Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve cAT-Triggered Tumor Cell Death
Abstract
1. Introduction
2. Results
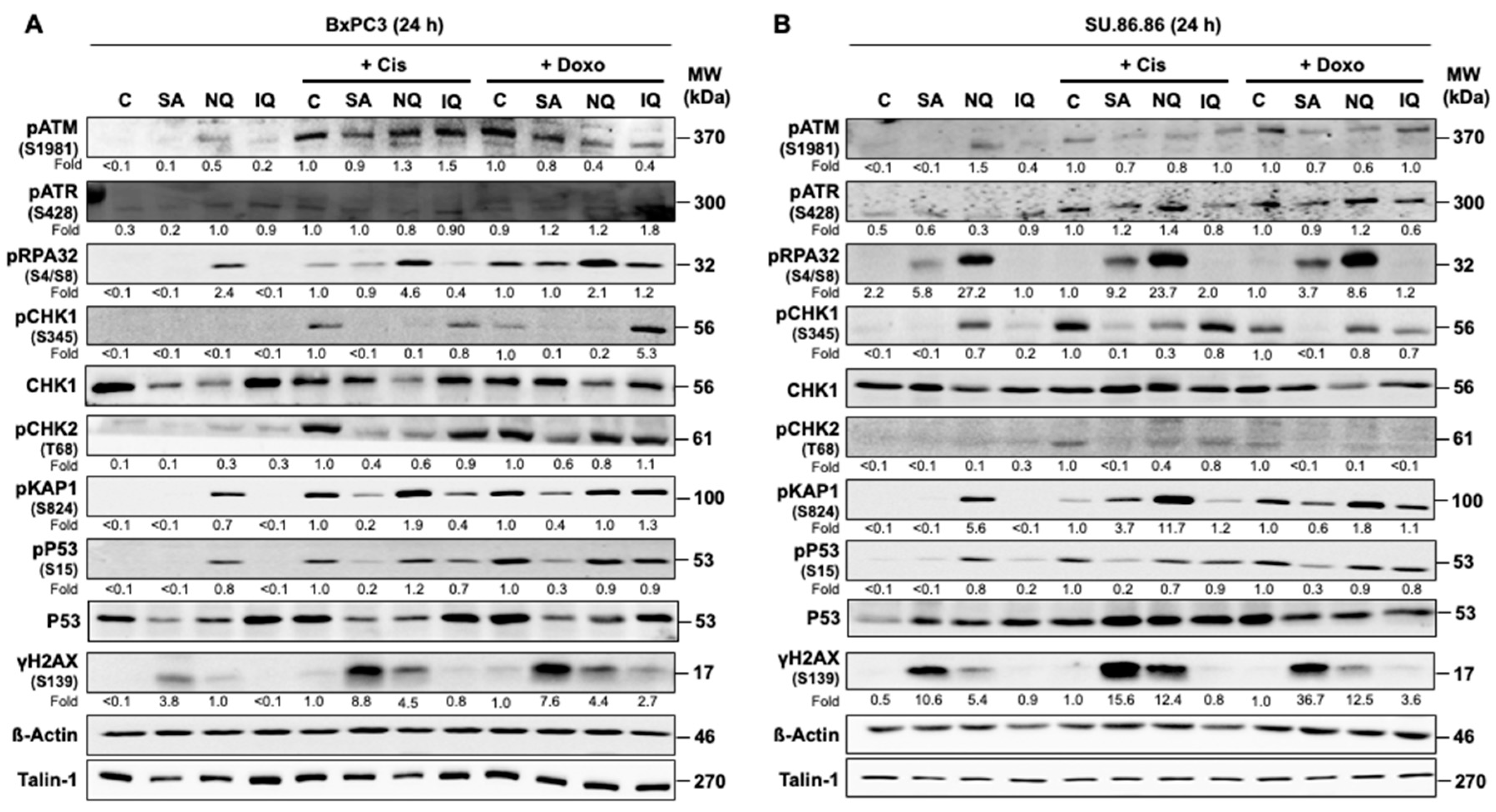
2.1. Influence of NC on Mechanisms of the DDR in Pancreatic Carcinoma Cells
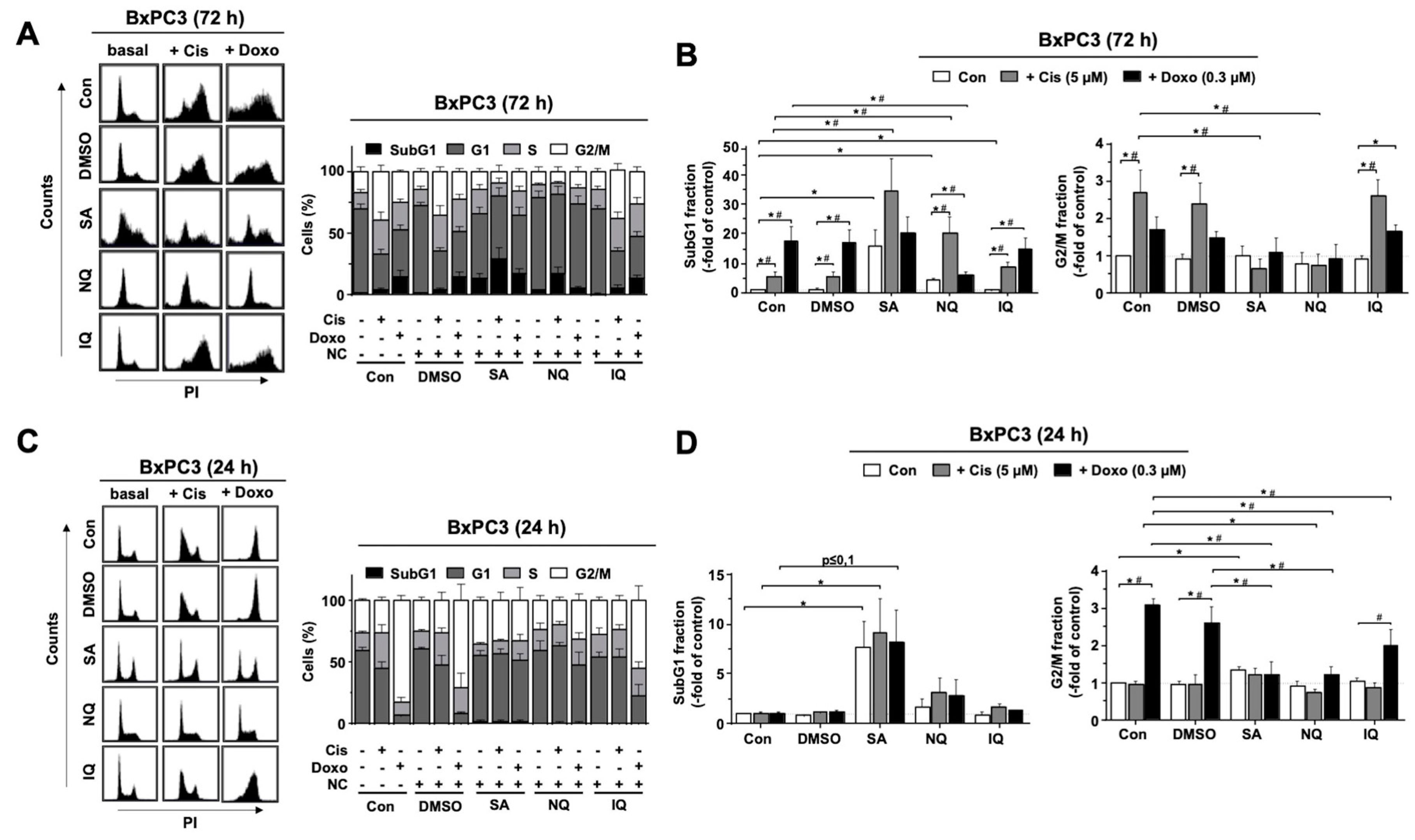
2.2. Cell Cycle Progression and Cell Death Induction Is Differently Affected by NC
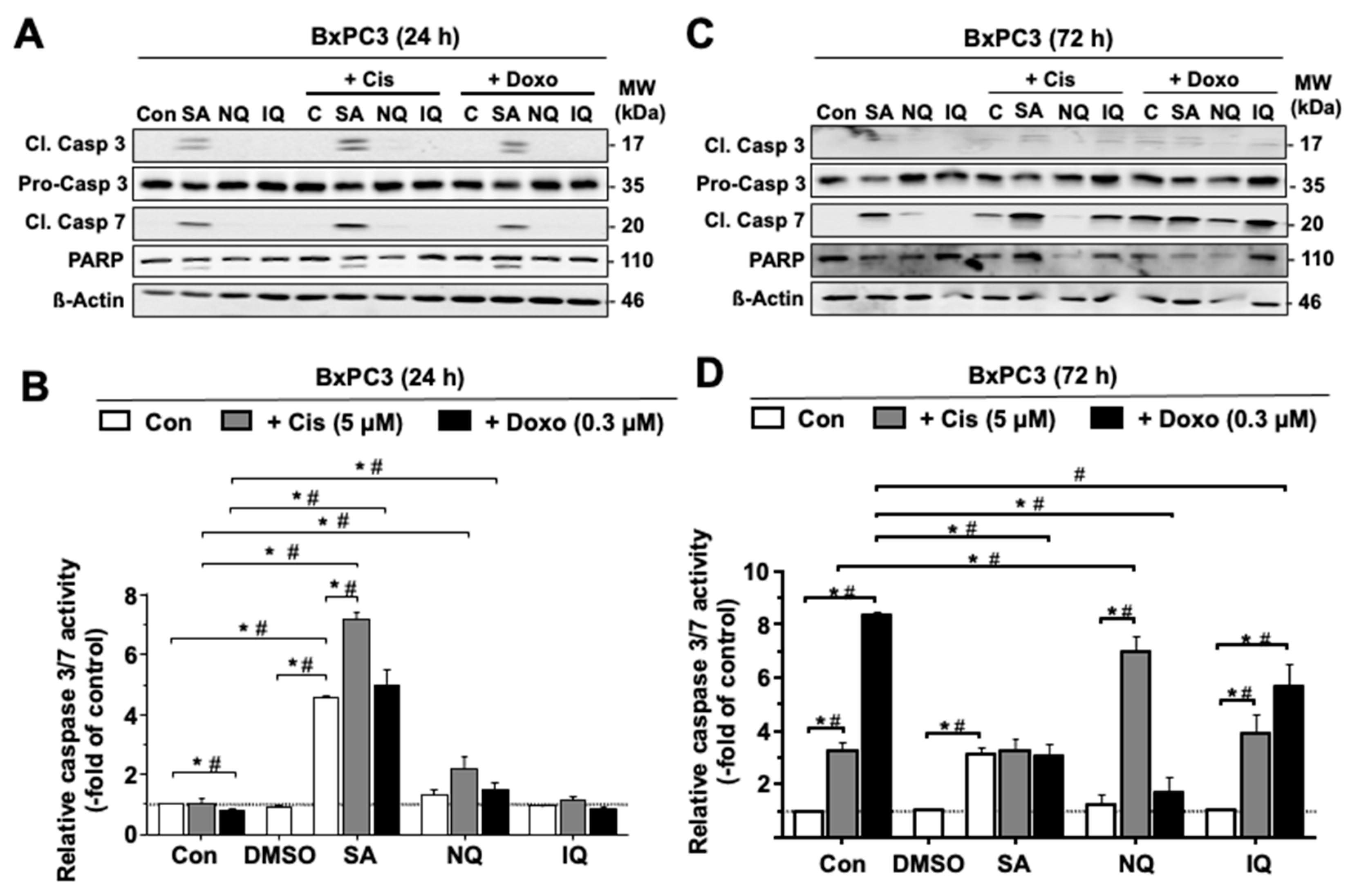
2.3. Influence of NC on the Activation of Caspase-Related Death Mechanisms
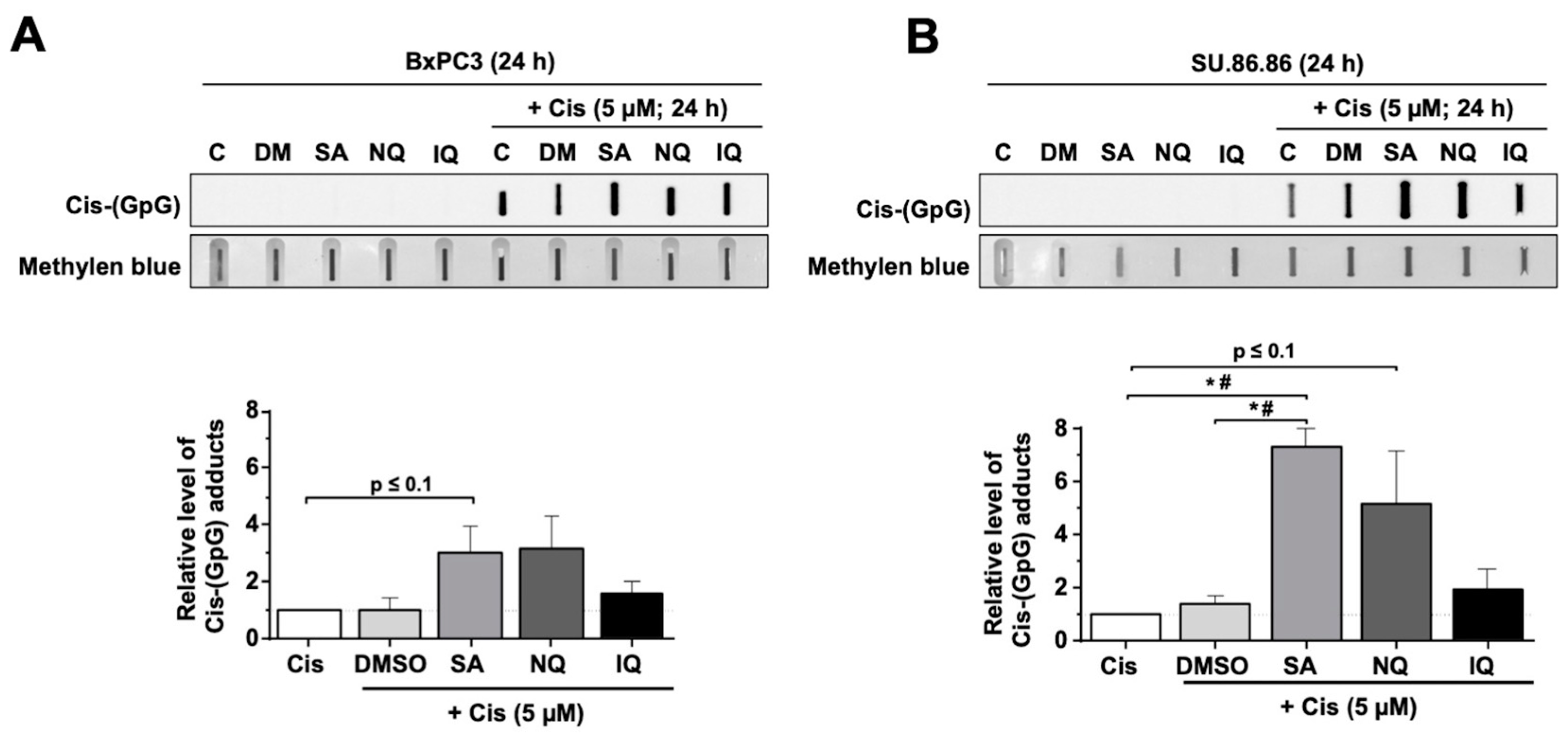
2.4. Impact of NC on the Formation of CisPt-Induced DNA Intrastrand Crosslinks
2.5. Formation and Repair of cAT-Induced DSB Is Affected by NC
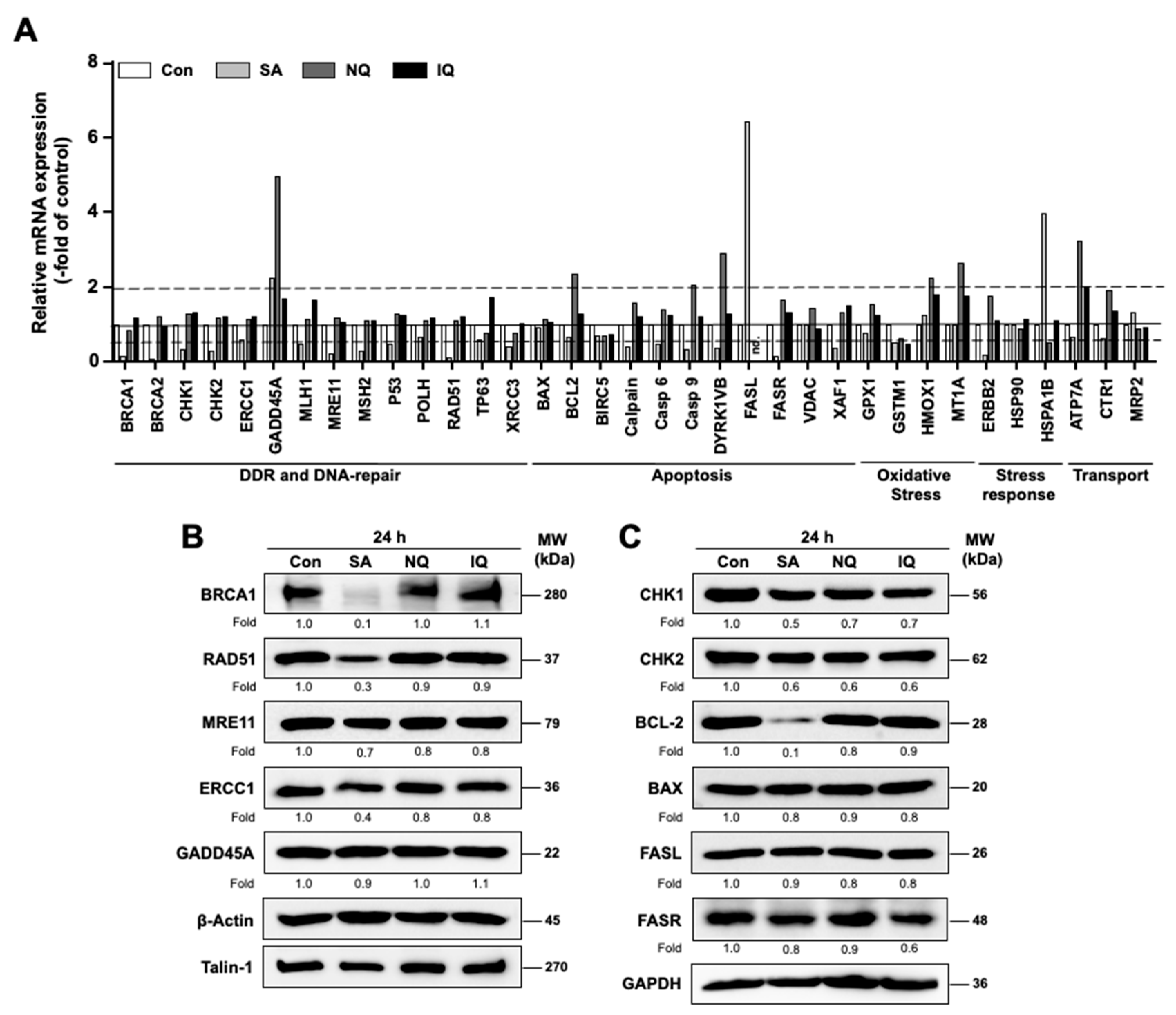
2.6. Impact of NC on the mRNA and Protein Expression of Susceptibility-Related Factors
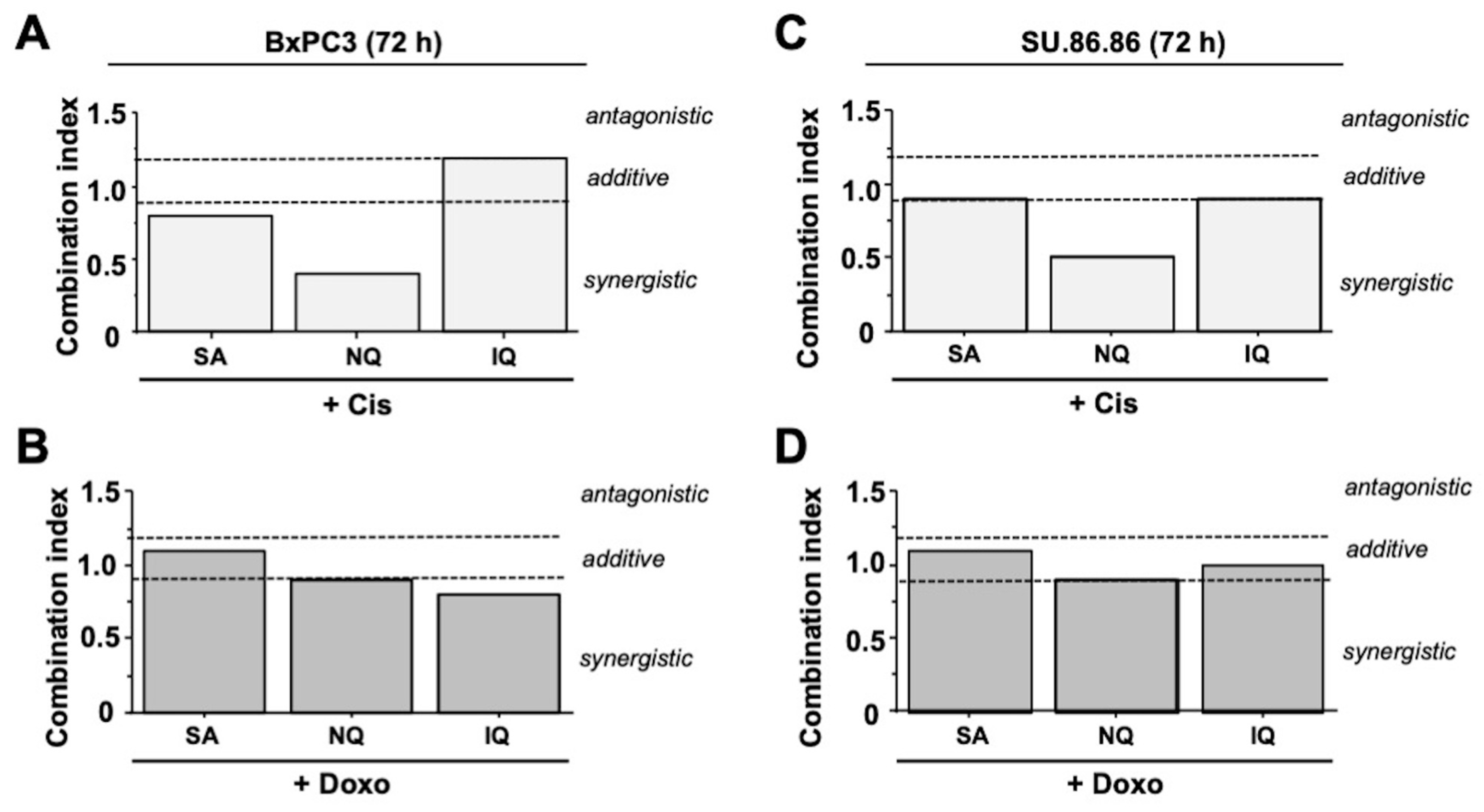
2.7. Additive and Synergistic Cytotoxic Effects of NC
2.8. Cytotoxic Activity of NC on Various Tumor Cell Lines
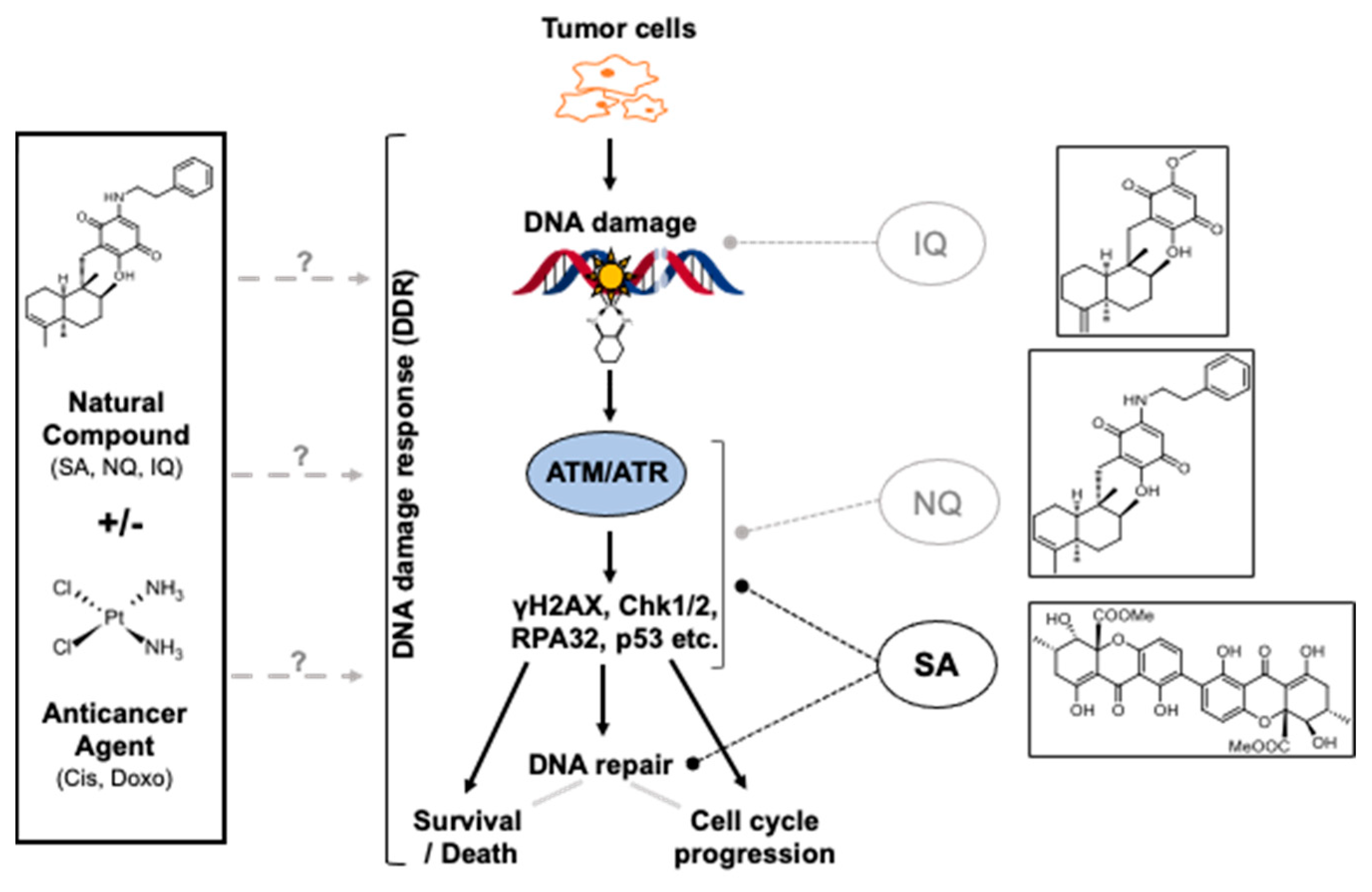
3. Discussion
3.1. Influence of NQ, IQ and SA on Mechanisms of the DDR
3.2. Modulation of Cell Cycle Progression and Cell Death Pathways by NC
3.3. Inhibitory Effects of SA on Mechanisms of DNA DSB Repair
4. Materials and Methods
4.1. Materials
4.2. Cell Culture and Drug Treatments
4.3. Determination of Cell Viability
4.4. Combination Index
4.5. Analysis of Nuclear Foci Formation
4.6. Western Blot Analysis
4.7. Analyses of Cell Cycle Progression by Flow Cytometry
4.8. Determination of Apoptotic Cell Death
4.9. Analysis of Platinum-Induced DNA Intrastrand Crosslinks
4.10. Quantitative Real-Time PCR-Based mRNA Expression Analyses
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y. ATM and ATR: Networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001, 11, 71–77. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Durocher, D.; Jackson, S.P. DNA-PK, ATM and ATR as sensors of DNA damage: Variations on a theme? Curr. Opin. Cell Biol. 2001, 13, 225–231. [Google Scholar] [CrossRef]
- Kumagai, A.; Dunphy, W.G. How cells activate ATR. Cell Cycle 2006, 5, 1265–1268. [Google Scholar] [CrossRef][Green Version]
