Biochemometry-Based Discovery of Phenylpropanoids from Azadirachta indica Fruits as Inhibitors of In Vitro Osteoclast Formation
Abstract
:1. Introduction
2. Results
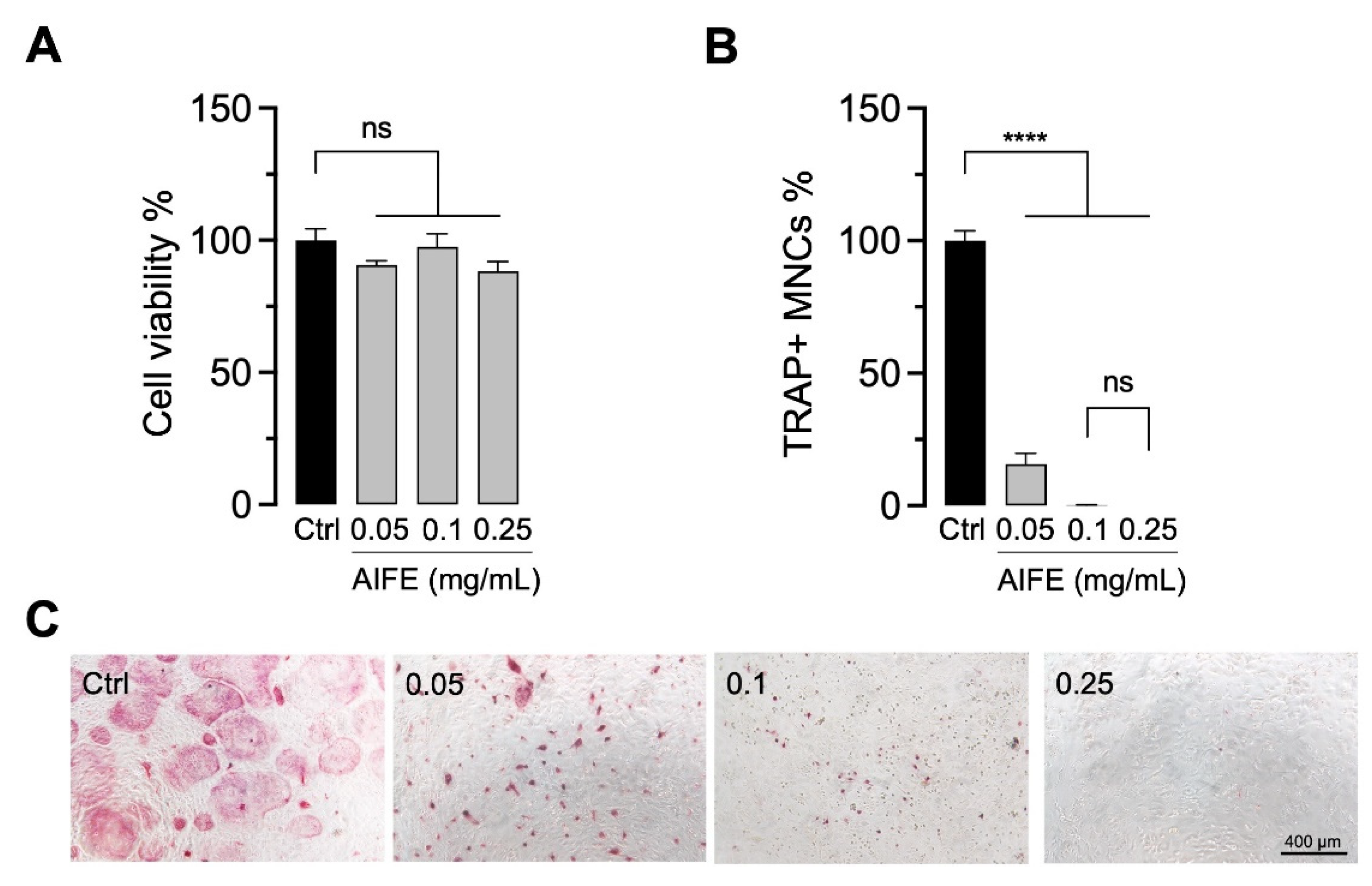
2.1. Extract Screening
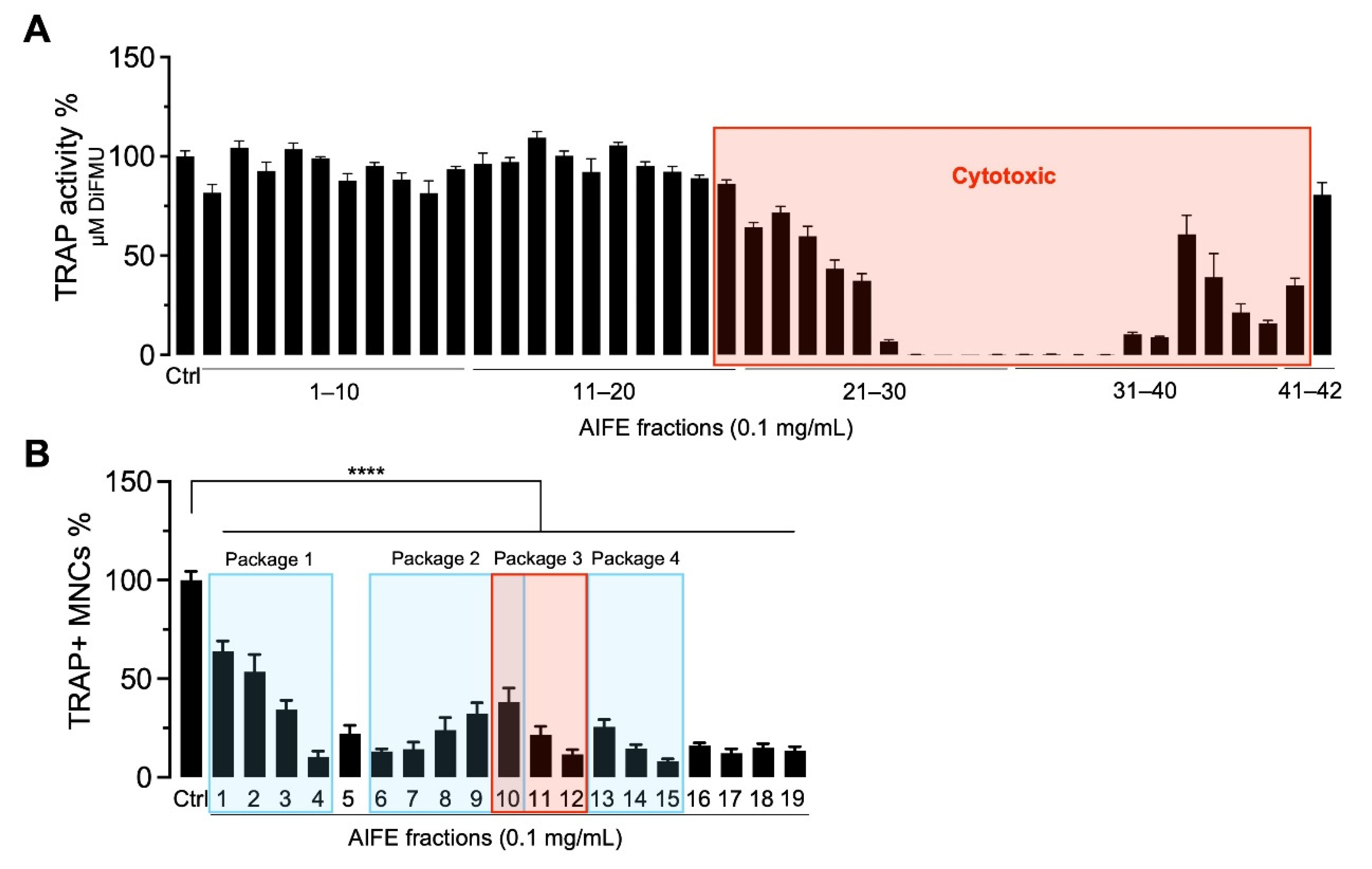
2.2. Biochemometry to Track Bioactive Constituents
- The HetCA plot of package 1 shows signals with a strong correlation of 1H NMR data with TRAP data (red signals); dereplication showed that the main compound in Fr 4 with MW = 452 Da matches the mass of many neem specific tetranortriterpenoids, limonoids (such as nimonol, 1,2-dehydromeldenin, 23-deoxyazadironolide and 1,2-dehydromeldenin). Limonoids are already well known to have anti-osteoporotic activity [24,25,26]. Consequently, microfractions of this package were excluded from further isolation steps.
- The HetCA plot of package 2 displays signals with a weak correlation of 1H NMR data with TRAP data; thus, this package was excluded from further steps.
