Chlorpromazine, a Clinically Approved Drug, Inhibits SARS-CoV-2 Nucleocapsid-Mediated Induction of IL-6 in Human Monocytes
Abstract
:1. Introduction
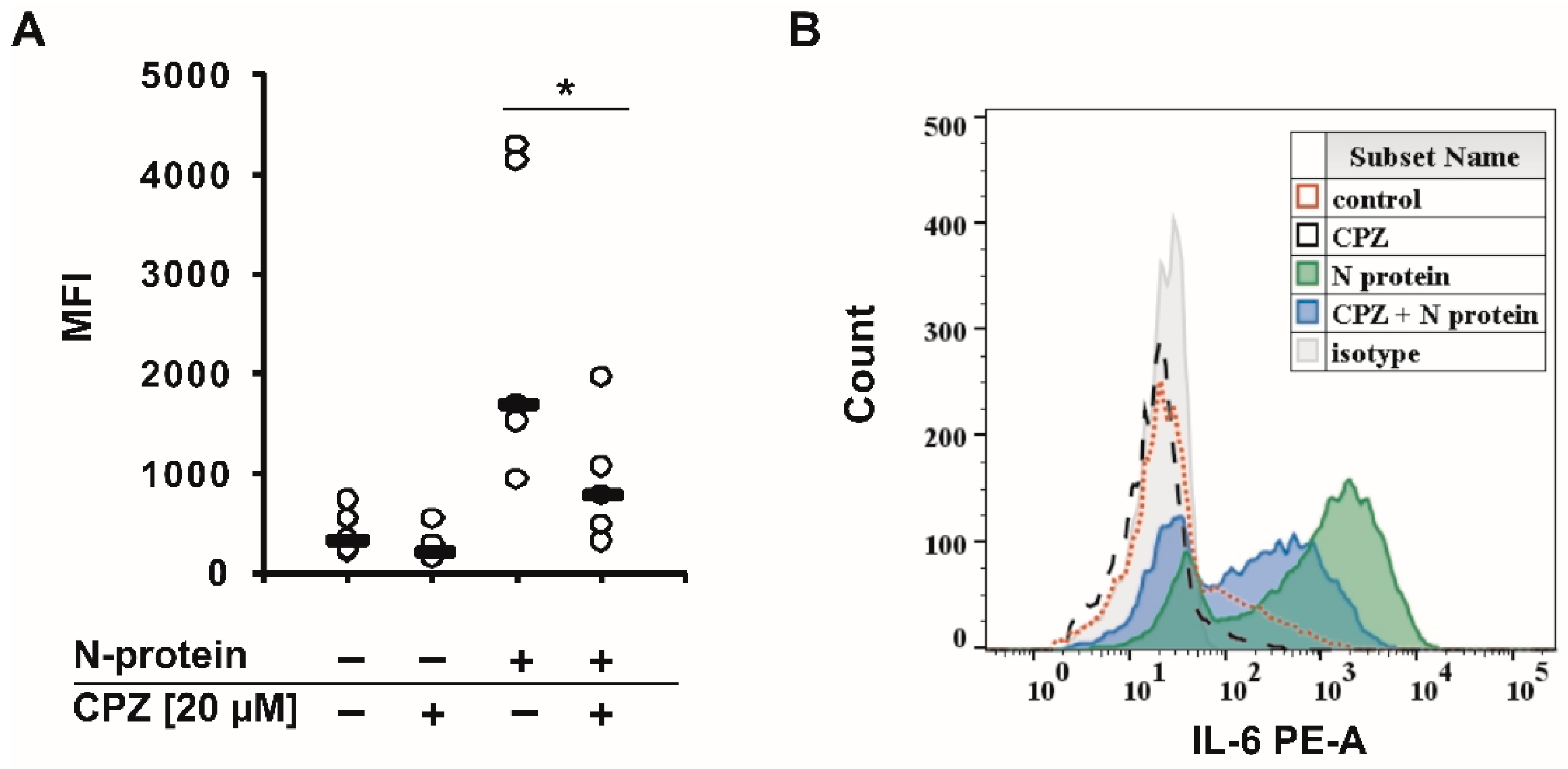
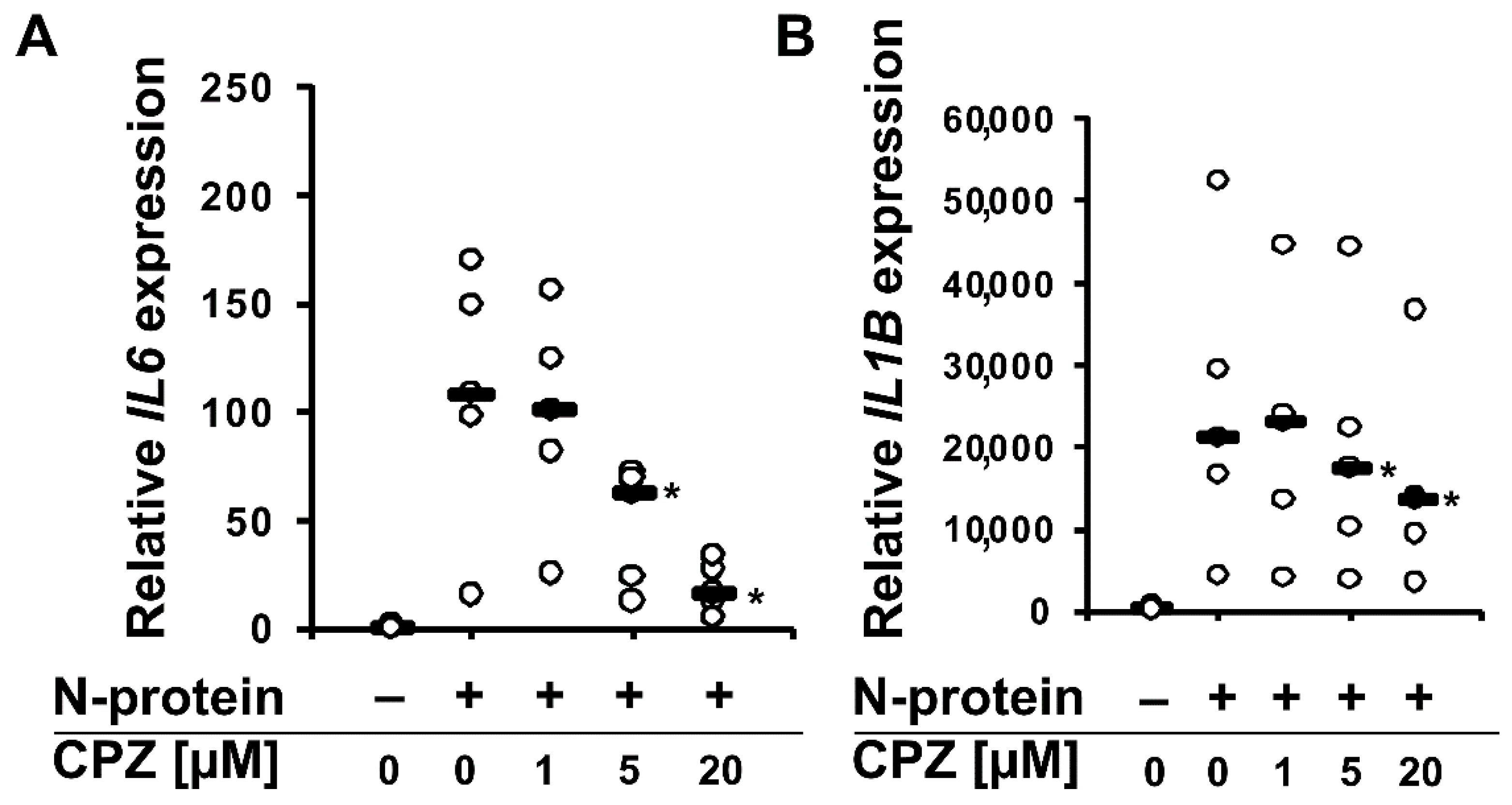