- Petermann, E.; Caldecott, K.W. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle 2006, 5, 2203–2209. [Google Scholar] [CrossRef]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Batista, L.F.; Roos, W.P.; Christmann, M.; Menck, C.F.; Kaina, B. Differential Sensitivity of Malignant Glioma Cells to Methylating and Chloroethylating Anticancer Drugs: p53 Determines the Switch by Regulating xpc, ddb2, and DNA Double-Strand Breaks. Cancer Res. 2007, 67, 11886–11895. [Google Scholar] [CrossRef]
- Matt, S.; Hofmann, T.G. The DNA damage-induced cell death response: A roadmap to kill cancer cells. Cell Mol. Life Sci. 2016, 73, 2829–2850. [Google Scholar] [CrossRef]
- Conrad, E.U.; Poloniovallon, T.; Meister, M.; Matt, S.; Bitomsky, N.; Herbel, C.; Liebl, M.; Greiner, V.J.; Kriznik, B.; Schumacher, S.E.; et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 2016, 23, 110–122. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Möller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Dröge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Eich, M.; Roos, W.P.; Nikolova, T.; Kaina, B. Contribution of ATM and ATR to the resistance of glioblastoma and malignant melanoma cells to the methylating anticancer drug temozolomide. Mol. Cancer Ther. 2013, 12, 2529–2540. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.M.; Ryan, A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015, 149, 124–138. [Google Scholar] [CrossRef]
- Carrassa, L.; Damia, G. DNA damage response inhibitors: Mechanisms and potential applications in cancer therapy. Cancer Treat. Rev. 2017, 60, 139–151. [Google Scholar] [CrossRef]
- Zhou, B.-B.S.; Bartek, J. Targeting the checkpoint kinases: Chemosensitization versus chemoprotection. Nat. Rev. Cancer 2004, 4, 216–225. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. The DNA damage response and cancer therapy. Nature 2012, 481, 287–294. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Ozleyen, A.; Tumer, T.B.; Adetunji, C.O.; El Omari, N.; Balahbib, A.; Taheri, Y.; Bouyahya, A.; Martorell, M.; Martins, N.; et al. Natural Products and Synthetic Analogs as a Source of Antitumor Drugs. Biomolecules 2019, 9, 679. [Google Scholar] [CrossRef]
