NMR Spectroscopy Applied to the Metabolic Analysis of Natural Extracts of Cannabis sativa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Extraction Procedure
2.2. Chemicals and Solvents
2.3. NMR Sample Preparation, Experimentation, and Data Processing
2.4. Statistical Analysis: Principal Component Analysis (PCA)
2.5. Chromatographic Experiments
3. Results and Discussion
3.1. NMR Characterization of Seeds Extracts
3.2. NMR Characterization of Flower Extracts
3.3. Multivariate Analysis
3.4. Quantitative Analysis of Inflorescences
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Small, E. Classification of Cannabis sativa L. In Relation to Agricultural, Biotechnological, Medical and Recreational Utilization. In Cannabis sativa L.—Botany and Biotechnology, 1st ed.; Chandra, S., Hemant, L., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–62. [Google Scholar]
- Li, H.L. An archaeological and historical account of Cannabis in China. Econ. Bot. 1974, 28, 437–447. [Google Scholar] [CrossRef]
- ElSolhy, M.A.; Tadwan, M.M.; Gul, W.; Chandra, S.; Galal, A. Phytochemistry of Cannabis sativa L. Prog. Chem. Org. Nat. Prod. 2017, 103, 1–36. [Google Scholar]
- Crescente, G.; Piccolella, S.; Esposito, A.; Scognamiglio, M.; Fiorentino, A.; Pacifico, S. Chemical composition and nutraceutical properties of hemp seeds: An ancient food with actual functional value. Phytochem. Rev. 2018, 17, 733–749. [Google Scholar] [CrossRef]
- Tura, M.; Ansorena, D.; Astiasarán, I.; Mandrioli, M.; Toschi, T.G. Evaluation of Hemp Seed Oils Stability under Accelerated Storage Test. Antioxidants 2022, 11, 490. [Google Scholar] [CrossRef]
- Banskota, A.H.; Jones, A.; Hui, J.P.M.; Stefanova, R. Triacylglycerols and Other Lipids Profiling of Hemp By-Products. Molecules 2022, 27, 2339. [Google Scholar] [CrossRef]
- Spano, M.; Di Matteo, G.; Rapa, M.; Ciano, S.; Ingallina, C.; Cesa, S.; Menghini, L.; Carradori, S.; Giusti, A.M.; Di Sotto, A.; et al. Commercial Hemp Seed Oils: A Multimethodological Characterization. Appl. Sci. 2020, 10, 6933. [Google Scholar] [CrossRef]
- Teleszko, M.; Zając, A.; Rusak, T. Hemp Seeds of the Polish ‘Bialobrzeskie’ and ‘Henola’ Varieties (Cannabis sativa L. var. sativa) as Prospective Plant Sources for Food Production. Molecules 2022, 27, 1448. [Google Scholar] [CrossRef]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic disease. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Ministero della Salute. Produzione e Commercializzazione di Prodotti a Base di Semi di Canapa Per L’utilizzo nci Settori Dell’alimentazione Umana; Ministero della Salute: Roma, Italy, 2009; pp. 1–4.
- Mercuri, A.M.; Accorsi, C.A.; Bandini Mazzanti, M. The long history of Cannabis and its cultivation by Romans in central Italy, shown by pollen records from Lago Albano and Lago di Nemi. Veget. Hist. Archaeobot. 2002, 11, 263–276. [Google Scholar] [CrossRef]
- ElSolhy, M.A.; Slade, D. Chemical Constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Radwan, M.M.; Chandra, S.; Gul, S.; Elsohly, M.A. Cannabinoids, phenolics, terpenes and alkaloids of cannabis. Molecules 2021, 26, 2774. [Google Scholar] [CrossRef] [PubMed]
- Bautista, J.L.; Yu, S.; Tian, L. Flavonoids in Cannabis sativa: Biosynthesis, bioactivities, and biotechnology. ACS Omega 2021, 6, 5119–5123. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.; Pryce, G.; Giovannoni, G.; Thompson, A.J. The therapeutic potential of Cannabis. Lancet. Neurol. 2003, 2, 191–298. [Google Scholar] [CrossRef]
- Izzo, A.A.; Borrelli, F.; Capasso, R.; Di Marzo, V.; Mechoulam, R. Non-psychotropic plant cannabinoids: New therapeutic opportunities from an ancient herb. Trends Pharmacol. Sci. 2009, 30, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Odieka, A.E.; Obuzor, G.U.; Oyedeji, O.O.; Gondwe, M.; Hosu, Y.S.; Oyedeji, A.O. The Medicinal Natural Products of Cannabis sativa Linn.: A Review. Molecules 2022, 27, 1689. [Google Scholar] [CrossRef]
- Iftikhar, A.; Zafar, U.; Ahmed, W.; Shabbir, M.A.; Sameen, A.; Sahar, A.; Bhat, Z.F.; Kowalczewski, P.Ł.; Jarzębski, M.; Aadil, R.M. Applications of Cannabis sativa L. in Food and Its Therapeutic Potential: From a Prohibited Drug to a Nutritional Supplement. Molecules 2021, 26, 7699. [Google Scholar] [CrossRef]
- Russo, E.B. Cannabis Therapeutics and the Future of Neurology. Front. Integr. Neurosci. 2018, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Stasiłowicz, A.; Tomala, A.; Podolak, I.; Cielecka-Piontek, J. Cannabis sativa L. as a Natural Drug Meeting the Criteria of a Multitarget Approach to Treatment. Int. J. Mol. Sci. 2021, 22, 778. [Google Scholar] [CrossRef]
- Kopustinskiene, D.M.; Masteikova, R.; Lazauskas, R.; Bernatoniene, J. Cannabis sativa L. Bioactive Compounds and Their Protective Role in Oxidative Stress and Inflammation. Antioxidants 2022, 11, 660. [Google Scholar] [CrossRef]
- Pisanti, S.; Bifulco, M. Medical Cannabis: A plurimillennial history of an evergreen. J. Cell. Physiol. 2019, 234, 8342–8351. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC. Off. J. Eur. Union 2009, 309, 1–50. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32009R1107 (accessed on 14 April 2022).
- Schachtsiek, J.; Warzecha, H.; Kayser, O.; Stehle, F. Current Perspectives on Biotechnological Cannabinoid Production in Plants. Planta Med. 2018, 84, 214–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetti, P.; Pichini, S.; Pacifici, R.; Busardò, F.P.; del Rio, A. Herbal preparations of medical cannabis: A vademecum for prescribing doctors. Medicina 2020, 56, 237. [Google Scholar] [CrossRef]
- Gazzetta Ufficiale. Legge 2 Dicembre 2016, n.242. Disposizioni per la Promozione della Coltivazione e della Filiera Agroindustriale della Canapa (16G00258), GU Serie Generale n. 304 del 30-12-2016. Available online: https://www.gazzettaufficiale.it/eli/id/2016/12/30/16G00258/sg (accessed on 20 April 2022).