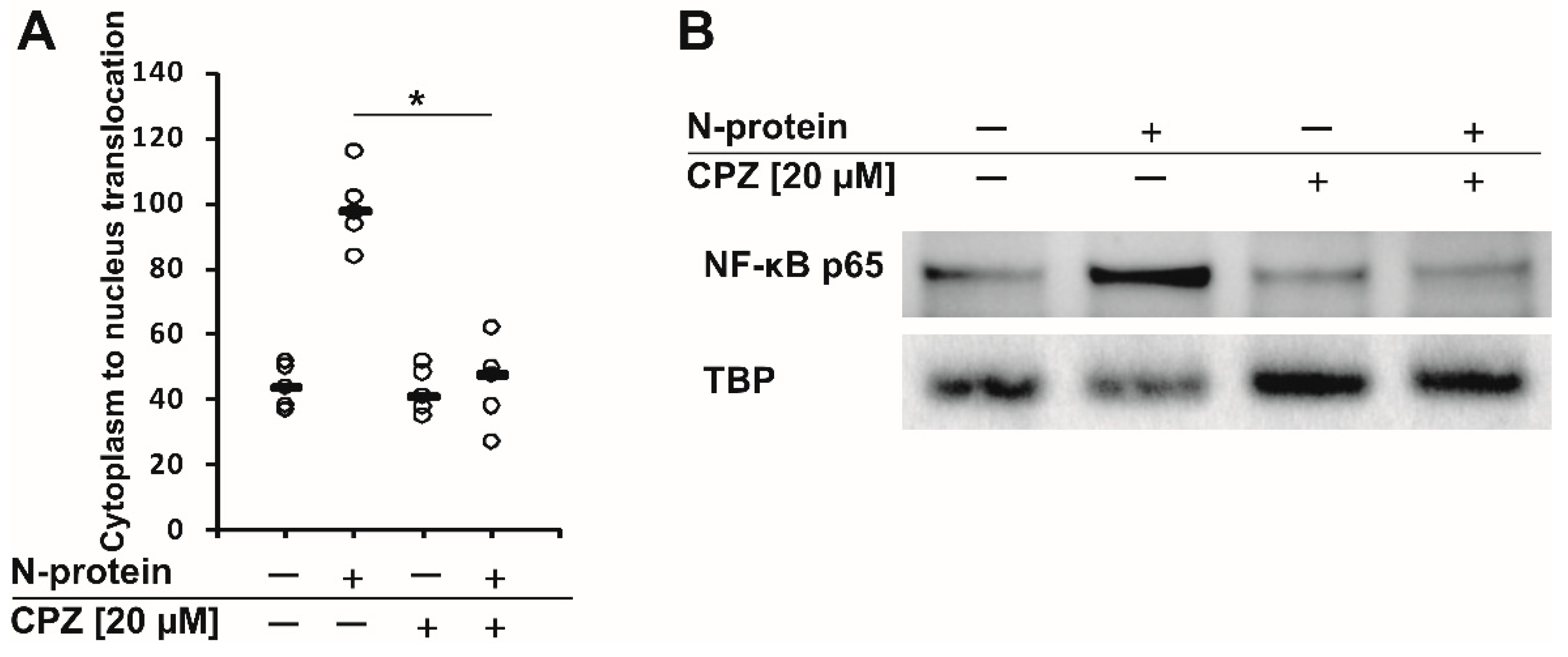
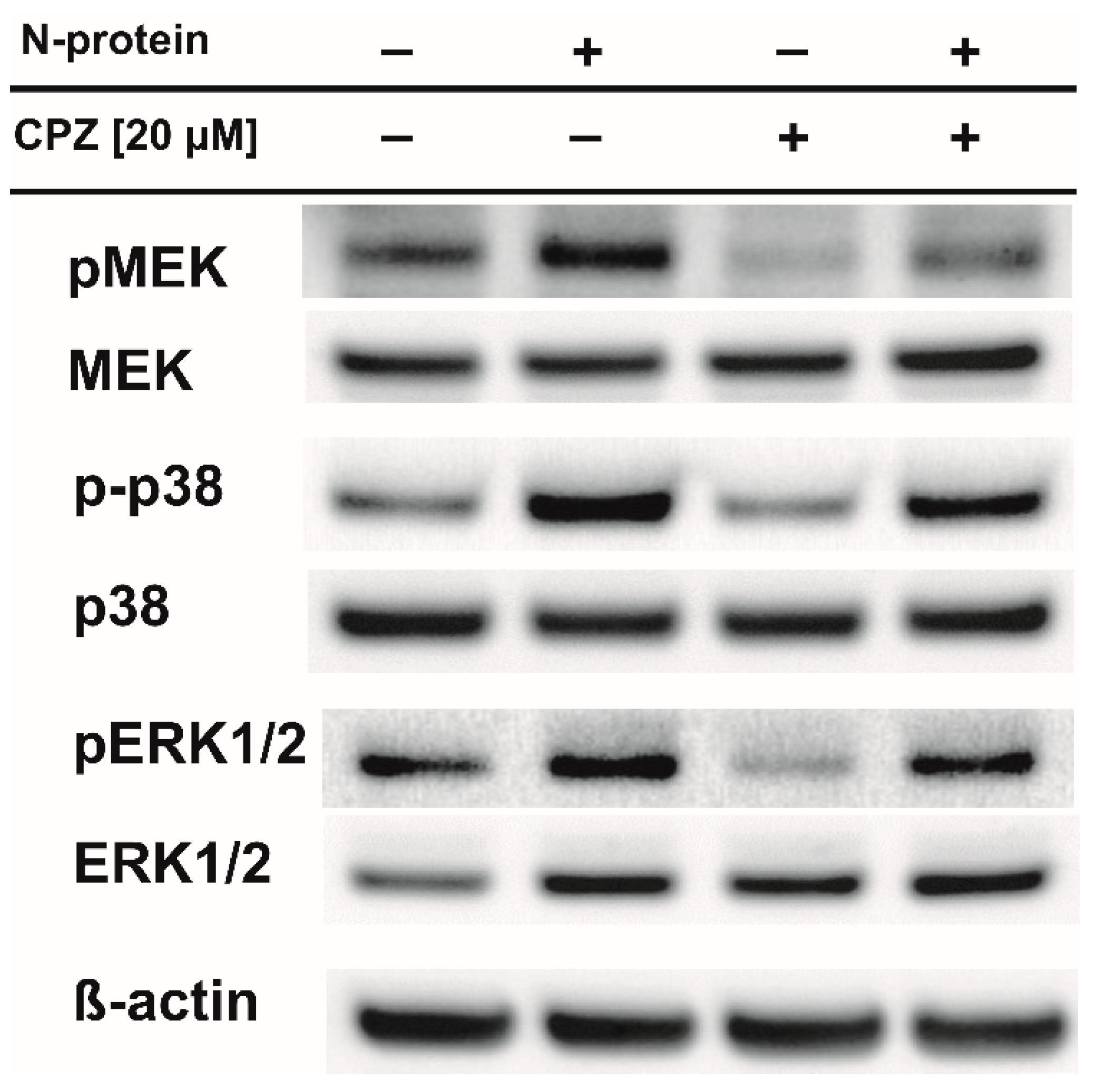
2. Results
3. Discussion
4. Materials and Methods
4.1. Monocyte Isolation
4.2. Proteins and Chemicals
4.3. Intracellular IL-6 Staining
4.4. Gene Expression Analysis
4.5. ELISA for the Detection of IL-6
4.6. NF-κB Translocation
4.7. Western Blotting
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://www.worldometers.info/coronavirus/ (accessed on 31 May 2022).