- Dehelean, C.A.; Marcovici, I.; Soica, C.; Mioc, M.; Coricovac, D.; Iurciuc, S.; Cretu, O.M.; Pinzaru, I. Plant-Derived Anticancer Compounds as New Perspectives in Drug Discovery and Alternative Therapy. Molecules 2021, 26, 1109. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Das, J.; Lalhlenmawia, H.; Tonk, R.K.; Singh, L.; Kumar, D. Targeting Cancer Using Phytoconstituents-Based Drug Delivery; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Ren, Y.; De Blanco, E.J.C.; Fuchs, J.R.; Soejarto, D.D.; Burdette, J.E.; Swanson, S.M.; Kinghorn, A.D. Potential Anticancer Agents Characterized from Selected Tropical Plants. J. Nat. Prod. 2019, 82, 657–679. [Google Scholar] [CrossRef] [PubMed]
- Seca, A.M.L.; Pinto, D.C.G.A. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef] [PubMed]
- Lichota, A.; Gwozdzinski, K. Anticancer Activity of Natural Compounds from Plant and Marine Environment. Int. J. Mol. Sci. 2018, 19, 3533. [Google Scholar] [CrossRef]
- Shao Rong, G.; Cao Chun, X.; Zhang, H.; Kohn Kurt, W.; Wold Marc, S.; Pommier, Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO Eur. Mol. Biol. Organ. J. 1999, 18, 1397–1406. [Google Scholar] [CrossRef]
- Ferrante, K.; Winograd, B.; Canetta, R. Promising new developments in cancer chemotherapy. Cancer Chemother. Pharmacol. 1999, 43, S61–S68. [Google Scholar] [CrossRef]
- Mohan, A.; Narayanan, S.; Sethuraman, S.; Krishnan, U.M. Combinations of plant polyphenols & anti-cancer molecules: A novel treatment strategy for cancer chemotherapy. Anti-Cancer Agents Med. Chem. 2013, 13, 281–295. [Google Scholar] [CrossRef]
- Spatafora, C.; Tringali, C. Natural-derived polyphenols as potential anticancer agents. Anti-Cancer Agents Med. Chem. 2012, 12, 902–918. [Google Scholar] [CrossRef]