- The HetCA plot of package 3 demonstrates signals with a strong correlation of 1H NMR data with TRAP data. Since no MS/MS-based annotation to main features of known A. indica constituents was achieved for the signals obtained in Fr12, thus assuming novel metabolites, we focused on this package for further isolation steps.
- The HetCA plot of package 4 shows a strong correlation of 1H NMR data with TRAP data. The dereplication showed that the main compound in Fr 15 with an MW of 720 Da matches the mass of a neem-specific triterpenoid, namely the limonoid azadirachtin, which has already been reported with anti-osteoporotic activity [27,28]. Consequently, this package was excluded from further steps.
2.3. Phytochemical Workup of Package 3
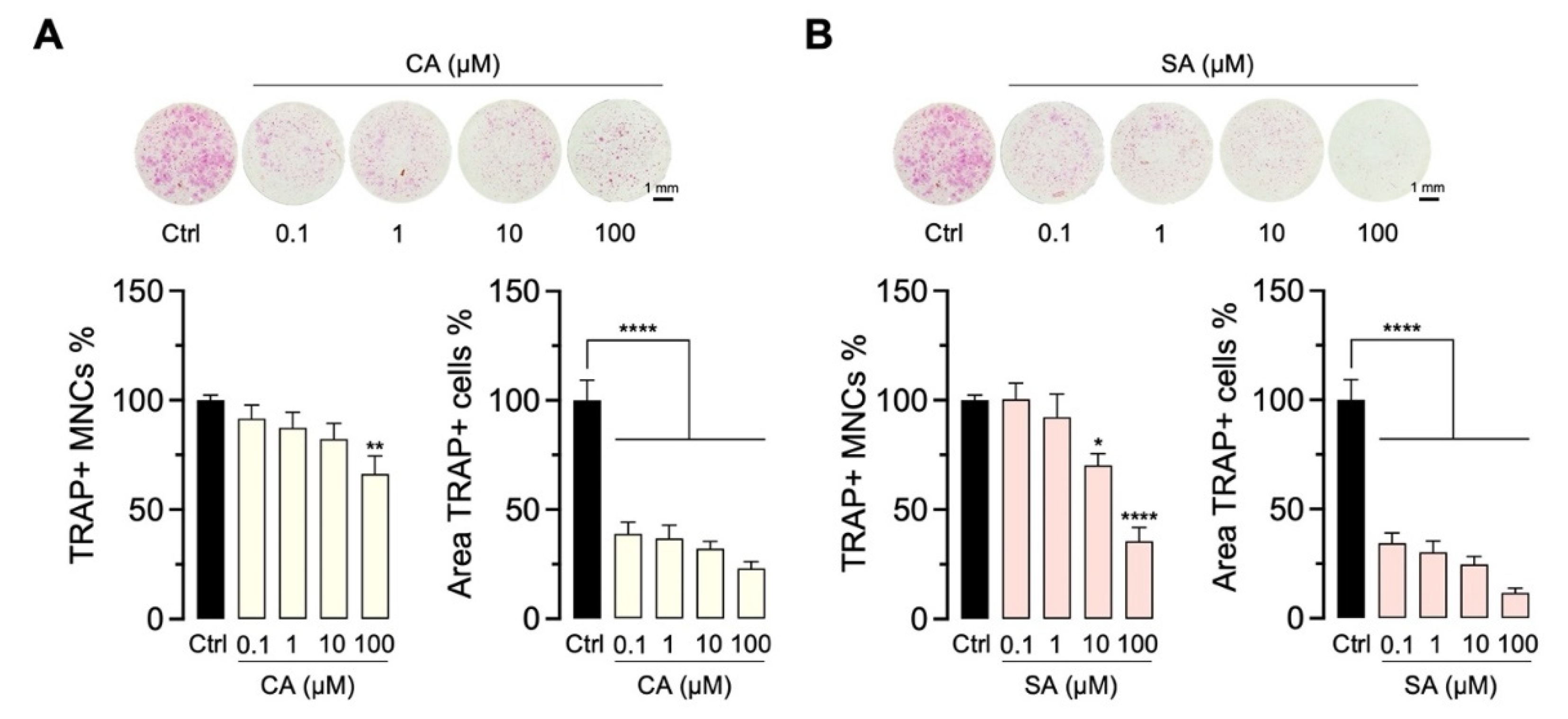
2.4. Bioactivity of CA and SA
3. Discussion
4. Materials and Methods
4.1. Plant Material, Chemicals and Reagents
4.2. Mouse co-Culture Model
4.3. Cell Viability and Osteoclast Formation
4.4. Nominal Mass Dereplication Using LC-MS
4.5. Crude Extract Generation and Flash Chromatography
4.6. Semi-Preparative SFC
4.7. NMR Measurements
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Cappariello, A.; Maurizi, A.; Veeriah, V.; Teti, A. The Great Beauty of the osteoclast. Arch. Biochem. Biophys. 2014, 558, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.; Neff, L.; Van, P.T.; Nefussi, J.R.; Vignery, A. Kinetic and cytochemical identification of osteoclast precursors and their differentiation into multinucleated osteoclasts. Am. J. Pathol. 1986, 122, 363–378. [Google Scholar]
- Fleisch, H. Bisphosphonates: Mechanisms of action. Endocr. Rev. 1998, 19, 80–100. [Google Scholar] [CrossRef]
- Ruggiero, S.L.; Fantasia, J.; Carlson, E. Bisphosphonate-related osteonecrosis of the jaw: Background and guidelines for diagnosis, staging and management. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2006, 102, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Levin, V.A.; Jiang, X.; Kagan, R. Estrogen therapy for osteoporosis in the modern era. Osteoporos. Int. 2018, 29, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, J.E.; Anderson, G.L.; Prentice, R.L.; LaCroix, A.Z.; Kooperberg, C.; Stefanick, M.L.; Jackson, R.D.; Beresford, S.A.; Howard, B.V.; Johnson, K.C.; et al. Risks and Benefits of Estrogen Plus Progestin in Healthy Postmenopausal WomenPrincipal Results From the Women’s Health Initiative Randomized Controlled Trial. JAMA 2002, 288, 321–333. [Google Scholar]
- An, J.; Yang, H.; Zhang, Q.; Liu, C.; Zhao, J.; Zhang, L.; Chen, B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast-mediated bone formation. Life Sci. 2016, 147, 46–58. [Google Scholar] [CrossRef]
- Sethi, G.; Aggarwal, B.B. Mending the Bones with Natural Products. Chem. Biol. 2007, 14, 738–740. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, J.P.; Xia, Y. Chinese herbal medicines for treating osteoporosis. Cochrane Database Syst. Rev. 2014, CD005467. [Google Scholar] [CrossRef] [PubMed]