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Wassenaar, T.M.; Zou, Y. 2019_nCoV/SARS-CoV-2: Rapid classification of betacoronaviruses and identification of Traditional Chinese Medicine as potential origin of zoonotic coronaviruses. Lett. Appl. Microbiol. 2020, 70, 342–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Wang, B.; Mao, J.; Fu, J.; Shang, S.; Shu, Q.; Zhang, T. Epidemiological analysis of COVID-19 and practical experience from China. J. Med. Virol. 2020, 92, 755–769. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, U. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ní Choileáin, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [Green Version]
- Conti, P.; Ronconi, G.; Caraffa, A.; Gallenga, C.; Ross, R.; Frydas, I.; Kritas, S. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents 2020, 34, 327–331. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Kurzrock, R. Repurposing Interleukin-6 Inhibitors to Combat COVID-19. J. Immunother. Precis. Oncol. 2020, 3, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, E.; Larionova, I.; Choinzonov, E.; Kzhyshkowska, J. Monocytes and Macrophages as Viral Targets and Reservoirs. Int. J. Mol. Sci. 2018, 19, 2821. [Google Scholar] [CrossRef] [Green Version]
- Cory, T.J.; Emmons, R.S.; Yarbro, J.R.; Davis, K.L.; Pence, B.D. Metformin Suppresses Monocyte Immunometabolic Activation by SARS-CoV-2 Spike Protein Subunit 1. Front. Immunol. 2021, 12, 733921. [Google Scholar] [CrossRef]
- Pence, B.D. Severe COVID-19 and aging: Are monocytes the key? GeroScience 2020, 42, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, A.; Chauhan, P.; Saha, B.; Jafarzadeh, S.; Nemati, M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020, 257, 118102. [Google Scholar] [CrossRef]
- Karwaciak, I.; Sałkowska, A.; Karaś, K.; Dastych, J.; Ratajewski, M. Nucleocapsid and Spike Proteins of the Coronavirus SARS-CoV-2 Induce IL6 in Monocytes and Macrophages-Potential Implications for Cytokine Storm Syndrome. Vaccines 2021, 9, 54. [Google Scholar] [CrossRef]
- Sałkowska, A.; Karwaciak, I.; Karaś, K.; Dastych, J.; Ratajewski, M. SARS-CoV-2 Proteins Induce IFNG in Th1 Lymphocytes Generated from CD4+ Cells from Healthy, Unexposed Polish Donors. Vaccines 2020, 8, 673. [Google Scholar] [CrossRef]
- Plaze, M.; Attali, D.; Petit, A.-C.; Blatzer, M.; Simon-Loriere, E.; Vinckier, F.; Cachia, A.; Chrétien, F.; Gaillard, R. Repurposing chlorpromazine to treat COVID-19: The reCoVery study. L’encephale 2020, 46, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Nobile, B.; Durand, M.; Courtet, P.; Van de Perre, P.; Nagot, N.; Molès, J.P.; Olié, E. Could the antipsychotic chlorpromazine be a potential treatment for SARS-CoV-2? Schizophr. Res. 2020, 223, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Otręba, M.; Kośmider, L.; Rzepecka-Stojko, A. Antiviral activity of chlorpromazine, fluphenazine, perphenazine, prochlorperazine, and thioridazine towards RNA-viruses. A review. Eur. J. Pharmacol. 2020, 887, 173553. [Google Scholar] [CrossRef]
- Masuda, K.; Ripley, B.; Nishimura, R.; Mino, T.; Takeuchi, O.; Shioi, G.; Kiyonari, H.; Kishimoto, T. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 9409–9414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodavanti, U.P.; Lockard, V.G.; Mehendale, H.M. In vivo toxicity and pulmonary effects of promazine and chlorpromazine in rats. J. Biochem. Toxicol. 1990, 5, 245–251. [Google Scholar] [CrossRef]
- Liao, Q.-J.; Ye, L.-B.; Timani, K.A.; Zeng, Y.-C.; She, Y.-L.; Ye, L.; Wu, Z.-H. Activation of NF-kappaB by the full-length nucleocapsid protein of the SARS coronavirus. Acta Biochim. Biophys. Sin. 2005, 37, 607–612. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Hao, Q.; Mu, Y.; Timani, K.A.; Ye, L.; Zhu, Y.; Wu, J. Nucleocapsid protein of SARS-CoV activates the expression of cyclooxygenase-2 by binding directly to regulatory elements for nuclear factor-kappa B and CCAAT/enhancer binding protein. Int. J. Biochem. Cell Biol. 2006, 38, 1417–1428. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, K.; Wang, D.; Yue, X.; Song, D.; Zhu, Y.; Wu, J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-kappaB. Virology 2007, 365, 324–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Ma, L.; Cai, S.; Zhuang, Z.; Zhao, Z.; Jin, S.; Xie, W.; Zhou, L.; Zhang, L.; Zhao, J.; et al. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-κB hyper-activation and inflammation. Signal Transduct. Target. Ther. 2021, 6, 167. [Google Scholar] [CrossRef]
