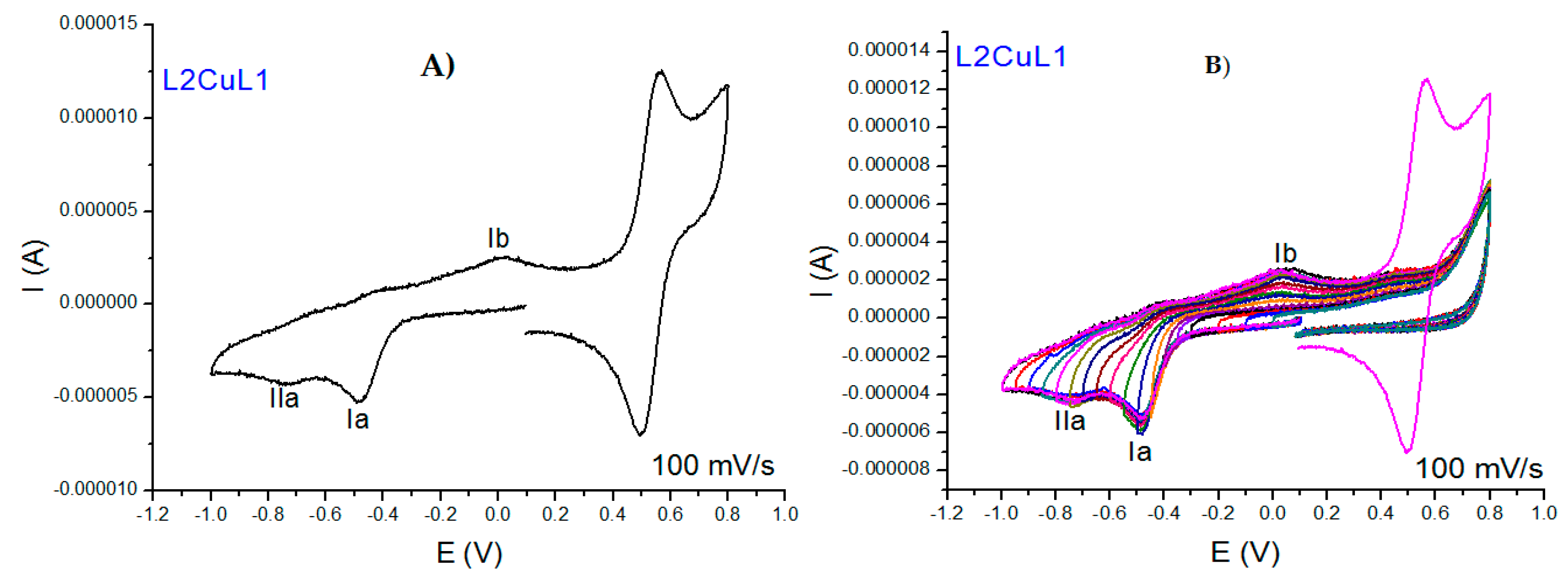
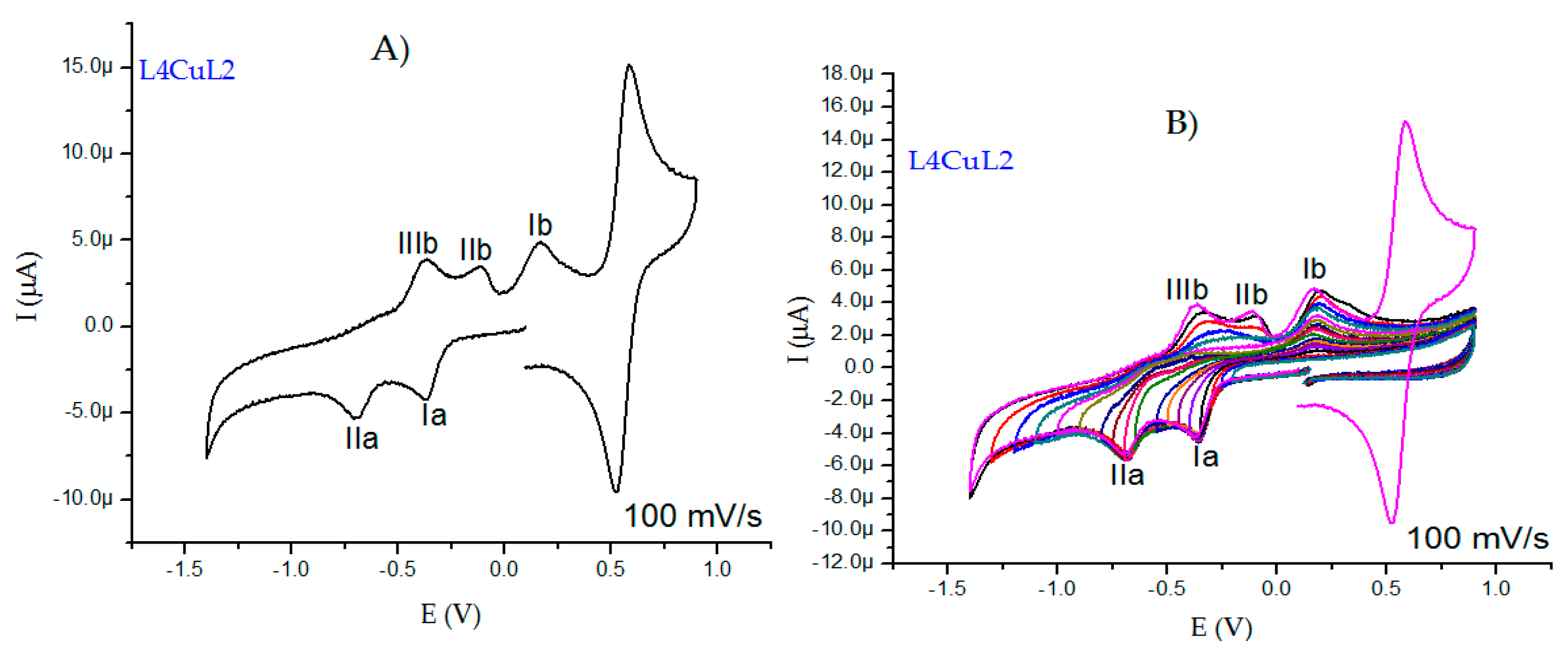



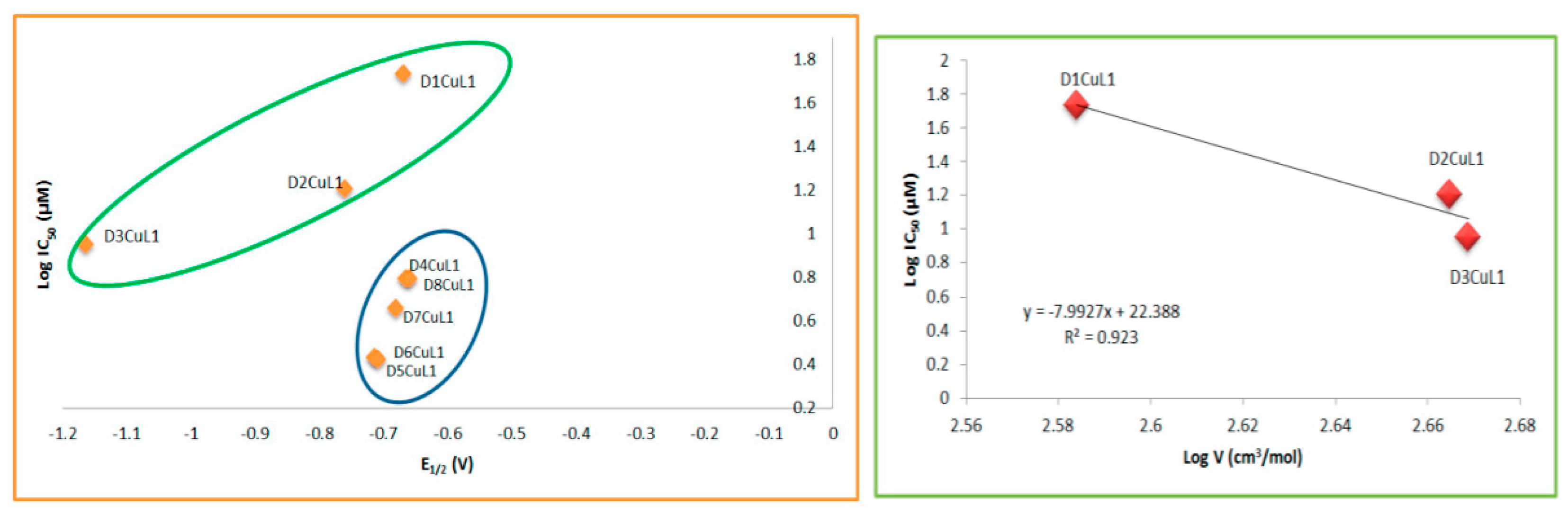
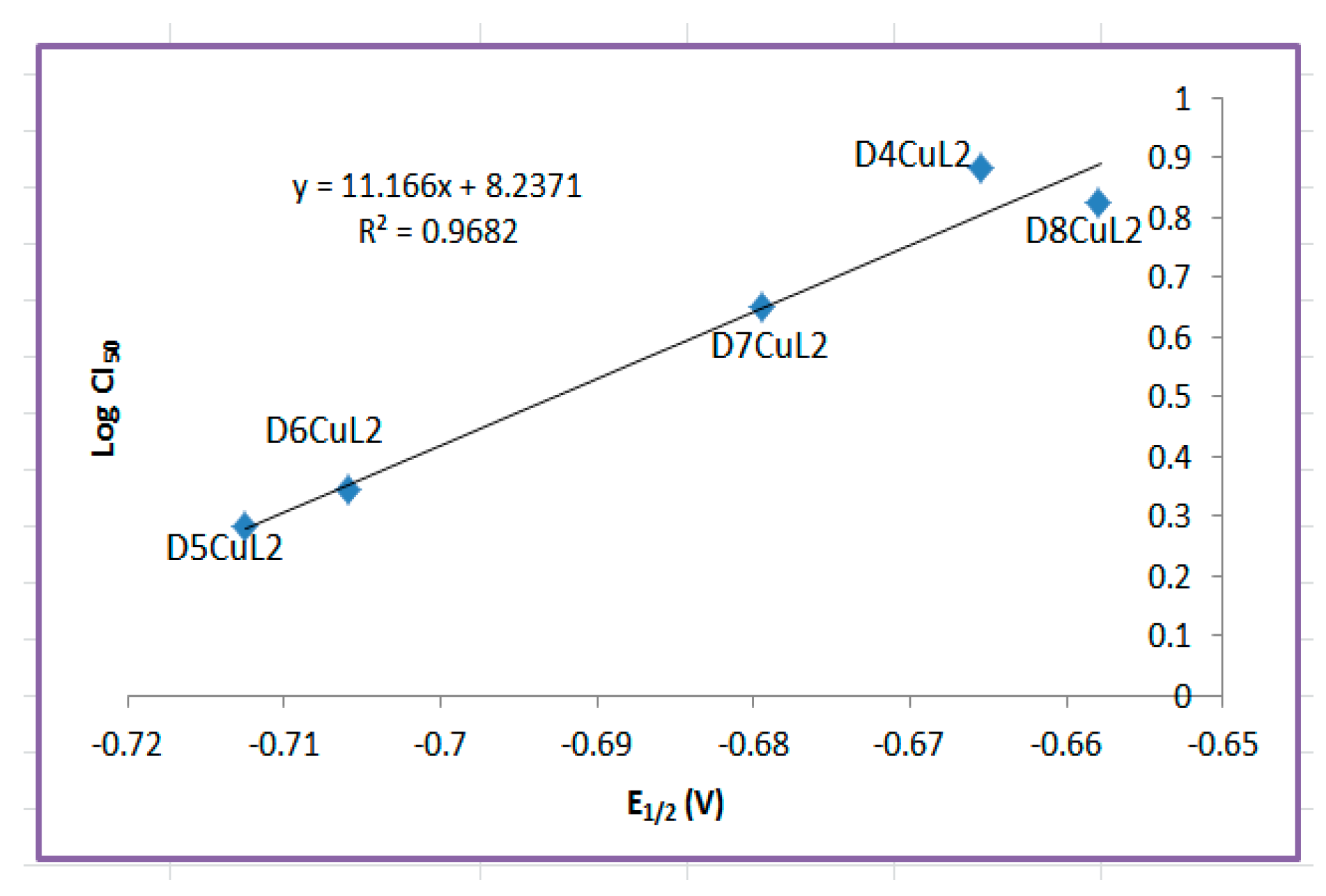
| Compound | λmax(nm) π-π* | ε (L/mol cm) | λmax(nm) TCLM | ε (L/mol cm) | λmax(nm) TCML | ε (L/mol cm) |
|---|---|---|---|---|---|---|
| D1CuL1 | 294 | 31,451 | 430 | 64,287 | 454 | 62,853 |
| D2CuL1 | 292 | 22,776 | 430 | 42,317 | 454 | 41,947 |
| D3CuL1 | 304 | 28,563 | 426 | 36,928 | 448 | 33,804 |
| D4CuL1 | 266 | 48,706 | 430 | 57,630 | 454 | 56,402 |
| D5CuL1 | 268 | 51,897 | 430 | 59,041 | 454 | 56,908 |
| D6CuL1 | 276 | 42,400 | 428 | 35,698 | 452 | 32,659 |
| D7CuL1 | 274 | 34,157 | 432 | 33,142 | 456 | 32,775 |
| D8CuL1 | 282 | 53,109 | 430 | 42,733 | 454 | 40,475 |
| D1CuL2 | 313 | 22,461 | 430 | 24,832 | 458 | 23,216 |
| D2CuL2 | 310 | 25,161 | 430 | 23,350 | 458 | 21,809 |
| D3CuL2 | 321 | 17,513 | 430 | 20,741 | 458 | 19,205 |
| D4CuL2 | 275 | 42,754 | 433 | 33,220 | 458 | 30,807 |
| D5CuL2 | 276 | 52,001 | 433 | 34,631 | 458 | 32,386 |
| D6CuL2 | 284 | 44,217 | 433 | 33,226 | 458 | 31,128 |
| D7CuL2 | 282 | 49,471 | 433 | 32,936 | 458 | 30,671 |
| D8CuL2 | 290 | 53,569 | 436 | 11,448 | 460 | 10,582 |
| Compound | Molecular Weight (g/mol) | Molecular Ion (Theoretical m/z) | Molecular Ion (Obtained m/z) |
|---|---|---|---|