- Bharti, A.C.; Takada, Y.; Aggarwal, B.B. Curcumin (Diferuloylmethane) Inhibits Receptor Activator of NF-κB Ligand-Induced NF-κB Activation in Osteoclast Precursors and Suppresses Osteoclastogenesis. J. Immunol. 2004, 172, 5940–5947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.P.; Nishishita, K.; Sakai, E.; Yoshida, H.; Kato, Y.; Tsukuba, T.; Okamoto, K. Berberine inhibits RANKL-induced osteoclast formation and survival through suppressing the NF-κB and Akt pathways. Eur. J. Pharmacol. 2008, 580, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Ballanti, P.; Minisola, S.; Pacitti, M.T.; Scarnecchia, L.; Rosso, R.; Mazzuoli, G.F.; Bonucci, E. Tartrate-resistant acid phosphate activity as osteoclastic marker: Sensitivity of cytochemical assessment and serum assay in comparison with standardized osteoclast histomorphometry. Osteoporos. Int. 1997, 7, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Puri, H.S. Neem: The Divine Tree Azadirachta Indica; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Drabu, S.; Khatri, S.; Babu, S. Neem: Healer of all ailments. Res. J. Pharm. Biol. Chem. Sci. 2012, 3, 120–126. [Google Scholar]
- Kirtikar, K.R.; Basu, B.D. Indian medicinal plants. In Sudhindra Nath Basu; Pâninî Office: Bahadurganj, India, 1918. [Google Scholar]
- Islas, J.F.; Acosta, E.; G-Buentello, Z.; Delgado-Gallegos, J.L.; Moreno-Treviño, M.G.; Escalante, B.; Moreno-Cuevas, J.E. An overview of Neem (Azadirachta indica) and its potential impact on health. J. Funct. Foods 2020, 74, 104171. [Google Scholar] [CrossRef]
- Akhila, A.; Rani, K. Chemistry of the Neem Tree (Azadirachta indica A. Juss.). In Fortschritte der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products; Herz, W., Falk, H., Kirby, G.W., Moore, R.E., Tamm, C., Eds.; Springer: Vienna, Austria, 1999; pp. 47–149. [Google Scholar]
- Biswas, K.; Chattopadhyay, I.; Banerjee, R.K.; Bandyopadhyay, U. Biological activities and medicinal properties of neem (Azadirachta indica). Curr. Sci. 2002, 82, 1336–1345. [Google Scholar]
- Gupta, S.C.; Prasad, S.; Tyagi, A.K.; Kunnumakkara, A.B.; Aggarwal, B.B. Neem (Azadirachta indica): An indian traditional panacea with modern molecular basis. Phytomedicine 2017, 34, 14–20. [Google Scholar] [CrossRef]
- Grienke, U.; Foster, P.A.; Zwirchmayr, J.; Tahir, A.; Rollinger, J.M.; Mikros, E. 1H NMR-MS-based heterocovariance as a drug discovery tool for fishing bioactive compounds out of a complex mixture of structural analogues. Sci. Rep. 2019, 9, 11113. [Google Scholar] [CrossRef] [Green Version]
- Zwirchmayr, J.; Grienke, U.; Hummelbrunner, S.; Seigner, J.; De Martin, R.; Dirsch, V.M.; Rollinger, J.M. A Biochemometric Approach for the Identification of In Vitro Anti-Inflammatory Constituents in Masterwort. Biomolecules 2020, 10, 679. [Google Scholar] [CrossRef]
- Nagini, S. Chapter Seven—Neem Limonoids as Anticancer Agents: Modulation of Cancer Hallmarks and Oncogenic Signaling. In The Enzymes; Bathaie, S.Z., Tamanoi, F., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 131–147. [Google Scholar]