- Kreuter, M.-H.; Leake, R.E.; Rinaldi, F.; Müller-Klieser, W.; Maidhof, A.; Müller, W.E.; Schröder, H.C. Inhibition of intrinsic protein tyrosine kinase activity of EGF-receptor kinase complex from human breast cancer cells by the marine sponge metabolite (+)-aeroplysinin-1. Comp. Biochem. Physiol. Part B Comp. Biochem. 1990, 97, 151–158. [Google Scholar] [CrossRef]
- Jin, M.; Zhao, W.; Zhang, Y.; Kobayashi, M.; Duan, H.; Kong, D. Antiproliferative effect of aaptamine on human chronic myeloid leukemia k562 cells. Int. J. Mol. Sci. 2011, 12, 7352–7359. [Google Scholar] [CrossRef]
- Stuhldreier, F.; Kassel, S.; Schumacher, L.; Wesselborg, S.; Proksch, P.; Fritz, G. Pleiotropic effects of spongean alkaloids on mechanisms of cell death, cell cycle progression and DNA damage response (DDR) of acute myeloid leukemia (AML) cells. Cancer Lett. 2015, 361, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Li, P.C.; Lam, E.; Roos, W.P.; Zdzienicka, M.Z.; Kaina, B.; Efferth, T. Artesunate Derived from Traditional Chinese Medicine Induces DNA Damage and Repair. Cancer Res. 2008, 68, 4347–4351. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hou, D.; Liu, Q.; Wu, T.; Guo, H.; Zhang, X.; Zou, Y.; Liu, Z.; Liu, J.; Wei, J.; et al. Artesunate sensitizes ovarian cancer cells to cisplatin by downregulating RAD51. Cancer Biol. Ther. 2015, 16, 1548–1556. [Google Scholar] [CrossRef]
- Berdelle, N.; Nikolova, T.; Quiros, S.; Efferth, T.; Kaina, B. Artesunate Induces Oxidative DNA Damage, Sustained DNA Double-Strand Breaks, and the ATM/ATR Damage Response in Cancer Cells. Mol. Cancer Ther. 2011, 10, 2224–2233. [Google Scholar] [CrossRef] [PubMed]
- Berte, N.; Lokan, S.; Eich, M.; Kim, E.; Kaina, B. Artesunate enhances the therapeutic response of glioma cells to temozolomide by inhibition of homologous recombination and senescence. Oncotarget 2016, 7, 67235–67250. [Google Scholar] [CrossRef] [PubMed]
- Gatz, S.A.; Keimling, M.; Baumann, C.; Dörk, T.; Debatin, K.-M.; Fulda, S.; Wiesmüller, L. Resveratrol modulates DNA double-strand break repair pathways in an ATM/ATR–p53- and –Nbs1-dependent manner. Carcinogenesis 2008, 29, 519–527. [Google Scholar] [CrossRef]