- Nilsson-Payant, B.E.; Uhl, S.; Grimont, A.; Doane, A.S.; Cohen, P.; Patel, R.S.; Higgins, C.A.; Acklin, J.A.; Bram, Y.; Chandar, V.; et al. The NF-κB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J. Virol. 2021, 95, e0125721. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Feng, N.; Fan, W.; Ma, X.; Yan, Q.; Lv, Z.; Zeng, Y.; Zhu, J.; Lu, C. Activation of PI3K/AKT and ERK MAPK signal pathways is required for the induction of lytic cycle replication of Kaposi’s sarcoma-associated herpesvirus by herpes simplex virus type 1. BMC Microbiol. 2011, 11, 240. [Google Scholar] [CrossRef] [Green Version]
- DuShane, J.K.; Wilczek, M.P.; Mayberry, C.L.; Maginnis, M.S. ERK Is a Critical Regulator of JC Polyomavirus Infection. J. Virol. 2018, 92, e01529-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Gong, R.; Qu, J.; Zhou, Y.; Liu, W.; Chen, M.; Liu, Y.; Zhu, Y.; Wu, J. Activation of the Ras/Raf/MEK pathway facilitates hepatitis C virus replication via attenuation of the interferon-JAK-STAT pathway. J. Virol. 2012, 86, 1544–1554. [Google Scholar] [CrossRef] [Green Version]
- Albarnaz, J.D.; De Oliveira, L.C.; Torres, A.A.; Palhares, R.M.; Casteluber, M.C.; Rodrigues, C.M.; Cardozo, P.L.; De Souza, A.M.R.; Pacca, C.C.; Ferreira, P.C.P.; et al. MEK/ERK activation plays a decisive role in yellow fever virus replication: Implication as an antiviral therapeutic target. Antivir. Res. 2014, 111, 82–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghasemnejad-Berenji, M.; Pashapour, S. SARS-CoV-2 and the Possible Role of Raf/MEK/ERK Pathway in Viral Survival: Is This a Potential Therapeutic Strategy for COVID-19? Pharmacology 2021, 106, 119–122. [Google Scholar] [CrossRef]
- Schreiber, A.; Viemann, D.; Schöning, J.; Schloer, S.; Mecate Zambrano, A.; Brunotte, L.; Faist, A.; Schöfbänker, M.; Hrincius, E.; Hoffmann, H.; et al. The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses. Cell. Mol. Life Sci. 2022, 79, 65. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [Green Version]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.; Yang, Z. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016, 13, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.P.; Viswanathan, S.; Wang, M.; Sun, L.-Q.; Clark, G.C.; D’Elia, R.V. Current and future developments in the treatment of virus-induced hypercytokinemia. Future Med. Chem. 2017, 9, 169–178. [Google Scholar] [CrossRef]
- Crayne, C.B.; Albeituni, S.; Nichols, K.E.; Cron, R.Q. The Immunology of Macrophage Activation Syndrome. Front. Immunol. 2019, 10, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef] [Green Version]
- Peiris, J.S.M.; Cheung, C.Y.; Leung, C.Y.H.; Nicholls, J.M. Innate immune responses to influenza A H5N1: Friend or foe? Trends Immunol. 2009, 30, 574–584. [Google Scholar] [CrossRef] [Green Version]
- De Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.D.; Chau, T.N.B.; Hoang, D.M.; Chau, N.V.V.; Khanh, T.H.; Dong, V.C.; et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.C.; Hoiland, R.L.; Stukas, S.; Wellington, C.L.; Sekhon, M.S. Confronting the controversy: Interleukin-6 and the COVID-19 cytokine storm syndrome. Eur. Respir. J. 2020, 56, 2003006. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [Green Version]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Qi, F.; Li, H.; Yang, Q.; Wang, H.; Wang, X.; Liu, X.; Zhao, J.; Liao, X.; Liu, Y.; et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020, 6, 73. [Google Scholar] [CrossRef]