- Wisutsitthiwong, C.; Buranaruk, C.; Pudhom, K.; Palaga, T. The plant limonoid 7-oxo-deacetoxygedunin inhibits RANKL-induced osteoclastogenesis by suppressing activation of the NF-κB and MAPK pathways. Biochem. Biophys. Res. Commun. 2011, 415, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Kimira, Y.; Taniuchi, Y.; Nakatani, S.; Sekiguchi, Y.; Kim, H.J.; Shimizu, J.; Ebata, M.; Wada, M.; Matsumoto, A.; Mano, H. Citrus limonoid nomilin inhibits osteoclastogenesis in vitro by suppression of NFATc1 and MAPK signaling pathways. Phytomedicine 2015, 22, 1120–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushwaha, P.; Khedgikar, V.; Haldar, S.; Gautam, J.; Mulani, F.A.; Thulasiram, H.V.; Trivedi, R. Azadirachta indica triterpenoids promote osteoblast differentiation and mineralization in vitro and in vivo. Bioorganic Med. Chem. Lett. 2016, 26, 3719–3724. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, P.; Ahmad, N.; Dhar, Y.V.; Verma, A.; Haldar, S.; Mulani, F.A.; Trivedi, P.K.; Mishra, P.R.; Thulasiram, H.V.; Trivedi, R. Estrogen receptor activation in response to Azadirachtin A stimulates osteoblast differentiation and bone formation in mice. J. Cell. Physiol. 2019, 234, 23719–23735. [Google Scholar] [CrossRef] [PubMed]
- Lancefield, C.S.; Westwood, N.J. The synthesis and analysis of advanced lignin model polymers. Green Chem. 2015, 17, 4980–4990. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, N.; Nakatsubo, T.; Suzuki, S.; Shibata, D.; Shimada, M.; Umezawa, T. Metabolic analysis of the cinnamate/monolignol pathway in Carthamus tinctorius seeds by a stable-isotope-dilution method. Org. Biomol. Chem. 2007, 5, 802–815. [Google Scholar] [CrossRef]
- Li, H.; Miyahara, T.; Tezuka, Y.; Namba, T.; Nemoto, N.; Tonami, S.; Seto, H.; Tada, T.; Kadota, S. The Effect of Kampo Formulae on Bone Resorption in Vitro and in Vivo. I. Active Constituents of Tsu-Kan-gan. Biol. Pharm. Bull. 1998, 21, 1322–1326. [Google Scholar] [CrossRef] [Green Version]
- Kartika, I.G.A.A.; Bang, I.J.; Riani, C.; Insanu, M.; Kwak, J.H.; Chung, K.H.; Adnyana, I.K. Isolation and Characterization of Phenylpropanoid and Lignan Compounds from Peperomia pellucida [L.] Kunth with Estrogenic Activities. Molecules 2020, 25, 4914. [Google Scholar] [CrossRef]
- Wang, X.-L.; Wang, N.-L.; Gao, H.; Zhang, G.; Qin, L.; Wong, M.-S.; Yao, X.-S. Phenylpropanoid and flavonoids from osteoprotective fraction of Drynaria fortunei. Nat. Prod. Res. 2010, 24, 1206–1213. [Google Scholar] [CrossRef]
- Aehle, E.; Müller, U.; Eklund, P.C.; Willför, S.; Sippl, W.; Dräger, B. Lignans as food constituents with estrogen and antiestrogen activity. Phytochemistry 2011, 72, 2396–2405. [Google Scholar] [CrossRef]