- Circolare—Camera dei Deputati. Circolare 31 Luglio 2018, Prot. 2018/43586. Aspetti Giuridico-Operativi Connessi al Fenomeno della Commercializzazione delle Infiorescenze della Canapa Tessile a Basso Tenore di THC e Relazioni con la Normativa Sugli Stupefacenti. Available online: https://www.camera.it/temiap/2018/12/14/OCD177-3851.pdf (accessed on 2 April 2022).
- Gazzetta Ufficiale. Decreto 4 Novembre 2019 Definizione di Livelli Massimi di Tetraidrocannabinolo (THC) Negli Alimenti. (20A00016) (GU Serie Generale n.11 del 15-01-2020). Available online: https://www.gazzettaufficiale.it/eli/id/2020/01/15/20A00016/sg (accessed on 20 April 2022).
- European Union. Annex III, Commission Delegated Regulation (EU) 2017/1155 of 15 February 2017 Amending Delegated Regulation (EU) No 639/2014 as Regards the Control Measures Relating to the Cultivation of Hemp, Certain Provisions on the Greening Payment, the Payment for Young Farmers in Control of a Legal Person, the Calculation of the per Unit Amount in the Framework of Voluntary Coupled Support, the Fractions of Payment Entitlements and Certain Notification Requirements Relating to the Single Area Payment Scheme and the Voluntary Coupled Support, and Amending Annex X to Regulation (EU) No 1307/2013 of the European Parliament and of the Council. Available online: https://eur-lex.europa.eu/eli/reg_del/2017/1155/oj (accessed on 2 April 2022).
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Eisenmann, P.; Ehlers, M.; Weinert, C.H.; Tzvetkova, P.; Silber, M.; Rist, M.J.; Luy, B.; Muhle-Goll, C. Untargeted NMR Spectroscopic Analysis of the Metabolic Variety of New Apple Cultivars. Metabolites 2016, 6, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, S.; Lata, H.; Khan, I.A.; ElSohly, M.A. Cannabis sativa L.—Botany and Horticulture. In Cannabis sativa L.—Botany and Biotechnology, 1st ed.; Chandra, S., Hemant, L., ElSohly, M.A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 79–100. [Google Scholar]
- Aizpurua-Olaizola, O.; Soydaner, U.; Öztürk, E.; Schibano, D.; Simsir, Y.; Navarro, P.; Etxebarria, N.; Usobiaga, A. Evolution of the Cannabinoid and Terpene Content during the Growth of Cannabis sativa Plants from Different Chemotypes. J. Nat. Prod. 2016, 79, 324–331. [Google Scholar] [CrossRef]
- Brighenti, V.; Protti, M.; Anceschi, L.; Zanardi, C.; Mercolini, L.; Pellati, F. Emerging challenges in the extraction, analysis and bioanalysis of cannabidiol and related compounds. J. Pharm. Biomed. Anal. 2021, 192, 113633. [Google Scholar] [CrossRef]
- Ohtsuki, T.; Friesen, J.B.; Chen, S.N.; McAlpine, J.B.; Pauli, G.F. Selective Preparation and High Dynamic-Range Analysis of Cannabinoids in “CBD Oil” and Other Cannabis sativa Preparations. J. Nat. Prod. 2022, 85, 634–646. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.-Y.; Li, S.-H.; Ma, W.; Wu, D.-T.; Li, H.-B.; Xiao, A.-P.; Liu, L.-L.; Zhu, F.; Gan, R.-Y. Cannabis sativa bioactive compounds and their extraction, separation, purification, and identification technologies: An updated review. TrAC 2022, 149, 116554. [Google Scholar] [CrossRef]
- Rezvankhah, A.; Emam-Djomeh, Z.; Safari, M.; Askari, G.; Salami, M. Investigation on the extraction yield, quality, and thermal properties of hemp seed oil during ultrasound-assisted extraction: A comparative study. J. Food Process. Preserv. 2018, 42, e13766. [Google Scholar] [CrossRef]
- Siudem, P.; Wawer, I.; Paradowska, K. Rapid evaluation of edible hemp oil quality using NMR and FT-IR spectroscopy. J. Mol. Struct. 2019, 1177, 204–208. [Google Scholar] [CrossRef]
- Hazekamp, A.; Choi, Y.H.; Verpoorte, R. Quantitative Analysis of Cannabinoids from Cannabis sativa using 1H-NMR. Chem. Pharm. Bull. 2004, 52, 718–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, L.; Brighenti, V.; Rossi, M.C.; Sperlea, J.; Pellati, F.; Bertelli, D. Use of 13C-qNMR Spectroscopy for the Analysis of Non-Psychoactive Cannabinoids in Fibre-Type Cannabis sativa L. (Hemp). Molecules 2019, 24, 1138. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.H.; Kim, H.K.; Hazekamp, A.; Erkelens, C.; Lefeber, A.W.M.; Verpoorte, R. Metabolomic differentiation of Cannabis sativa cultivars using 1H NMR spectroscopy and principal component analysis. J. Nat. Prod. 2004, 67, 953–957. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; Viant, M.R. Two-dimensional J-resolved NMR spectroscopy: Review of a key methodology in the metabolomics toolbox. Phytochem. Anal. 2010, 21, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, Z.; Chen, H.; Feng, J.; Cai, S.; Chen, Z. A high-resolution 2D J-resolved NMR detection technique for metabolite analyses of biological samples. Sci. Rep. 2015, 5, 8390. [Google Scholar] [CrossRef] [Green Version]
- Brighenti, V.; Marchetti, L.; Anceschi, L.; Protti, M.; Verri, P.; Pollastro, F.; Mercolini, L.; Bertelli, D.; Zanardi, C.; Pellati, F. Separation and non-separation methods for the analysis of cannabinoids in Cannabis sativa L. J. Pharm. Biomed. 2021, 206, 114346. [Google Scholar] [CrossRef]
- Kumar Bharti, S.; Roy, R. Quantitative 1H NMR Spectroscopy. TrAC 2012, 35, 5–26. [Google Scholar]
- Araneda, J.F.; Chu, T.; Leclerc, M.C.; Riegel, S.D.; Spingarn, N. Quantitative analysis of cannabinoids using benchtop NMR instruments. Anal. Methods 2020, 12, 4853–4857. [Google Scholar] [CrossRef]
- TopSpin. 2018. Available online: http://www.bruker-biospin.com/topspin.html (accessed on 15 March 2022).