- Song, J.-W.; Zhang, C.; Fan, X.; Meng, F.-P.; Xu, Z.; Xia, P.; Cao, W.-J.; Yang, T.; Dai, X.-P.; Wang, S.-Y.; et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020, 11, 3410. [Google Scholar] [CrossRef]
- Nienhold, R.; Ciani, Y.; Koelzer, V.H.; Tzankov, A.; Haslbauer, J.D.; Menter, T.; Schwab, N.; Henkel, M.; Frank, A.; Zsikla, V.; et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat. Commun. 2020, 11, 5086. [Google Scholar] [CrossRef]
- Li, S.; Jiang, L.; Li, X.; Lin, F.; Wang, Y.; Li, B.; Jiang, T.; An, W.; Liu, S.; Liu, H.; et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight 2020, 5, e138070. [Google Scholar] [CrossRef]
- Strohbehn, G.W.; Heiss, B.L.; Rouhani, S.J.; Trujillo, J.A.; Yu, J.; Kacew, A.J.; Higgs, E.F.; Bloodworth, J.C.; Cabanov, A.; Wright, R.C.; et al. COVIDOSE: A Phase II Clinical Trial of Low-Dose Tocilizumab in the Treatment of Noncritical COVID-19 Pneumonia. Clin. Pharmacol. Ther. 2021, 109, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.N.; Hernández-Sánchez, J.; Nagel, S.; Feng, Y.; Cai, F.; Rabin, J.; Morse, C.G.; Nadig, N.R.; Ashraf, O.; Gotur, D.B.; et al. Safety and Efficacy of Tocilizumab 4 or 8 mg/kg in Hospitalized Patients with Moderate to Severe Coronavirus Disease 2019 Pneumonia: A Randomized Clinical Trial. Open Forum Infect. Dis. 2022, 9, ofab608. [Google Scholar] [CrossRef] [PubMed]
- Merchante, N.; Cárcel, S.; Garrido-Gracia, J.C.; Trigo-Rodríguez, M.; Moreno, M.Á.E.; León-López, R.; Espíndola-Gómez, R.; Alonso, E.A.; García, D.V.; Romero-Palacios, A.; et al. Early Use of Sarilumab in Patients Hospitalized with COVID-19 Pneumonia and Features of Systemic Inflammation: The SARICOR Randomized Clinical Trial. Antimicrob. Agents Chemother. 2022, 66, e0210721. [Google Scholar] [CrossRef]
- López-Muñoz, F.; Alamo, C.; Cuenca, E.; Shen, W.W.; Clervoy, P.; Rubio, G. History of the discovery and clinical introduction of chlorpromazine. Ann. Clin. Psychiatry 2005, 17, 113–135. [Google Scholar] [CrossRef]
- Libermann, T.A.; Baltimore, D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 1990, 10, 2327–2334. [Google Scholar] [CrossRef] [PubMed]
- Matsusaka, T.; Fujikawa, K.; Nishio, Y.; Mukaida, N.; Matsushima, K.; Kishimoto, T.; Akira, S. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. USA 1993, 90, 10193–10197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, Y.; Moldawer, L.L.; Marano, M.; Wei, H.; Tatter, S.B.; Clarick, R.H.; Santhanam, U.; Sherris, D.; May, L.T.; Sehgal, P.B. Endotoxemia elicits increased circulating beta 2-IFN/IL-6 in man. J. Immunol. 1989, 142, 2321–2324. [Google Scholar]
- Sehgal, P.B.; Helfgott, D.C.; Santhanam, U.; Tatter, S.B.; Clarick, R.H.; Ghrayeb, J.; May, L.T. Regulation of the acute phase and immune responses in viral disease. Enhanced expression of the beta 2-interferon/hepatocyte-stimulating factor/interleukin 6 gene in virus-infected human fibroblasts. J. Exp. Med. 1988, 167, 1951–1956. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Narazaki, M.; Masuda, K.; Kishimoto, T. Regulation of IL-6 in Immunity and Diseases. Adv. Exp. Med. Biol. 2016, 941, 79–88. [Google Scholar] [CrossRef]
- Vanden Berghe, W.; Plaisance, S.; Boone, E.; De Bosscher, K.; Schmitz, M.L.; Fiers, W.; Haegeman, G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J. Biol. Chem. 1998, 273, 3285–3290. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Liu, M.; Kirkwood, K.L. p38alpha stabilizes interleukin-6 mRNA via multiple AU-rich elements. J. Biol. Chem. 2008, 283, 1778–1785. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.; Ryan, M.P.; Watson, A.J.; Schramek, H.; Healy, E. Role of MAP kinase pathways in mediating IL-6 production in human primary mesangial and proximal tubular cells. Kidney Int. 1999, 56, 1366–1377. [Google Scholar] [CrossRef] [Green Version]
- Bertini, R.; Garattini, S.; Delgado, R.; Ghezzi, P. Pharmacological activities of chlorpromazine involved in the inhibition of tumour necrosis factor production in vivo in mice. Immunology 1993, 79, 217–219. [Google Scholar]