- Li, B.; Hou, D.; Guo, H.; Zhou, H.; Zhang, S.; Xu, X.; Liu, Q.; Zhang, X.; Zou, Y.; Gong, Y.; et al. Resveratrol sequentially induces replication and oxidative stresses to drive p53-CXCR2 mediated cellular senescence in cancer cells. Sci. Rep. 2017, 7, 208. [Google Scholar] [CrossRef]
- Rejhová, A.; Opattová, A.; Čumová, A.; Slíva, D.; Vodička, P. Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 2018, 144, 582–594. [Google Scholar] [CrossRef]
- Efferth, T. Cancer combination therapy of the sesquiterpenoid artesunate and the selective EGFR-tyrosine kinase inhibitor erlotinib. Phytomedicine 2017, 37, 58–61. [Google Scholar] [CrossRef]
- Efferth, T. Cancer combination therapies with artemisinin-type drugs. Biochem. Pharmacol. 2017, 139, 56–70. [Google Scholar] [CrossRef]
- Van Stuijvenberg, J.; Proksch, P.; Fritz, G. Targeting the DNA damage response (DDR) by natural compounds. Bioorganic Med. Chem. 2020, 28, 115279. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Balancing life-or-death decisions. Science 2006, 314, 261–262. [Google Scholar] [CrossRef] [PubMed]
- Toledo, L.I.; Altmeyer, M.; Rask, M.-B.; Lukas, C.; Larsen, D.H.; Povlsen, L.K.; Bekker-Jensen, S.; Mailand, N.; Bartek, J.; Lukas, J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell 2013, 155, 1088–1103. [Google Scholar] [CrossRef] [PubMed]
- De Feraudy, S.; Revet, I.; Bezrookove, V.; Feeney, L.; Cleaver, J.E. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2010, 107, 6870–6875. [Google Scholar] [CrossRef] [PubMed]
- Marti, T.M.; Hefner, E.; Feeney, L.; Natale, V.; Cleaver, J.E. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2006, 103, 9891–9896. [Google Scholar] [CrossRef]
- Chou, T.C. Comparison of drug combinations in vitro, in animals, and in clinics by using the combination index method via computer simulation. Cancer Res. 2009, 69, 4752. [Google Scholar]
- Chou, T.C.; Chou, L.H. Computerized indexing of drug combinations: Prediction of synergism and antagonism of more than two drugs by polygonogram. FASEB J. 1998, 12, A143. [Google Scholar]