- Varmuz, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics, 1st ed.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2009; pp. 59–101. [Google Scholar]
- Ebrahimi, P.; Viereck, N.; Bro, R.; Engelsen, S.B. Chemometric Analysis of NMR Spectra. In Modern Magnetic Resonance, 2nd ed.; Webb, A.G., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1649–1668. [Google Scholar]
- Ren, S.; Hinzman, A.A.; Kang, E.L.; Szczesniak, R.; Lu, L.J. Computational and Statistical Analysis of Metabolomics Data. Metabolomics 2015, 11, 1492–1513. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 21 March 2022).
- Popescu, R.; Costinel, D.; Dinca, O.R.; Marinescu, A.; Stefanescu, I.; Ionete, R.E. Discrimination of vegetable oils using NMR spectroscopy and chemometrics. Food Control 2015, 48, 84–90. [Google Scholar] [CrossRef]
- Siudem, P.; Wawer, I.; Kowalska, V.; Paradowska, K. Rapid 1H NMR and chemometric methods in verification of hemp-seed oil quality. J. Pharm. Biomed. 2022, 212, 114650. [Google Scholar] [CrossRef] [PubMed]
- Mikulcová, V.; Kašpárková, V.; Humpolíček, P.; Bunková, L. Formulation, Characterization and Properties of Hemp Seed Oil and Its Emulsions. Molecules 2017, 22, 700. [Google Scholar] [CrossRef] [PubMed]
- Farinon, B.; Costantini, L.; Molinari, R.; Di Matteo, G.; Garzoli, S.; Ferri, S.; Ceccantoni, B.; Mannina, L.; Merendino, N. Effect of malting on nutritional and antioxidant properties of the seeds of two industrial hemp (Cannabis sativa L.) cultivars. Food Chem. 2022, 370, 131348. [Google Scholar] [CrossRef]
- Borges, G.R.; Birk, L.; Scheid, C.; Morés, L.; Carasek, E.; Kitamura, R.O.S.; Roveri, F.; Eller, S.; de Oliveira Merib, J.; de Oliveria, T.F. Simple and straightforward analysis of cannabinoids in medicinal products by fast-GC–FID. Forensic Toxicol. 2020, 38, 531–535. [Google Scholar] [CrossRef]
