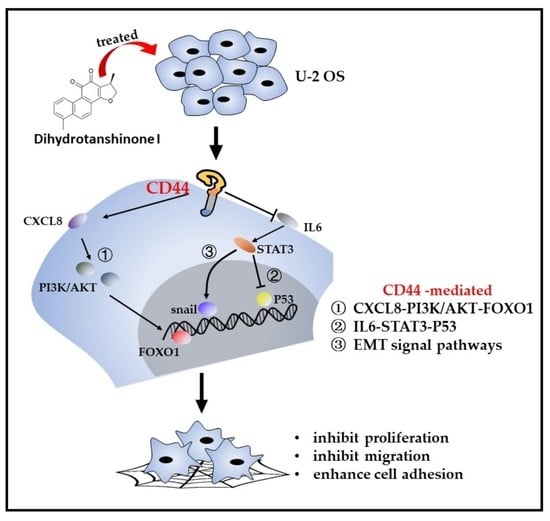
Dihydrotanshinone I Enhances Cell Adhesion and Inhibits Cell Migration in Osteosarcoma U−2 OS Cells through CD44 and Chemokine Signaling
Abstract
:1. Introduction
2. Results
2.1. Tanshinones Inhibited Cells’ Proliferation
2.2. DS−1 Inhibited the Expression of Transcription Factor FOXO1 in U−2 OS Cells
2.3. RNA-Seq Gene Expression Profiling in U−2 OS Cells Treated with DS−1
2.4. DS−1 Downregulated Cell Cycle Regulators of U−2OS Cells
2.5. DS−1 Inhibited U−2OS Cell Migration
2.6. DS−1 Enhanced U−2 OS Cells’ Adhesion
2.7. DS−1 Activated CXCL8-PI3K/AKT Signaling Pathway
2.8. DS−1 Changed IL6-STAT3-P53 Signal
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Procedures
4.2.1. Cell Proliferation Assay (MTT)
4.2.2. Transcriptome Analysis
4.2.3. RT-PCR
4.2.4. Scratch Motility (Wound-Healing) Assay
4.2.5. Immunofluorescence
4.2.6. Western Blot
4.2.7. Transient Transfection to Knock Down CD44 in U−2 OS Cell
4.2.8. Antibodies
4.2.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Jemal, A. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar]
- Jonathan, G.; Richard, G. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021, 18, 609–624. [Google Scholar]
- Misaghi, A.; Goldin, A.; Awad, M.; Kulidjian, A.A. Osteosarcoma: A comprehensive review. Sicot-J. 2018, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolati, S.; Sanaie, S.; Naseri, A.; Rostamzadeh, D. Osteosarcoma: A comprehensive review of management and treatment strategies. Ann. Diagn. Pathol. 2020, 49, 151654. [Google Scholar]
- Chen, Y.; Cao, J.; Zhang, N.; Yang, B.; Ying, M. Advances in differentiation therapy for osteosarcoma. Drug Discov. Today 2020, 25, 497–504. [Google Scholar] [CrossRef]
- Atik, S.; Li, F.; Li, M.; He, B.; He, X.; Chen, G.; Guo, B.; Li, D.; Jiang, F.; Dang, L.; et al. Present advances and future perspectives of molecular targeted therapy for Osteosarcoma. Int. J. Mol. Sci. 2016, 17, 506. [Google Scholar]
- Chen, Y.; Liu, R.; Wang, W.; Wang, C.; Ying, M. Advances in targeted therapy for osteosarcoma based on molecular classification. Pharmacol. Res. 2021, 169, 105684. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012, 75, 311–335. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.A.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B. A new golden age of natural products drug discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, G.D.; Brooks, L.; Zhang, X.; Mahoney, J.M.; Martyanov, V.; Wood, T.A.; Sherlockb, G.; Chenga, C.; Whitfielda, M.L. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell 2013, 24, 3634–3650. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Chondrogianni, N.; Amarantos, I.; Blake, J.; Schwager, C.; Wirkner, U.; Ansorge, W.; Gonos, E.S. Genome-wide transcriptome profile of the human osteosarcoma Sa OS and U−2 OS cell lines. Cancer Genet. Cytogen. 2010, 196, 109–118. [Google Scholar] [CrossRef]
- Wang, X.; Morris-Natschke, S.L.; Lee, K.H. New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med. Res. Rev. 2007, 27, 133–148. [Google Scholar] [CrossRef]
- Nwafor, E.O.; Lu, P.; Li, J.W.; Zhang, Q.Q.; Qi, D.L.; Liu, Z.D.; Peng, H.; Qin, H.; Gao, Y.Q.; Wang, J.L. Traditional Chinese medicine of Salvia miltiorrhiza Bunge: A review of phytochemistry, pharmacology and pharmacokinetics. Tradit. Med. Res. 2021, 6, 35. [Google Scholar] [CrossRef]