| Curcumin (L1) | 368.37 | 368 | 369.2 |
| [Cu(2,2′-bipyridine)(curcumina)]NO3 (D1CuL1) | 649.01 | 586.1 | 586.9 |
| [Cu(4,4′-dimethyl-2,2′-bipyridine)(curcumin)]NO3 (D2CuL1) | 695.17 | 614.4 | 615.0 |
| [Cu(5,5′-dimethyl-2,2′-bipyridine)(curcumin)]NO3 (D3CuL1) | 695.17 | 614.4 | 614.0 |
| [Cu(1,10-phenanthroline)(curcumin)]NO3 (D4CuL1) | 673.12 | 610.1 | 610.9 |
| [Cu(4,7-dimethyl-1,10-phenanthroline)(curcumin)]NO3 (D5CuL1) | 719.19 | 638.1 | 638.0 |
| [Cu(5,6-dimetyl-1,10-phenanthroline)(curcumin)]NO3 (D6CuL1) | 701.18 | 638.1 | 638.0 |
| [Cu(3,4,7,8-tetrametyl-1,10-phenanthroline)(curcumin)]NO3 (D7CuL1) | 747.24 | 666.1 | 667.1 |
| [Cu(4,7-diphenyl-1,10-phenanthroline)(curcumin)]NO3 (D8CuL1) | 825.32 | 762.1 | 762.1 |
| Dimethoxycurcumin (L2) | 396.43 | 396.4 | 397.7 |
| [Cu(2,2′-bipyridine)(dimethoxycurcumin)]NO3 (D1CuL2) | 677.16 | 614.1 | 615.3 |
| [Cu(4,4′-dimethyl-2,2′-bipyridine)(dimethoxycurcumin)]NO3 (D2CuL2) | 723.22 | 642.1 | 643.2 |
| [Cu(5,5′-dimethyl-2,2′-bipyridine)(dimethoxycurcumin)]NO3 (D3CuL2) | 723.22 | 642.1 | 642.1 |
| [Cu(1,10-phenanthroline)(dimethoxycurcumin)]NO3 (D4CuL2) | 737.21 | 638.1 | 638.0 |
| [Cu(4,7-dimethyl-1,10-phenanthroline)(dimethoxycurcumin)]NO3 (D5CuL2) | 729.23 | 666.1 | 668.2 |
| [Cu(5,6-dimethyl-1,10-phenanthroline)(dimethoxycurcumin)]NO3 (D6CuL2) | 729.23 | 666.1 | 668.2 |