- Daletos, G.; de Voogd, N.J.; Müller, W.E.G.; Wray, V.; Lin, W.; Feger, D.; Kubbutat, M.; Aly, A.H.; Proksch, P. Cytotoxic and Protein Kinase Inhibiting Nakijiquinones and Nakijiquinols from the Sponge Dactylospongia Metachromia. J. Nat. Prod. 2014, 77, 218–226. [Google Scholar] [CrossRef]
- Amaral, N.; Vendrell, A.; Funaya, C.; Idrissi, F.-Z.; Maier, M.; Kumar, A.; Neurohr, G.; Colomina, N.; Torres-Rosell, J.; Geli, M.-I.; et al. The Aurora-B-dependent NoCut checkpoint prevents damage of anaphase bridges after DNA replication stress. Nat. Cell Biol. 2016, 18, 516–526. [Google Scholar] [CrossRef]
- Pelegrini, A.L.; Moura, D.J.; Brenner, B.L.; Ledur, P.F.; Maques, G.P.; Henriques, J.A.P.; Saffi, J.; Lenz, G. Nek1 silencing slows down DNA repair and blocks DNA damage-induced cell cycle arrest. Mutagenesis 2010, 25, 447–454. [Google Scholar] [CrossRef]
- Stahl, P.; Kissau, L.; Mazitschek, R.; Huwe, A.; Furet, P.; Giannis, A.; Waldmann, H. Total Synthesis and Biological Evaluation of the Nakijiquinones. J. Am. Chem. Soc. 2001, 123, 11586–11593. [Google Scholar] [CrossRef] [PubMed]
- Toulany, M.; Kasten-Pisula, U.; Brammer, I.; Wang, S.; Chen, J.; Dittmann, K.; Baumann, M.; Dikomey, E.; Rodemann, H.P. Blockage of Epidermal Growth Factor Receptor-Phosphatidylinositol 3-Kinase-AKT Signaling Increases Radiosensitivity of K-RAS Mutated Human Tumor Cells In Vitro by Affecting DNA Repair. Clin. Cancer Res. 2006, 12, 4119–4126. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Chung, K.J.; Hwang, I.H.; Gwak, J.; Park, S.; Ju, B.G.; Yun, E.; Kim, D.-E.; Chung, Y.-H.; Na, M.; et al. Activation of p53 with ilimaquinone and ethylsmenoquinone, marine sponge metabolites, induces apoptosis and autophagy in colon cancer cells. Mar. Drugs 2015, 13, 543–557. [Google Scholar] [CrossRef]
- Xie, L.; Li, M.; Liu, D.; Wang, X.; Wang, P.; Dai, H.; Yang, W.; Liu, W.; Hu, X.; Zhao, M. Secalonic Acid-F, a Novel Mycotoxin, Represses the Progression of Hepatocellular Carcinoma via MARCH1 Regulation of the PI3K/AKT/beta-catenin Signaling Pathway. Molecules 2019, 24, 393. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sun, H.L.; Liu, D.S.; Zhang, J.R.; Yan, M.M.; Pan, X.H. Secalonic acid- F inhibited cell growth more effectively than 5-fluorouracil on hepatocellular carcinoma in vitro and in vivo. Neoplasma 2017, 64, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.-H.; Chueh, S.-C.; Kung, F.-L.; Pan, S.-L.; Shen, Y.-C.; Guh, J.-H. Ilimaquinone, a marine sponge metabolite, displays anticancer activity via GADD153-mediated pathway. Eur. J. Pharmacol. 2007, 556, 45–54. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013, 332, 237–248. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- Bellmunt, J.; Paz-Ares, L.; Cuello, M.; Cecere, F.L.; Albiol, S.; Guillem, V.; Gallardo, E.; Carles, J.; Mendez, P.; de la Cruz, J.J.; et al. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann. Oncol. 2007, 18, 522–528. [Google Scholar] [CrossRef]
- Olaussen, K.A.; Dunant, A.; Fouret, P.; Brambilla, E.; Andre, F.; Haddad, V.; Taranchon, E.; Filipits, M.; Pirker, R.; Popper, H.H.; et al. DNA Repair by ERCC1 in Non–Small-Cell Lung Cancer and Cisplatin-Based Adjuvant Chemotherapy. N. Engl. J. Med. 2006, 355, 983–991. [Google Scholar] [CrossRef]
- Friboulet, L.; Barrios-Gonzales, D.; Commo, F.; Olaussen, K.A.; Vagner, S.; Adam, J.; Goubar, A.; Dorvault, N.; Lazar, V.; Job, B.; et al. Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clin. Cancer Res. 2011, 17, 5562–5572. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-W.; Bai, L.-Y.; Su, J.-H.; Chiu, C.-F.; Lin, W.-Y.; Huang, W.-T.; Shih, M.-C.; Huang, Y.-T.; Hu, J.-L.; Weng, J.-R. Ilimaquinone Induces Apoptosis and Autophagy in Human Oral Squamous Cell Carcinoma Cells. Biomedicines 2020, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Kwak, C.-H.; Jin, L.; Han, J.H.; Han, C.W.; Kim, E.; Cho, M.; Chung, T.-W.; Bae, S.-J.; Jang, S.B.; Ha, K.-T. Ilimaquinone Induces the Apoptotic Cell Death of Cancer Cells by Reducing Pyruvate Dehydrogenase Kinase 1 Activity. Int. J. Mol. Sci. 2020, 21, 6021. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Olive, P.L. Detection of DNA damage in individual cells by analysis of histone H2AX phosphorylation. Methods Cell Biol. 2004, 75, 355–373. [Google Scholar] [CrossRef]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008, 36, 5678–5694. [Google Scholar] [CrossRef]