- Lee, W.; Ko, K.R.; Kim, H.-K.; Lim, S.; Kim, S. Dehydrodiconiferyl alcohol promotes BMP-2-induced osteoblastogenesis through its agonistic effects on estrogen receptor. Biochem. Biophys. Res. Commun. 2018, 495, 2242–2248. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-R.; Kim, S.; Cho, M.; Kang, S.W.; Yun, H.-M. Effects of PIN on Osteoblast Differentiation and Matrix Mineralization through Runt-Related Transcription Factor. Int. J. Mol. Sci. 2020, 21, 9579. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Prasain, J.K.; Tezuka, Y.; Namba, T.; Miyahara, T.; Tonami, S.; Seto, H.; Tada, T.; Kadota, S. Antiosteoporotic activity of the stems of Sambucus sieboldiana. Biol. Pharm. Bull. 1998, 21, 594–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhang, Z.; Guo, Q.; Dong, Y.; Zhao, Q.; Ma, X. Syringin prevents bone loss in ovariectomized mice via TRAF6 mediated inhibition of NF-κB and stimulation of PI3K/AKT. Phytomedicine 2018, 42, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Thummuri, D.; Guntuku, L.; Challa, V.S.; Ramavat, R.N.; Naidu, V.G.M. Abietic acid attenuates RANKL induced osteoclastogenesis and inflammation associated osteolysis by inhibiting the NF-KB and MAPK signaling. J. Cell. Physiol. 2018, 234, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Wang, G.; Xu, W.; Tao, P.; Lv, X.; Wang, Y. Tartrate-resistant acid phosphatase 5b is a marker of osteoclast number and volume in RAW 264.7 cells treated with receptor-activated nuclear κB ligand. Exp. Ther. Med. 2015, 9, 143–146. [Google Scholar] [CrossRef] [Green Version]
- Kampleitner, C.; Obi, J.; Vassilev, N.; Epstein, M.M.; Hoffmann, O. Biological Compatibility Profile on Biomaterials for Bone Regeneration. J. Vis. Exp. 2018, e58077. [Google Scholar] [CrossRef]
- Takahashi, N.; Akatsu, T.; Udagawa, N.; Sasaki, T.; Yamaguchi, A.; Moseley, J.M.; Martin, T.J.; Suda, T. Osteoblastic cells are involved in osteoclast formation. Endocrinology 1988, 123, 2600–2602. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahir, A.; Kampleitner, C.; Wirglauer, T.; Grienke, U.; Hoffmann, O.; Rollinger, J.M. Biochemometry-Based Discovery of Phenylpropanoids from Azadirachta indica Fruits as Inhibitors of In Vitro Osteoclast Formation. Molecules 2022, 27, 3611. https://doi.org/10.3390/molecules27113611
Tahir A, Kampleitner C, Wirglauer T, Grienke U, Hoffmann O, Rollinger JM. Biochemometry-Based Discovery of Phenylpropanoids from Azadirachta indica Fruits as Inhibitors of In Vitro Osteoclast Formation. Molecules. 2022; 27(11):3611. https://doi.org/10.3390/molecules27113611
Chicago/Turabian StyleTahir, Ammar, Carina Kampleitner, Theresa Wirglauer, Ulrike Grienke, Oskar Hoffmann, and Judith M. Rollinger. 2022. "Biochemometry-Based Discovery of Phenylpropanoids from Azadirachta indica Fruits as Inhibitors of In Vitro Osteoclast Formation" Molecules 27, no. 11: 3611. https://doi.org/10.3390/molecules27113611
APA StyleTahir, A., Kampleitner, C., Wirglauer, T., Grienke, U., Hoffmann, O., & Rollinger, J. M. (2022). Biochemometry-Based Discovery of Phenylpropanoids from Azadirachta indica Fruits as Inhibitors of In Vitro Osteoclast Formation. Molecules, 27(11), 3611. https://doi.org/10.3390/molecules27113611