- Mazzara, E.; Torresi, J.; Fico, G.; Papini, A.; Kulbaka, N.; Dall’Acqua, S.; Sut, S.; Garzoli, S.; Mustafa, A.M.; Cappellacci, L.; et al. A Comprehensive Phytochemical Analysis of Terpenes, Polyphenols and Cannabinoids, and Micromorphological Characterization of 9 Commercial Varieties of Cannabis sativa L. Plants 2022, 11, 891. [Google Scholar] [CrossRef]
- Brighenti, V.; Pellati, F.; Steinbach, M.; Maran, D.; Benvenuti, S. Development of a new extraction technique and HPLC method for the analysis of non-psychoactive cannabinoids in Cannabis sativa L. fibosa (hemp). J. Pharm. Biomed. Anal. 2017, 143, 228–236. [Google Scholar] [CrossRef]
- Pellati, F.; Brighenti, V.; Sperlea, J.; Marchetti, L.; Bertelli, D.; Benvenuti, S. New methods for the comprehensive analysis of bioactive compounds in Cannabis sativa L. (hemp). Molecules 2018, 23, 2639. [Google Scholar] [CrossRef] [Green Version]
- Valizadehderakhshan, M.; Shahbazi, A.; Kazem-Rostami, M.; Todd, M.S.; Bhowmik, A.; Wang, L. Extraction of Cannabinoids from Cannabis sativa L. (Hemp)—Review. Agriculture 2021, 11, 384. [Google Scholar] [CrossRef]
- Peschel, W.; Politi, M. 1H NMR and HPLC/DAD for Cannabis sativa L. Chemotype distinction, extract profiling and specification. Talanta 2015, 140, 150–165. [Google Scholar] [CrossRef]
- Spano, M.; Di Matteo, G.; Ingallina, C.; Botta, B.; Quaglio, D.; Ghirga, F.; Balducci, S.; Cammarone, S.; Campiglia, E.; Giusti, A.M.; et al. Emerging challenges in the extraction, analysis and bioanalysis of cannabidiol and related compounds. Molecules 2021, 26, 2912. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.C.; Frei, P.; Weber, S.; Scheurer, E.; · Mercer-Chalmers-Bender, K. Beyond Δ9-tetrahydrocannabinol and cannabidiol: Chemical differentiation of cannabis varieties applying targeted and untargeted analysis. Anal. Bioanal. Chem. 2022, 414, 3847–3862. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, S.; Mascini, M.; Ricci, A.; Fanti, F.; Ottaviani, C.; Lo Sterzo, C.; Sergi, M. Identification of Cannabis sativa L. (hemp) Retailers by Means of Multivariate Analysis of Cannabinoids. Molecules 2019, 24, 3602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudge, E.M.; Murch, S.J.; Brown, P.N. Chemometric Analysis of Cannabinoids: Chemotaxonomy and Domestication Syndrome. Sci. Rep. 2018, 8, 13090. [Google Scholar] [CrossRef] [PubMed]
- Ingallina, C.; Sobolev, A.P.; Circi, S.; Spano, M.; Fraschetti, C.; Filippi, A.; Di Sotto, A.; Di Giacomo, S.; Mazzoccanti, G.; Gasparrini, F.; et al. Cannabis sativa L. Inflorescences from Monoecious Cultivars Grown in Central Italy: An Untargeted Chemical Characterization from Early Flowering to Ripening. Molecules 2020, 25, 1908. [Google Scholar] [CrossRef] [PubMed]
- Olasehinde, T.A.; Olaniran, A.O. Neurotoxicity of anthracene and benz[a]anthracene involves oxidative stress-induced neuronal damage, cholinergic dysfunction and disruption of monoaminergic and purinergic enzymes. Toxicol. Res. 2022, 80, 105312. [Google Scholar] [CrossRef]
- Nagy, D.U.; Cianfaglione, K.; Maggi, F.; Sut, S.; Dall’Acqua, S. Chemical Characterization of Leaves, Male and Female Flowers from Spontaneous Cannabis (Cannabis sativa L.) Growing in Hungary. Chem. Biodivers. 2019, 16, e1800562. [Google Scholar] [CrossRef]
- Kostas Ioannidis, K.; Dadiotis, E.; Mitsis, V.; Melliou, E.; Magiatis, P. Biotechnological Approaches on Two High CBD and CBG Cannabis sativa L. (Cannabaceae) Varieties: In Vitro Regeneration and Phytochemical Consistency Evaluation of Micropropagated Plants Using Quantitative 1H-NMR. Molecules 2020, 25, 5928. [Google Scholar] [CrossRef]
- Siciliano, C.; Bartella, L.; Mazzotti, F.; Aiello, D.; Napoli, A.; De Luca, P.; Temperini, A. 1H NMR quantification of cannabidiol (CBD) in industrial products derived from Cannabis sativa L. (hemp) seeds. IOP Conf. Ser. Mater. Sci. Eng. 2019, 572, 012010. [Google Scholar] [CrossRef]