- Bertini, R.; Mengozzi, M.; Bianchi, M.; Sipe, J.D.; Ghezzi, P. Chlorpromazine protection against interleukin-1 and tumor necrosis factor-mediated activities in vivo. Int. J. Immunopharmacol. 1991, 13, 1085–1090. [Google Scholar] [CrossRef]
- Labuzek, K.; Kowalski, J.; Gabryel, B.; Herman, Z.S. Chlorpromazine and loxapine reduce interleukin-1beta and interleukin-2 release by rat mixed glial and microglial cell cultures. Eur. Neuropsychopharmacol. 2005, 15, 23–30. [Google Scholar] [CrossRef]
- Zucker, S.; Zarrabi, H.M.; Schubach, W.H.; Varma, A.; Derman, R.; Lysik, R.M.; Habicht, G.; Seitz, P.M. Chlorpromazine-induced immunopathy: Progressive increase in serum IgM. Medicine 1990, 69, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Kimura, A.; Hanieh, H.; Nguyen, N.T.; Nakahama, T.; Chinen, I.; Otoyo, Y.; Murotani, T.; Yamatodani, A.; Kishimoto, T. Aryl hydrocarbon receptor negatively regulates LPS-induced IL-6 production through suppression of histamine production in macrophages. Int. Immunol. 2011, 23, 637–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boraschi, D.; Villa, L.; Ghiara, P.; Tagliabue, A.; Mengozzi, M.; Solito, E.; Parente, L.; Silvestri, S.; Van Damme, J.; Ghezzi, P. Mechanism of acute toxicity of IL-1 beta in mice. Eur. Cytokine Netw. 1991, 2, 61–67. [Google Scholar]
- Mengozzi, M.; Fantuzzi, G.; Faggioni, R.; Marchant, A.; Goldman, M.; Orencole, S.; Clark, B.D.; Sironi, M.; Benigni, F.; Ghezzi, P. Chlorpromazine specifically inhibits peripheral and brain TNF production, and up-regulates IL-10 production, in mice. Immunology 1994, 82, 207–210. [Google Scholar]
- Baumeister, D.; Ciufolini, S.; Mondelli, V. Effects of psychotropic drugs on inflammation: Consequence or mediator of therapeutic effects in psychiatric treatment? Psychopharmacology 2016, 233, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.P.; Chesney, E.; Oliver, D.; Pollak, T.A.; McGuire, P.; Fusar-Poli, P.; Zandi, M.S.; Lewis, G.; David, A.S. Psychiatric and neuropsychiatric presentations associated with severe coronavirus infections: A systematic review and meta-analysis with comparison to the COVID-19 pandemic. Lancet Psychiatry 2020, 7, 611–627. [Google Scholar] [CrossRef]
- Mueller, J.K.; Riederer, P.; Müller, W.E. Neuropsychiatric Drugs against COVID-19: What is the Clinical Evidence? Pharmacopsychiatry 2022, 55, 7–15. [Google Scholar] [CrossRef]
- Plaze, M.; Attali, D.; Prot, M.; Petit, A.-C.; Blatzer, M.; Vinckier, F.; Levillayer, L.; Chiaravalli, J.; Perin-Dureau, F.; Cachia, A.; et al. Inhibition of the replication of SARS-CoV-2 in human cells by the FDA-approved drug chlorpromazine. Int. J. Antimicrob. Agents 2021, 57, 106274. [Google Scholar] [CrossRef]
- Comar, D.; Zarifian, E.; Verhas, M.; Soussaline, F.; Maziere, M.; Berger, G.; Loo, H.; Cuche, H.; Kellershohn, C.; Deniker, P. Brain distribution and kinetics of 11C-chlorpromazine in schizophrenics: Positron emission tomography studies. Psychiatry Res. 1979, 1, 23–29. [Google Scholar] [CrossRef]
- Krizanová, O.; Ciampor, F.; Veber, P. Influence of chlorpromazine on the replication of influenza virus in chick embryo cells. Acta Virol. 1982, 26, 209–216. [Google Scholar] [PubMed]
- de Wilde, A.H.; Jochmans, D.; Posthuma, C.C.; Zevenhoven-Dobbe, J.C.; van Nieuwkoop, S.; Bestebroer, T.M.; van den Hoogen, B.G.; Neyts, J.; Snijder, E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014, 58, 4875–4884. [Google Scholar] [CrossRef] [Green Version]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, Y.; Hart, B.J.; Gross, R.; Zhou, H.; Frieman, M.; Bollinger, L.; Wada, J.; Hensley, L.E.; Jahrling, P.B.; Dyall, J.; et al. MERS-CoV pathogenesis and antiviral efficacy of licensed drugs in human monocyte-derived antigen-presenting cells. PLoS ONE 2018, 13, e0194868. [Google Scholar] [CrossRef]
- Weston, S.; Coleman, C.M.; Haupt, R.; Logue, J.; Matthews, K.; Li, Y.; Reyes, H.M.; Weiss, S.R.; Frieman, M.B. Broad Anti-coronavirus Activity of Food and Drug Administration-Approved Drugs against SARS-CoV-2 In Vitro and SARS-CoV In Vivo. J. Virol. 2020, 94, e01218-20. [Google Scholar] [CrossRef]