| [Cu(3,4,7,8-tetramethyl-1,10-phenanthroline)(dimethoxycurcumin)]NO3 (D7CuL2) | 757.28 | 694.2 | 696.1 |
| [Cu(4,7-diphenyl-1,10-phenanthroline)(dimethoxycurcumin)]NO3 (D8CuL2) | 853.37 | 790.2 | 791.5 |
| Compound | Epc (V) | Epa (V) | ΔE (V) | E1/2 (V) (Fc+/Fc) |
|---|---|---|---|---|
| L1 | −0.978 | 0.488 | 1.467 | --- |
| D1CuL1 | −0.429 | 0.130 | 0.560 | −0.668 |
| D2CuL1 | −0.484 | 0.023 | 0.507 | −0.758 |
| D3CuL1 | −0.474 | 0.067 | 0.542 | −1.164 |
| D4CuL1 | −0.407 | 0.140 | 0.547 | −0.665 |
| D5CuL1 | −0.440 | 0.123 | 0.563 | −0.712 |
| D6CuL1 | −0.611 | −0.04 | 0.571 | −0.705 |
| D7CuL1 | −0.405 | 0.154 | 0.560 | −0.679 |
| D8CuL1 | −0.491 | 0.126 | 0.617 | −0.657 |
| L2 | −1.05 | --- | --- | --- |
| D1CuL2 | −0.444 | 0.139 | 0.623 | −0.656 |
| D2CuL2 | −0.469 | 0.117 | 0.586 | −0.709 |
| D3CuL2 | −0.390 | 0.162 | 0.552 | −0.656 |
| D4CuL2 | −0.373 | 0.168 | 0.541 | −0.656 |
| D5CuL2 | −0.447 | 0.088 | 0.535 | −0.701 |
| D6CuL2 | −0.388 | 0.158 | 0.546 | −0.682 |
| D7CuL2 | −0.396 | 0.096 | 0.492 | −0.690 |
| D8CuL2 | −0.171 | 0.122 | 0.293 | −0.559 |
| Compound | Bond Atom-Atom (A°) | Lengths | Bond Atom-Atom (A°) | Angles (°) |
|---|---|---|---|---|
| D5CuL2 square planar geometry | Cu-N1 | 1.977 | O1-Cu-O2 | 94.68 |
| Cu-N2 | 1.993 | N1-Cu-N2 | 81.84 | |
| Cu-O1 | 1.906 | O1-Cu-N1 | 92.37 | |