- Dickey, J.S.; Redon, C.E.; Nakamura, A.J.; Baird, B.J.; Sedelnikova, O.A.; Bonner, W.M. H2AX: Functional roles and potential applications. Chromosoma 2009, 118, 683–692. [Google Scholar] [CrossRef]
- Redon, C.E.; Nakamura, A.J.; Martin, O.A.; Parekh, P.R.; Weyemi, U.S.; Bonner, W.M. Recent developments in the use of gamma-H2AX as a quantitative DNA double-strand break biomarker. Aging 2011, 3, 168–174. [Google Scholar] [CrossRef]
- Galluzzi, L.; Senovilla, L.; Vitale, I.; Michels, J.; Martins, I.; Kepp, O.; Castedo, M.; Kroemer, G. Molecular mechanisms of cisplatin resistance. Oncogene 2012, 31, 1869–1883. [Google Scholar] [CrossRef]
- Höhn, A.; Krüger, K.; Skowron, M.A.; Bormann, S.; Schumacher, L.; Schulz, W.A.; Hoffmann, M.J.; Niegisch, G.; Fritz, G. Distinct mechanisms contribute to acquired cisplatin resistance of urothelial carcinoma cells. Oncotarget 2016, 7, 41320–41335. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aengenvoort, J.; Sekeres, M.; Proksch, P.; Fritz, G. Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve cAT-Triggered Tumor Cell Death. Molecules 2022, 27, 3567. https://doi.org/10.3390/molecules27113567
Aengenvoort J, Sekeres M, Proksch P, Fritz G. Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve cAT-Triggered Tumor Cell Death. Molecules. 2022; 27(11):3567. https://doi.org/10.3390/molecules27113567
Chicago/Turabian StyleAengenvoort, Jana, Marlena Sekeres, Peter Proksch, and Gerhard Fritz. 2022. "Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve cAT-Triggered Tumor Cell Death" Molecules 27, no. 11: 3567. https://doi.org/10.3390/molecules27113567
APA StyleAengenvoort, J., Sekeres, M., Proksch, P., & Fritz, G. (2022). Targeting Mechanisms of the DNA Damage Response (DDR) and DNA Repair by Natural Compounds to Improve cAT-Triggered Tumor Cell Death. Molecules, 27(11), 3567. https://doi.org/10.3390/molecules27113567