- Chen, X.; Guo, J.; Bao, J.; Lu, J.; Wang, Y. The anticancer properties of Salvia Miltiorrhiza Bunge (Danshen): A systematic review. Med. Res. Rev. 2014, 34, 768–794. [Google Scholar] [CrossRef]
- Zhang, L.H.; Wang, Y.Y.; Guo, X.Y.; Wu, S.H. Concentrical coils counter-current chromatography for natural products isolation: Salvia miltiorrhiza Bunge as example. J. Chromatogr. A 2017, 1491, 108–116. [Google Scholar] [CrossRef]
- Meng, J.; Yang, Z.; Liang, J.; Guo, M.; Wu, S. Multi-channel recycling counter-current chromatography for natural product isolation: Tanshinones as examples. J. Chromatogr. A 2014, 1327, 27–38. [Google Scholar] [CrossRef]
- Zhou, L.M.; Zuo, Z.; Chow, M.S.S. Danshen: An overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J. Clin. Pharmacol. 2005, 45, 1345–1359. [Google Scholar] [CrossRef]
- Fu, L.; Han, B.; Zhou, Y.; Ren, J.; Zhang, J. The anticancer properties of tanshinones and the pharmacological effects of their active ingredients. Front. Pharmacol. 2020, 11, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, J.; Little, P.J.; Xu, S. Atheroprotective effects and molecular targets of tanshinones derived from herbal medicine Danshen. Med. Res. Rev. 2018, 38, 201–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Su, X.; Fang, J.; Xin, X.; Zhao, X.; Gaur, U.; Wen, Q.; Xu, J.; Little, P.J.; Zheng, W. Tanshinone IIA attenuates insulin like growth factor 1 -induced cell proliferation in PC12 Cells through the PI3K/Akt and MEK/ERK pathways. Int. J. Mol. Sci. 2018, 19, 2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.C.; Lin, Y.H. Tanshinone IIA inhibits human breast cancer cells through increased Bax to Bcl-xL ratios. Int. J. Mol. Med. 2008, 22, 357–361. [Google Scholar]
- Chen, J.; Shi, D.Y.; Liu, S.L.; Zhong, L. Tanshinone IIA induces growth inhibition and apoptosis in gastric cancer in vitro and in vivo. Oncol. Rep. 2012, 27, 523–528. [Google Scholar]
- Gong, Y.; Li, Y.; Lu, Y.; Li, L.; Abdolmaleky, H.; Blackburn, G.L.; Zhou, J.R. Bioactive tanshinones in Salvia miltiorrhiza inhibit the growth of prostate cancer cells in vitro and in mice. Int. J. Cancer 2011, 129, 1042–1052. [Google Scholar] [CrossRef]
- Liu, X.D.; Fan, R.F.; Zhang, Y.; Yang, H.Z.; Fang, Z.G.; Guan, W.B.; Lin, D.J.; Xiao, R.Z.; Huang, R.W.; Huang, H.Q.; et al. Down-regulation of telomerase activity and activation of caspase-3 are responsible for Tanshinone I-induced apoptosis in monocyte leukemia cells in vitro. Int. J. Mol. Sci. 2010, 11, 2267–2280. [Google Scholar] [CrossRef]
- Hong, H.J.; Liu, J.C.; Cheng, T.H.; Chan, P. Tanshinone IIA attenuates angiotensin II-induced apoptosis via Akt pathway in neonatal rat cardiomyocytes. Acta Pharmacol. Sin. 2010, 31, 1569–1575. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.H.; Wu, Y.R.; Li, B.; Yan, Z.Y. Cryptotanshinone: A review of its pharmacology activities and molecular mechanisms. Fitoterapia 2020, 145, 104633. [Google Scholar] [CrossRef]
- Chiu, S.C.; Huang, S.Y.; Chang, S.F.; Chen, S.P.; Chen, C.C.; Lin, T.H.; Liu, H.H.; Tsai, T.H.; Lee, S.S.; Pang, C.Y.; et al. Potential therapeutic roles of tanshinone IIA in human bladder cancer cells. Int. J. Mol. Sci. 2014, 15, 15622–15637. [Google Scholar] [CrossRef] [Green Version]