| Cu-O2 | 1.886 | O2-Cu-N2 | 89.45 | |
| Cu-O (H2O) | 5.456 | O1-Cu-N2 | 168.51 | |
| Cu-O (NO3) | 6.582 | O2-Cu-N1 | 166.09 | |
| D6CuL2 square-based pyramid geometry | Cu-N1 | 2.013 | O1-Cu-O2 | 94.68 |
| Cu-N2 | 2.022 | N1-Cu-N2 | 81.84 | |
| Cu-O1 | 1.921 | O1-Cu-N1 | 92.37 | |
| Cu-O2 | 1.911 | O2-Cu-N2 | 89.45 | |
| Cu-O (H2O) | 2.260 | O1-Cu-N2 | 168.51 | |
| Cu-O (NO3) | 4.958 | O2-Cu-N1 | 166.09 |
| Compound | Molar Volume (cm3/mol) | IE (eV) | EA (eV) | E1/2 (V) |
|---|---|---|---|---|
| D1CuL1 | 383.712 | 8.04749134 | 2.82789556 | −0.878171 |
| D2CuL1 | 462.274 | 7.99361656 | 2.76906755 | −0.930038 |
| D3CuL1 | 466.413 | 8.00534368 | 2.7998987 | −0.872582 |
| D4CuL1 | 410.031 | 8.10950385 | 2.87669331 | −0.725702 |
| D5CuL1 | 437.24 | 8.04221051 | 2.80906527 | −0.749246 |
| D6CuL1 | 417.529 | 8.07647188 | 2.85005565 | −0.849173 |
| D7CuL1 | 373.335 | 8.01205011 | 2.79094547 | −0.907115 |
| D8CuL1 | 492.824 | 8.07950481 | 2.86419938 | −0.873203 |
| D1CuL2 | 451.839 | 8.07429826 | 2.84794598 | −0.836537 |
| D2CuL2 | 450.172 | 8.01993014 | 2.78888151 | −0.858569 |
| D3CuL2 | 386.932 | 8.03231631 | 2.82004192 | −0.840938 |
| D4CuL2 | 393.699 | 8.13593635 | 2.8959002 | −0.728105 |
| D5CuL2 | 422.974 | 8.06659075 | 2.82811977 | −0.809915 |
| D6CuL2 | 466.821 | 8.10139715 | 2.86922906 | −0.782915 |