- Barthlott, I.; Scharinger, A.; Golombek, P.; Kuballa, T.; Lachenmeier, D.W. A Quantitative 1H NMR Method for Screening Cannabinoids in CBD Oils. Toxics 2021, 9, 136. [Google Scholar] [CrossRef]
- Risoluti, R.; Gullifa, G.; Battistini, A.; Materazzi, S. Monitoring of cannabinoids in hemp flours by MicroNIR/Chemometrics. Talanta 2020, 211, 120672. [Google Scholar] [CrossRef] [PubMed]
- Sgrò, S.; Lavezzi, B.; Caprari, C.; Polito, M.; D’Elia, M.; Lago, G.; Furlan, G.; Girotti, S.; Ferri, E.N. Delta9-THC determination by the EU official method: Evaluation of measurement uncertainty and compliance assessment of hemp samples. Anal. Bioanal. Chem. 2021, 413, 3399–3410. [Google Scholar] [CrossRef] [PubMed]
- Nahar, L.; Guo, M.; Sarker, S.D. Gas chromatographic analysis of naturally occurring cannabinoids: A review of literature published during the past decade. Phytochem. Anal. 2020, 31, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Dadiotis, E.; Mitsis, V.; Melliou, E.; Magiatis, P. Direct Quantitation of Phytocannabinoids by One-Dimensional 1H qNMR and Two-Dimensional 1H-1H COSY qNMR in Complex Natural Mixtures. Molecules 2022, 27, 2965. [Google Scholar] [CrossRef] [PubMed]


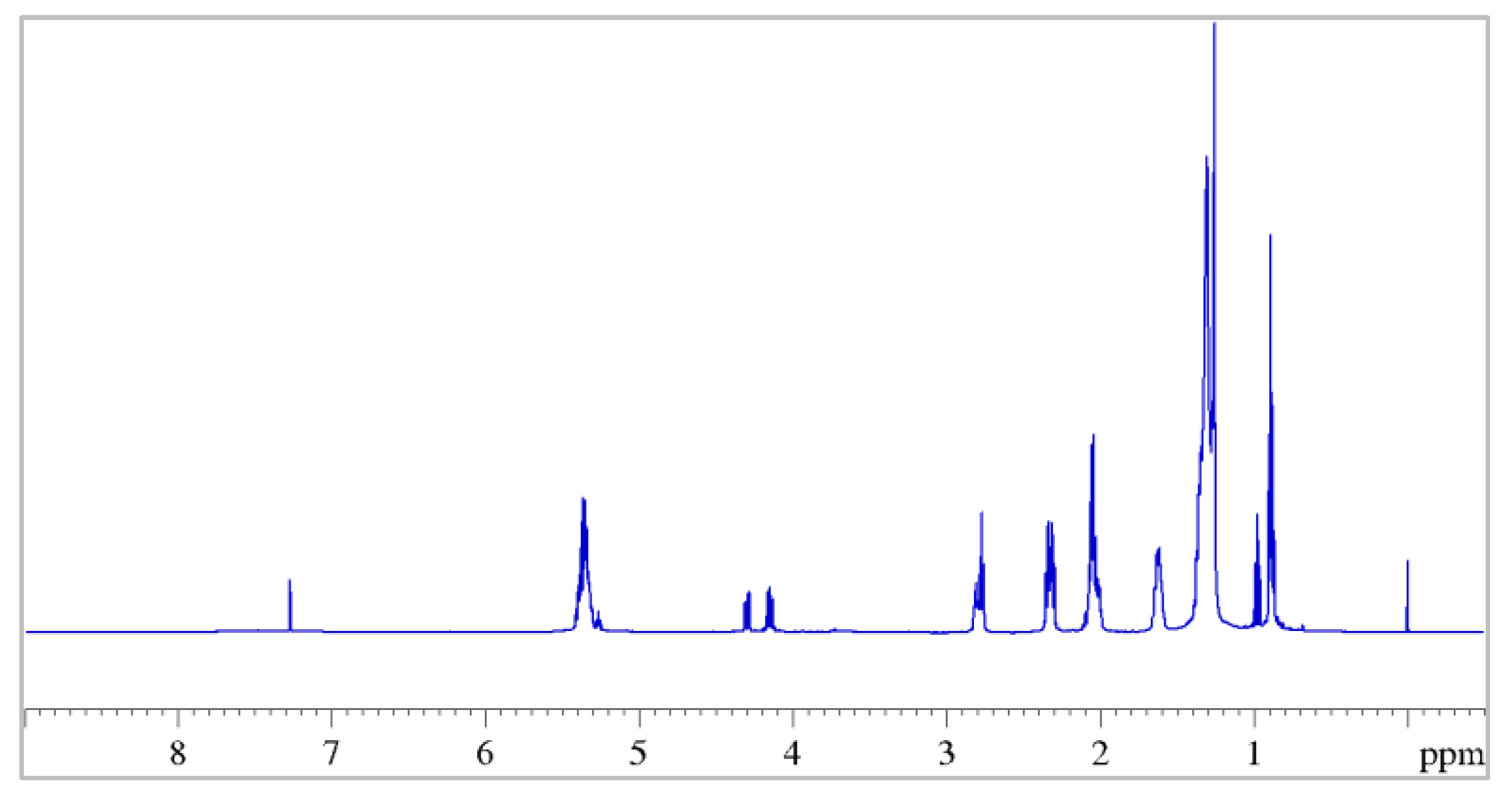
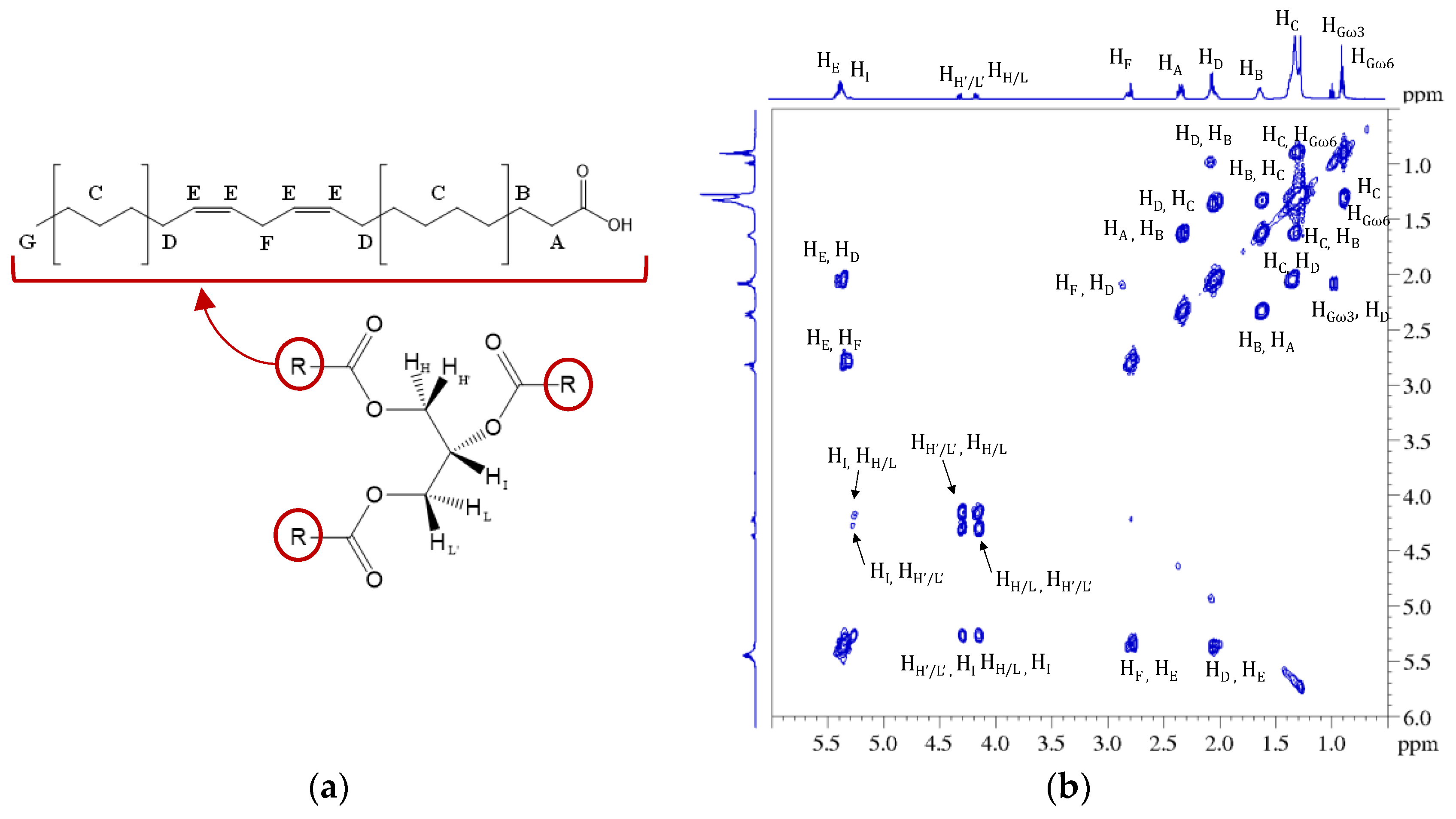




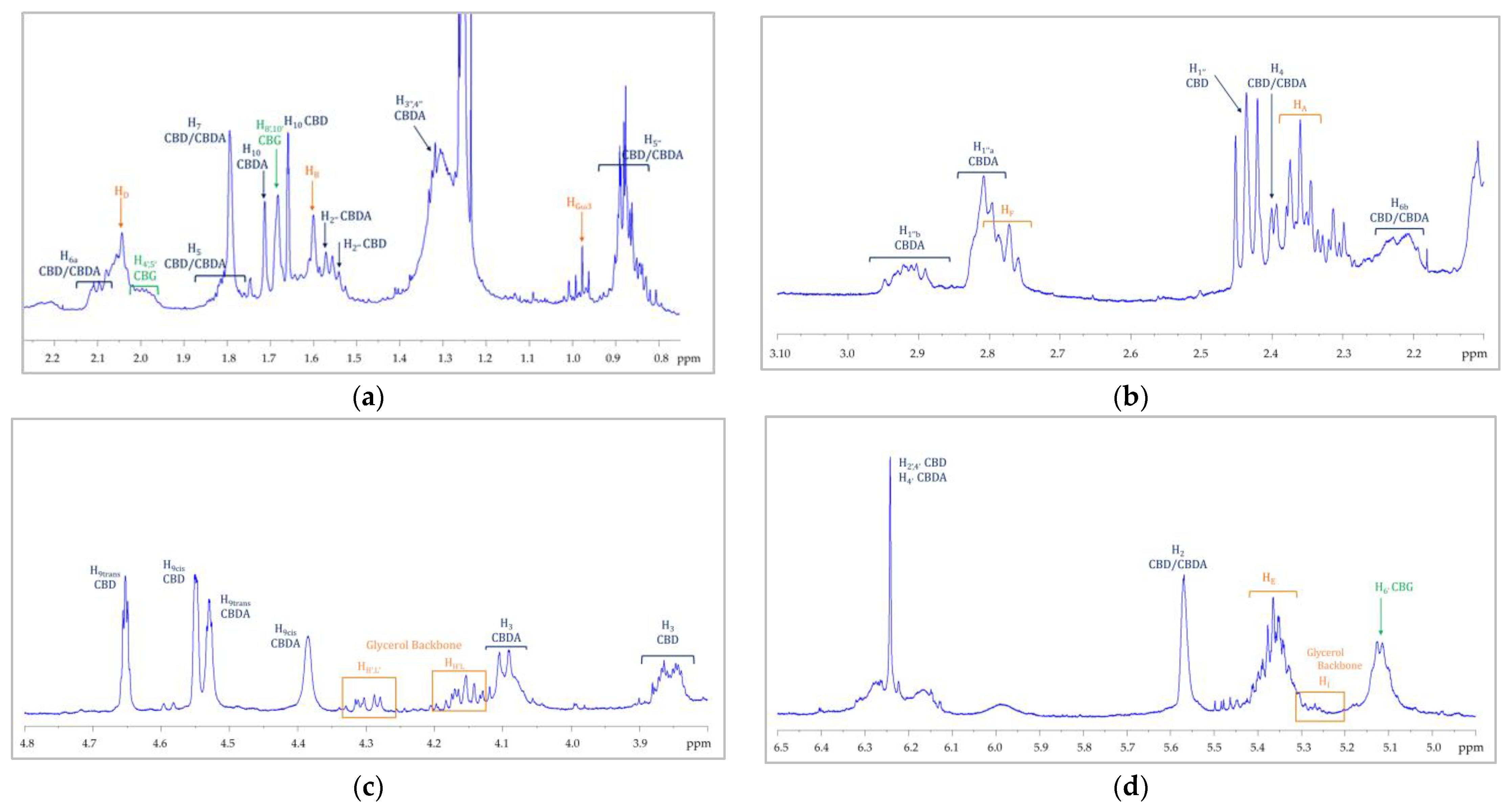
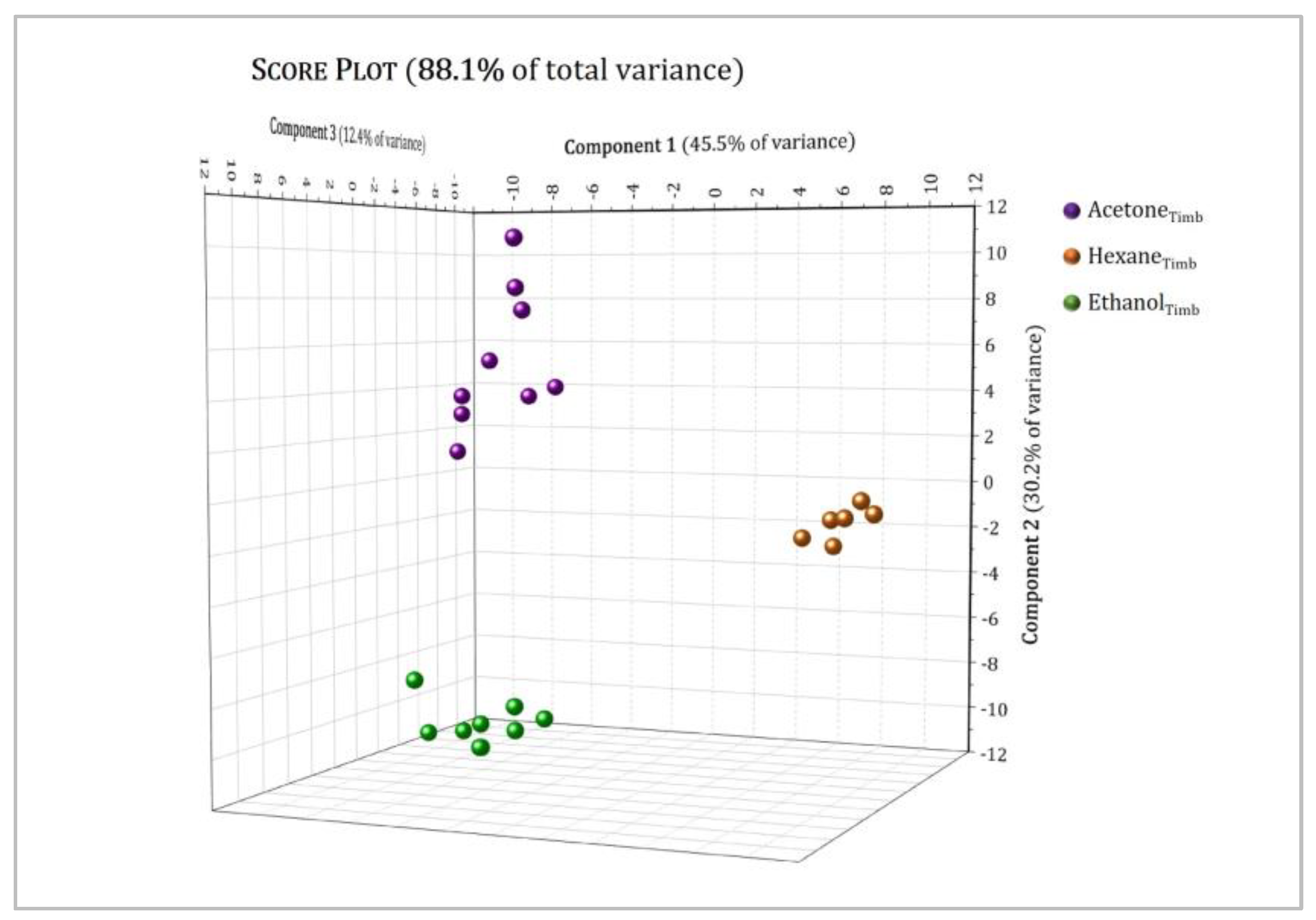
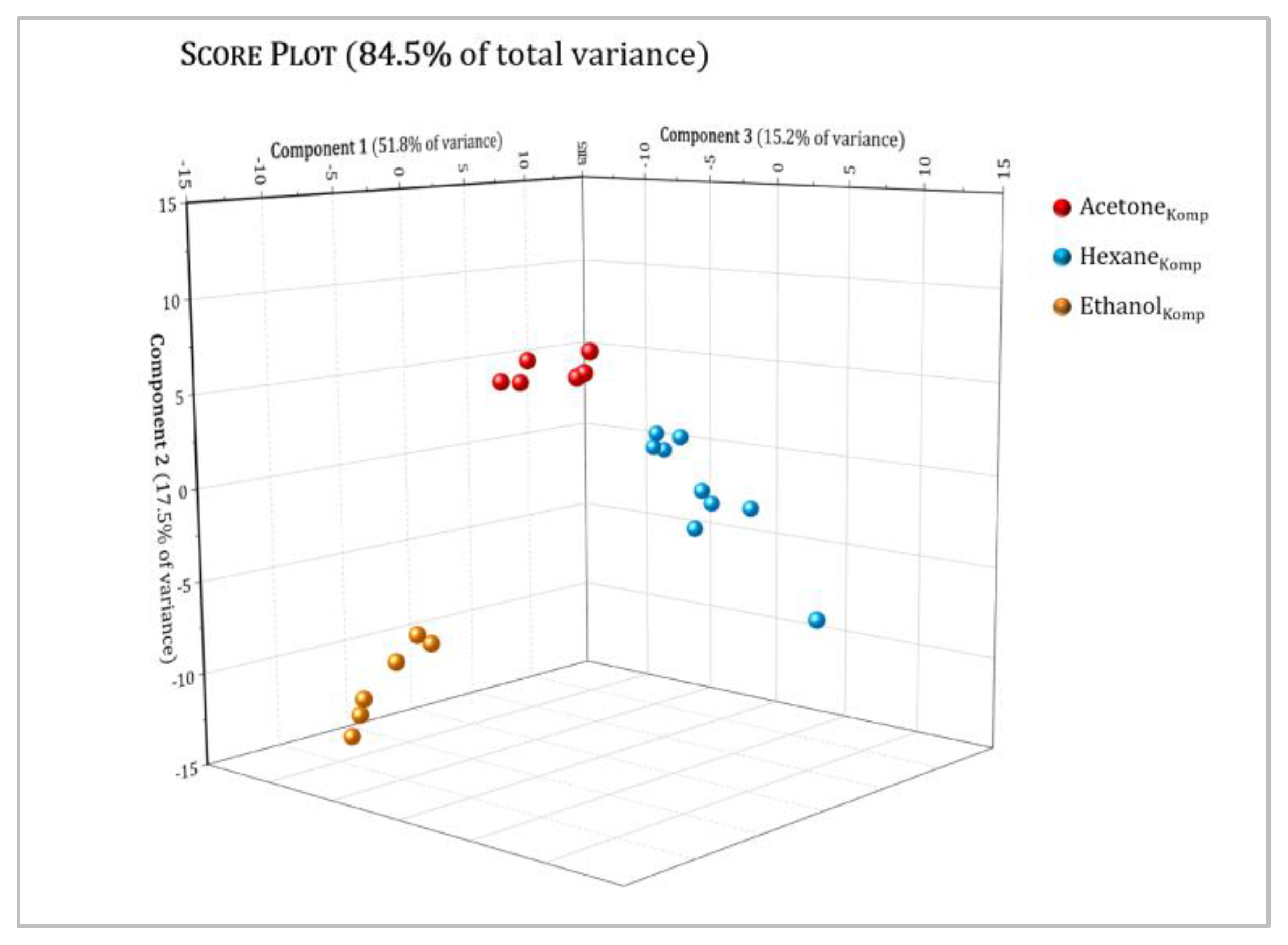
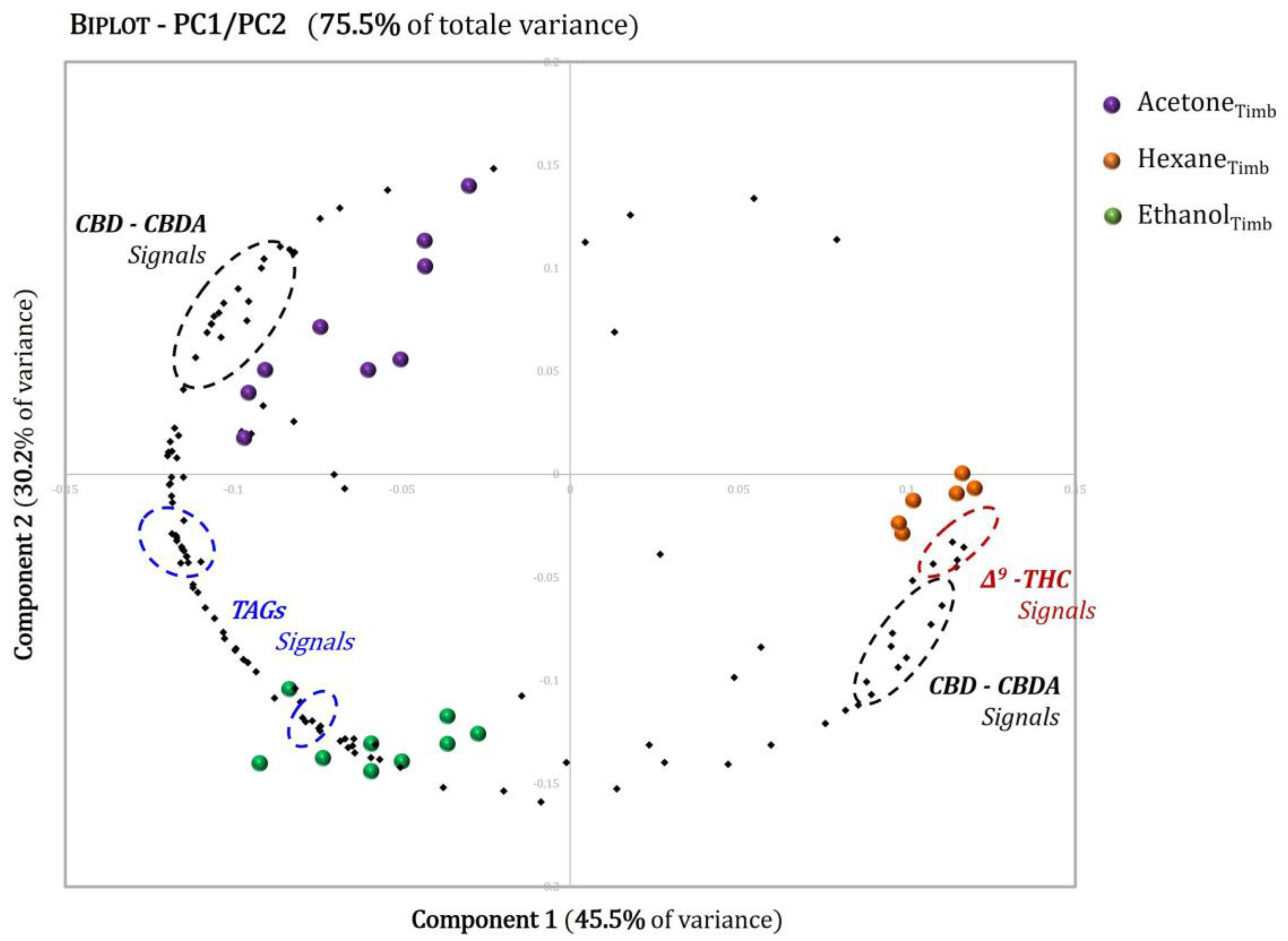
| Position | δH, Multiplicity a (J in Hz) | COSY |
|---|---|---|
| A | 2.27–2.37, m | E |
| B | 1.55–1.67, m | C, A |
| C | 1.23–1.39, m | Gω6 B, D |
| D | 1.98–2.11, m | Gω3, C, F, E |
| E | 5.28–5.42, m | D, F |
| F | 2.72–2.86, m | D, E |
| Gω3 | 0.97, t | D |
| Gω6 | 0.88, m | C |
| H, L (Gly a) | 4.14, dd (11.88, 5.93) | H’, L’, I |
| I (Gly a) | 5.26, m | H, H’, L, L’ |
| H’, L’ (Gly a) | 4.29, dd (11.88, 4.31) | H, L, I |
| Compound | δ 1H ppm (Multiplicity *, 1H-1H J-Coupling—Hz) | δ 13C ppm | ||