- Fu, P.F.; Du, F.Y.; Chen, W.; Yao, M.Y.; Lv, K.Z.; Liu, Y. Tanshinone IIA blocks epithelial-mesenchymal transition through HIF-1 alpha downregulation, reversing hypoxia-induced chemotherapy resistance in breast cancer cell lines. Oncol. Rep. 2014, 31, 2561–2568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.Y.; Chang, S.F.; Liao, K.F.; Chiu, S.C. Tanshinone IIA inhibits epithelial-mesenchymal transition in bladder cancer cells via modulation of STAT3-CCL2 signaling. Int. J. Mol. Sci. 2017, 18, 1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Q.; Xu, X.; Jiang, G.Q.; Zhang, C.L.; Liu, L.K.; Kang, J.; Wang, J.; Owusu, L.; Zhou, L.Y.; Zang, L.; et al. Dihydrotanshinone I inhibits ovarian cancer cell proliferation and migration by transcriptional repression of PIK3CA gene. J. Cell. Mol. Med. 2020, 24, 11177–11187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, D.; Zhang, X.; Liu, Y.; Yan, L. Tanshinone IIA regulates fibroblast proliferation and migration and post-surgery arthrofibrosis through the autophagy-mediated PI3K and AMPK-mTOR signaling pathway. Am. J. Transl. Res. 2021, 13, 565–584. [Google Scholar]
- Zhou, H.Y.; Jiang, S.; Li, P.F.; Shen, H.; Yang, H.; Xu, S.Q.; Ye, C.Y.; Chen, M.Q.; Lu, H. Improved tendon healing by a combination of Tanshinone IIA and miR-29b inhibitor treatment through preventing tendon adhesion and enhancing tendon strength. Int. J. Med. Sci. 2020, 17, 1083–1094. [Google Scholar] [CrossRef]
- Tang, C.; Xue, H.L.; Bai, C.L.; Fu, R. Regulation of adhesion molecules expression in TNF-alpha-stimulated brain microvascular endothelial cells by tanshinone IIA: Involvement of NF-kappa B and ROS generation. Phytother. Res. 2011, 25, 376–380. [Google Scholar]
- Kashyap, A.; Umar, S.; Arundhathi, D.; Prasad, C. Dihydrotanshinone-I modulates epithelial mesenchymal transition (EMT) thereby impairing migration and clonogenicity of triple negative breast cancer cells. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 2177–2184. [Google Scholar] [CrossRef]
- Tan, T.; Wang, N.; Lin, D.Y.; Yang, S.D.; Lin, H.; Deng, Z.Q.; Luo, J.Y.; Luo, X.J. Dihydrotanshinone Ⅰ inhibits proliferation, migration and invasion of human osteosarcoma 143B cells and promotes apoptosis. Tumor 2019, 39, 450–459. [Google Scholar]
- Chen, X.P.; Li, Q.H.; He, Y.; Du, H.J.; Zhan, Z.J.; Zhao, H.; Shi, J.J.; Ye, Q.S.; Hu, J.A. 15,16-dihydrotanshinone I induces apoptosis and inhibits the proliferation, migration of human Osteosarcoma cell line 143B in vitro. Anti-Cancer Agents Med. Chem. 2017, 17, 1234–1242. [Google Scholar] [CrossRef]
- Harjunp, H.; Asens, M.L.; Guenther, C.; Fagerholm, S.C. Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front. Immunol. 2019, 10, 1078. [Google Scholar] [CrossRef] [Green Version]
- Bendas, G.; Borsig, L. Cancer cell adhesion and metastasis: Selectins, integrins, and the inhibitory potential of heparins. Int. J. Cell Biol. 2012, 2012, 676731. [Google Scholar] [CrossRef]
- Parsons, J.T.; Horwitz, A.R.; Schwartz, M.A. Cell adhesion: Integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010, 11, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pan, L.; Liang, J.; Li, J.; Wu, S. Cryptotanshinone inhibits proliferation and induces apoptosis via mitochondria-derived reactive oxygen species involving FOXO1 in estrogen receptor-negative breast cancer Bcap37 cells. RSC Adv. 2016, 6, 22232–22243. [Google Scholar] [CrossRef]