| D7CuL2 | 545.631 | 8.03569891 | 2.8086884 | −0.799709 |
| D8CuL2 | 536.71 | 8.09949401 | 2.87311289 | −0.452801 |
| Collection Parameters | D5CuL2 | D6CuL2 |
|---|---|---|
| Empirical formula | C37H35N3O9Cu | C37H35N3O9Cu |
| Formula weight | 729.23 g/mol | 729.23 g/mol |
| Temperature (K) | 100 | 100 |
| Wavelength (Ǻ) | 1.5417 | 0.71073 |
| Crystal system | Triclinic | Triclinic |
| Space group | P-1 | P-1 |
| a (Ǻ) | 7.7004 (7) | 10.0604 (7) |
| b (Ǻ) | 13.7026 (12) | 13.6229 (9) |
| c (Ǻ) | 17.1812 (15) | 14.0129 (9) |
| α (°) | 110.132 (5) | 70.5193 (12) |
| β(°) | 94.503 (6) | 76.4453 (13) |
| γ(°) | 91.987 (6) | 71.7320 (13) |
| Volume (Ǻ3) | 1693.2 (3) | 1701.1 (2) |
| Z | 2 | 1 |
| Dcalc (mg/m3) | 1.466 | 1.485 |
| Absorption coefficient (mm−1) | 1.462 | 0.709 |
| F(000) | 778 | 793 |
| Crystal size (mm3) | 0.300 × 0.041 × 0.036 | 0.150 × 0.092 × 0.056 |
| Theta range for data collection (°) | 2.752 to 68.243 | 1.558 to 27.442 |
| Index ranges | −9 ≤ h ≤ 9, −16 ≤ k ≤ 16, −20 ≤ 1 ≤ 20 | −13 ≤ h ≤ 13, −17 ≤ k ≤ 17, −18 ≤ l ≤ 18 |
| Reflections collected | 45,131 | 27,234 |
| Independent reflections | 5817 [R(int) = 0.0466] | 7773 [R(int) = 0.0363] |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 5818/563/584 | 7773/360/582 |
| Goodness-of-fit on F2 | 1.086 | 1.040 |
| Final R indices (I > 2sigma(I)) | R1 = 0.0538, wR2 = 0.1580 | R1 = 0.0355, wR2 = 0.0889 |