|---|---|---|---|---|
| CBD | H3 | 3.86 (ddt; JH3-H4 = 13.00 Hz (d), JH3-H2 = 3.51 Hz (d), JH3-H5 = 2.51 Hz (t)) | C3 | 37.01 |
| H2 | 5.57 | C2 | 124.14 | |
| H6a | 2.05–2.09 | C6 | 30.36 | |
| H6b | 2.22 | |||
| H5 | 1.78–1.84 (ddd; JH5-H4 = 5.30 Hz (d), JH5-H6a = 1.30 Hz (d), JH5-H6b = 0.60 Hz (d)) | C5 | 28.35 | |
| H4 | 2.40 (dd; JH4-H3 = 13.00 Hz (d), JH4-H5 = 5.00 Hz (d)) | C4 | 46.16 | |
| H7 | 1.79 (d;3JH7-H2 = 0.50 Hz) | C7 | 23.69 | |
| H9trans | 4.64 (dq; J9trans-9cis = 2.65 Hz (d), 3J9trans-10 = 1.50 Hz (q)) | C9 | 110.81 | |
| H9cis | 4.53 (dq; J9cis-9trans = 2.65 Hz (d), 3J9cis-10 = 0.92 Hz (q)) | |||
| H10 | 1.66 (dd; 3J10-9cis = 0.92 Hz (d), 3J10-9trans = 1.50 Hz (d)) | C10 | 20.30 | |
| H2′ | 6.26 | C2′ | 109.56 | |
| H4′ | 6.16 | C4′ | 107.92 | |
| H1″ | 2.43 (t) | C1″ | 35.46 | |
| H2″ | 1.52–1.61 | C2″ | 30.65 | |
| H3″, H4″ | 1.27–1.32 | C3″ | 31.48 | |
| C4″ | 22.54 | |||
| H5″ | 0.86–0.88 | C5″ | 14.04 | |
| CBDA | H3 | 4.08 | C3 | 35.38 |
| H2 | 5.55 | C2 | 124.14 | |
| H6a | 2.05–2.09 | C6 | 30.36 | |
| H6b | 2.22 | |||
| H5 | 1.79 (ddd; JH5-H4 = 5.30 Hz (d), JH5-H6a = 1.30 Hz (d), JH5-H6b = 0.60 Hz (d)) | C5 | 28.35 | |
| H4 | 2.40 (dd; JH4-H3 = 13.00 Hz (d), JH4-H5 = 5.00 Hz (d)) | C4 | 46.45 | |
| H7 | 1.79 (d; 3JH7-H2 = 0.50 Hz) | C7 | 23.69 | |
| H9trans | 4.51 (dq; 3J9cis-9trans = 3.00 Hz (d); 3J9trans-10 = 1.76 Hz (q)) | C9 | 111.21–111.25 | |
| H9cis | 4.39 (dm; 3J9cis-9trans = 3.00 Hz (d)) | |||
| H10 | 1.70 | C10 | 18.91 | |
| H4′ | 6.21 | C4′ | 111.21–111.25 | |
| H1″a | 2.81 | C1″ | 36.68 | |
| H1″b | 2.92 | |||
| H2″ | 1.52–1.61 | C2″ | 31.24 | |
| H3″, H4″ | 1.27–1.32 | C3″ | 31.94 | |
| C4″ | 22.54 | |||
| H5″ | 0.86–0.88 | C5″ | 14.04 | |
| CBG | H2 | 6.24 | C2 | 108.25 |
| H5′, H4′ | 2.04 | C4′ | 32.28 | |
| C5″ | 26.51 | |||
| H6′ | 5.12 | C6′ | 125.08 | |
| H8′, H10′ | 1.68 | C8′ | 20.51 | |
| C10″ | 23.44 | |||