- Calissi, G.; Lam, E.W.F.; Link, W. Therapeutic strategies targeting FOXO transcription factors. Nat. Rev. Drug Discov. 2021, 20, 21–38. [Google Scholar] [CrossRef]
- Eijkelenboom, A.; Burgering, B. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Boehm, M.; Nabel, E.G. Cell cycle and cell migration—New pieces to the puzzle. Circulation 2001, 103, 2879–2881. [Google Scholar] [CrossRef] [PubMed]
- Hochegger, H.; Takeda, S.; Hunt, T. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all? Nat. Rev. Mol. Cell Biol. 2008, 9, 910-U26. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [Green Version]
- Caldon, C.E.; Daly, R.J.; Sutherland, R.L.; Musgrove, E.A. Cell cycle control in breast cancer cells. J. Cell. Biochem. 2006, 97, 261–274. [Google Scholar] [CrossRef]
- Tang, N.; Song, W.X.; Luo, J.Y.; Haydon, R.C.; He, T.C. Osteosarcoma development and stem cell differentiation. Clin. Orthop. Relat. Res. 2008, 466, 2114–2130. [Google Scholar] [CrossRef] [Green Version]
- Yeung, K.T.; Jing, Y. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, H.A.; Holly, J.M.P.; Bahl, A.; Perks, C.M. Inhibition of FASN and ER alpha signalling during hyperglycaemia-induced matrix-specific EMT promotes breast cancer cell invasion via a caveolin-1-dependent mechanism. Cancer Lett. 2018, 419, 187–202. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, H.; Li, J.R.; Fang, X.H.; Pan, H.; Yuan, X.L.; Zhang, P. Up-regulated FASN expression promotes transcoelomic metastasis of ovarian cancer cell through epithelial-mesenchymal transition. Int. J. Mol. Sci. 2014, 15, 11539–11554. [Google Scholar] [CrossRef] [PubMed]
- Vuoriluoto, K.; Haugen, H.; Kiviluoto, S.; Mpindi, J.P.; Nevo, J.; Gjerdrum, C.; Tiron, C.; Lorens, J.B.; Ivaska, J. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2011, 30, 1436–1448. [Google Scholar] [CrossRef] [Green Version]
- Kudo-Saito, C.; Shirako, H.; Takeuchi, T.; Kawakami, Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 2009, 15, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Niu, N.K.; Wang, Z.L.; Pan, S.T.; Ding, H.Q.; Au, G.H.T.; He, Z.X.; Zhou, Z.W.; Xiao, G.Z.; Yang, Y.X.; Zhang, X.J.; et al. Pro-apoptotic and pro-autophagic effects of the Aurora kinase A inhibitor alisertib (MLNLN8237) on human osteosarcoma U−2 OS and MG-63 cells through the activation of mitochondria-mediated pathway and inhibition of p38 MAPK/PI3K/Akt/mTOR signaling pathway. Drug Des. Dev. Ther. 2015, 9, 1555–1584. [Google Scholar]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A multifunctional cell surface adhesion receptor is a regulator of progression and metastasis of cancer cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Wolf, K.J.; Shukla, P.; Springer, K.; Lee, S.; Coombes, J.D.; Choy, C. A mode of cell adhesion and migration facilitated by CD44-dependent microtentacles. Proc. Natl. Acad. Sci. USA 2020, 117, 11432–11443. [Google Scholar] [CrossRef]
- Sato, H.; Okada, Y.; Seiki, M. Membrane-type matrix metalloproteinases (MT-MMPs) in cell invasion. Thromb. Haemost. 1997, 78, 497–500. [Google Scholar] [CrossRef]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Knall, C.; Worthen, G.S.; Johnson, G.L. Interleukin 8-stimulated phosphatidylinositol-3-kinase activity regulates the migration of human neutrophils independent of extracellular signal-regulated kinase and p38 mitogen-activated protein kinases. Proc. Natl. Acad. Sci. USA 1997, 94, 3052–3057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhar, D.; Antonucci, L.; Nakagawa, H.; Kim, J.Y.; Glitzner, E.; Caruso, S.; Shalapour, S.; Yang, L.; Valasek, M.A.; Lee, S.; et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell 2018, 33, 1061–1077.e6. [Google Scholar]