- Fred, S.M.; Kuivanen, S.; Ugurlu, H.; Casarotto, P.C.; Levanov, L.; Saksela, K.; Vapalahti, O.; Castrén, E. Antidepressant and Antipsychotic Drugs Reduce Viral Infection by SARS-CoV-2 and Fluoxetine Shows Antiviral Activity Against the Novel Variants in vitro. Front. Pharmacol. 2021, 12, 755600. [Google Scholar] [CrossRef]
- Hiemke, C.; Bergemann, N.; Clement, H.W.; Conca, A.; Deckert, J.; Domschke, K.; Eckermann, G.; Egberts, K.; Gerlach, M.; Greiner, C.; et al. Consensus Guidelines for Therapeutic Drug Monitoring in Neuropsychopharmacology: Update 2017. Pharmacopsychiatry 2018, 51, 9–62. [Google Scholar] [CrossRef] [Green Version]
- Forrest, I.S.; Bolt, A.G.; Serra, M.T. Distribution of chlorpromazine metabolites in selected organs of psychiatric patients chronically dosed up to the time of death. Biochem. Pharmacol. 1968, 17, 2061–2070. [Google Scholar] [CrossRef]
- May, P.R.; Van Putten, T.; Jenden, D.J.; Yale, C.; Dixon, W.J. Chlorpromazine levels and the outcome of treatment in schizophrenic patients. Arch. Gen. Psychiatry 1981, 38, 202–207. [Google Scholar] [CrossRef]
- Daniel, W.A.; Bickel, M.H.; Honegger, U.E. The contribution of lysosomal trapping in the uptake of desipramine and chloroquine by different tissues. Pharmacol. Toxicol. 1995, 77, 402–406. [Google Scholar] [CrossRef]
- Daniel, W.A.; Wójcikowski, J. Contribution of lysosomal trapping to the total tissue uptake of psychotropic drugs. Pharmacol. Toxicol. 1997, 80, 62–68. [Google Scholar] [CrossRef]
- Boer, F. Drug handling by the lungs. Br. J. Anaesth. 2003, 91, 50–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, W.A. Mechanisms of cellular distribution of psychotropic drugs. Significance for drug action and interactions. Prog. Neuropsychopharmacol. Biol. Psychiatry 2003, 27, 65–73. [Google Scholar] [CrossRef]
- Keeling, P.J.; Henson, P.M. Lysosomal enzyme release from human monocytes in response to particulate stimuli. J. Immunol. 1982, 128, 563–567. [Google Scholar] [PubMed]
- Odaka, C.; Mizuochi, T. Role of macrophage lysosomal enzymes in the degradation of nucleosomes of apoptotic cells. J. Immunol. 1999, 163, 5346–5352. [Google Scholar]
- Wang, L.H.; Rothberg, K.G.; Anderson, R.G. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 1993, 123, 1107–1117. [Google Scholar] [CrossRef]
- Vercauteren, D.; Vandenbroucke, R.E.; Jones, A.T.; Rejman, J.; Demeester, J.; De Smedt, S.C.; Sanders, N.N.; Braeckmans, K. The use of inhibitors to study endocytic pathways of gene carriers: Optimization and pitfalls. Mol. Ther. 2010, 18, 561–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sałkowska, A.; Karaś, K.; Walczak-Drzewiecka, A.; Dastych, J.; Ratajewski, M. Differentiation stage-specific effect of histone deacetylase inhibitors on the expression of RORγT in human lymphocytes. J. Leukoc. Biol. 2017, 102, 1487–1495. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwaciak, I.; Karaś, K.; Sałkowska, A.; Pastwińska, J.; Ratajewski, M. Chlorpromazine, a Clinically Approved Drug, Inhibits SARS-CoV-2 Nucleocapsid-Mediated Induction of IL-6 in Human Monocytes. Molecules 2022, 27, 3651. https://doi.org/10.3390/molecules27123651
Karwaciak I, Karaś K, Sałkowska A, Pastwińska J, Ratajewski M. Chlorpromazine, a Clinically Approved Drug, Inhibits SARS-CoV-2 Nucleocapsid-Mediated Induction of IL-6 in Human Monocytes. Molecules. 2022; 27(12):3651. https://doi.org/10.3390/molecules27123651
Chicago/Turabian StyleKarwaciak, Iwona, Kaja Karaś, Anna Sałkowska, Joanna Pastwińska, and Marcin Ratajewski. 2022. "Chlorpromazine, a Clinically Approved Drug, Inhibits SARS-CoV-2 Nucleocapsid-Mediated Induction of IL-6 in Human Monocytes" Molecules 27, no. 12: 3651. https://doi.org/10.3390/molecules27123651
APA StyleKarwaciak, I., Karaś, K., Sałkowska, A., Pastwińska, J., & Ratajewski, M. (2022). Chlorpromazine, a Clinically Approved Drug, Inhibits SARS-CoV-2 Nucleocapsid-Mediated Induction of IL-6 in Human Monocytes. Molecules, 27(12), 3651. https://doi.org/10.3390/molecules27123651