| Compound | qNMR on Flowers UAE Extracts | GC-FID | |||
|---|---|---|---|---|---|
| qNMR IS | Hexane | Acetone | Ethanol | ||
| CBDA content * | Anthracene | 6.3 ± 0.8 | 0.40 ± 0.1 | 0.31 ± 0.04 | Validated Laboratory Method 6.9 ± 0.2 Referred to CBD after decarbossilation |
| Benzoic acid | 6.5 ± 0.8 | 0.40 ± 0.1 | 0.39 ± 0.06 | ||
| TMSP-d4 | 6.4 ± 0.6 | 0.41 ± 0.1 | 0.41 ± 0.06 | ||
| CBD content * | Anthracene | 0.4 ± 0.1 | 4.60 ± 0.8 | 0.30 ± 0.1 | |
| Benzoic acid | 0.30 ± 0.06 | 4.54 ± 0.6 | 2.2 ± 0.1 | ||
| TMSP-d4 | 0.4 ± 0.1 | 4.59 ± 0.6 | 2.9 ± 0.1 | ||
| Δ9-THC content * | Anthracene | 0.11 ± 0.44 | <LOD | <LOD | Regulation (EU) N° 639/2014 [15] 0.09 ± 0.01 |
| Benzoic acid | 0.07 ± 0.02 | ||||
| TMSP-d4 | 0.10 ± 0.02 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colella, M.F.; Salvino, R.A.; Gaglianò, M.; Litrenta, F.; Oliviero Rossi, C.; Le Pera, A.; De Luca, G. NMR Spectroscopy Applied to the Metabolic Analysis of Natural Extracts of Cannabis sativa. Molecules 2022, 27, 3509. https://doi.org/10.3390/molecules27113509
Colella MF, Salvino RA, Gaglianò M, Litrenta F, Oliviero Rossi C, Le Pera A, De Luca G. NMR Spectroscopy Applied to the Metabolic Analysis of Natural Extracts of Cannabis sativa. Molecules. 2022; 27(11):3509. https://doi.org/10.3390/molecules27113509
Chicago/Turabian StyleColella, Maria Francesca, Rosachiara Antonia Salvino, Martina Gaglianò, Federica Litrenta, Cesare Oliviero Rossi, Adolfo Le Pera, and Giuseppina De Luca. 2022. "NMR Spectroscopy Applied to the Metabolic Analysis of Natural Extracts of Cannabis sativa" Molecules 27, no. 11: 3509. https://doi.org/10.3390/molecules27113509
APA StyleColella, M. F., Salvino, R. A., Gaglianò, M., Litrenta, F., Oliviero Rossi, C., Le Pera, A., & De Luca, G. (2022). NMR Spectroscopy Applied to the Metabolic Analysis of Natural Extracts of Cannabis sativa. Molecules, 27(11), 3509. https://doi.org/10.3390/molecules27113509