| R indices (all data) | R1 =0.0588, wR2 = 0.1637 | R1 = 0.0477, wR2 = 0.0889 |
| Largest diff. peak and hole e Ǻ−3 | 0.878 and −0.651 | 0.366 and −0.520 |
| Extinction coefficient | n/a | n/a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figueroa-DePaz, Y.; Pérez-Villanueva, J.; Soria-Arteche, O.; Martínez-Otero, D.; Gómez-Vidales, V.; Ortiz-Frade, L.; Ruiz-Azuara, L. Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and Structure–Activity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands. Molecules 2022, 27, 3504. https://doi.org/10.3390/molecules27113504
Figueroa-DePaz Y, Pérez-Villanueva J, Soria-Arteche O, Martínez-Otero D, Gómez-Vidales V, Ortiz-Frade L, Ruiz-Azuara L. Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and Structure–Activity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands. Molecules. 2022; 27(11):3504. https://doi.org/10.3390/molecules27113504
Chicago/Turabian StyleFigueroa-DePaz, Yeshenia, Jaime Pérez-Villanueva, Olivia Soria-Arteche, Diego Martínez-Otero, Virginia Gómez-Vidales, Luis Ortiz-Frade, and Lena Ruiz-Azuara. 2022. "Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and Structure–Activity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands" Molecules 27, no. 11: 3504. https://doi.org/10.3390/molecules27113504