- Collado, M.; Blasco, M.A.; Serrano, M. Cellular senescence in cancer and aging. Cell 2007, 130, 223–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldin, V.; Lukas, J.; Marcote, M.J.; Pagano, M.; Draetta, G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993, 7, 812–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.H.; Deng, M.; Wei, Q.; Liu, T.; Tong, X.M.; Ye, X. Phosphorylation of MCM3 protein by Cyclin E/Cyclin-dependent Kinase 2 (Cdk2) regulates its function in cell cycle. J. Biol. Chem. 2011, 286, 39776–39785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.J.; Lin, C.Y.; Liao, W.Y.; Hour, T.C.; Wang, H.D.; Chuu, C.P. CD44 promotes migration and invasion of docetaxel-resistant prostate cancer cells likely via induction of hippo-yap signaling. Cells 2019, 8, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miletti-Gonzalez, K.E.; Chen, S.; Muthukumaran, N.; Saglimbeni, G.N.; Wu, X.; Yang, J.; Apolito, K.; Shih, W.J.; Hait, W.N.; Rodriguez-Rodriguez, L. The CD44 receptor interacts with P-glycoprotein to promote cell migration and invasion in cancer. Cancer Res. 2005, 65, 6660–6667. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.X.; Niu, M.K.; Yuan, X.; Wu, K.M.; Liu, A.G. CD44 as a tumor biomarker and therapeutic target. Exp. Hematol. Oncol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Seo, J.M.; Park, S.; Kim, J.H. Leukotriene B-4 receptor-2 promotes invasiveness and metastasis of ovarian cancer cells through signal transducer and activator of transcription 3 (STAT3)-dependent up-regulation of matrix metalloproteinase 2. J. Biol. Chem. 2012, 287, 13840–13849. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.H.; Jia, Y.; Guo, F.J.; Chen, J.; Zhang, X.W.; Cui, M.H. Phosphorylation of STAT3 at Tyr705 regulates MMP-9 production in epithelial ovarian cancer. PLoS ONE 2017, 12, e0183622. [Google Scholar] [CrossRef] [Green Version]
- Weng, M.T.; Tsao, P.N.; Lin, H.L.; Tung, C.C.; Change, M.C.; Chang, Y.T.; Wong, J.M.; Wei, S.C. Hes1 increases the invasion ability of colorectal cancer cells via the STAT3-MMP14 pathway. PLoS ONE 2015, 10, e0144322. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pongcharoen, P.; Jinawath, A.; Tohtong, R. Silencing of CD44 by siRNA suppressed invasion, migration and adhesion to matrix, but not secretion of MMPs, of cholangiocarcinoma cells. Clin. Exp. Metastasis 2011, 28, 827–839. [Google Scholar] [CrossRef] [PubMed]











| IC50 (µM) | DS−1 | DS−2 | DS−3 | DS−4 | DS−5 |
|---|---|---|---|---|---|
| U−2 OS cells (24 h) | 3.83 ± 0.49 | 47.84 ± 4.96 | 17.75 ± 1.03 | 22.06 ± 1.31 | 25.40 ± 1.43 |
| (48 h) | 1.99 ± 0.37 | 25.60 ± 0.81 | 4.54 ± 1.00 | 5.89 ± 0.40 | 5.91 ± 1.08 |
| HeLa cells (24 h) | 15.48 ± 0.98 | 52.07 ± 3.20 | 20.92 ± 2.81 | 25.03 ± 2.03 | 32.09 ± 1.94 |
| NRK−49F cells (24 h) | 25.00 ± 1.98 | 35.39 ± 3.25 | 107.60 ± 28.85 | 31.84 ± 1.59 | / a |
| DS−1 (µM) | 0 | 2.5 | 5.0 | 7.5 |
|---|---|---|---|---|
| G0/G1 | 54.70 ± 2.21 | 61.07 ± 2.04 | 65.20 ± 4.35 | 65.65 ± 1.83 |
| S | 24.00 ± 2.15 | 20.10 ± 2.07 | 19.55 ± 3.08 | 14.20 ± 0.40 |
| G2/M | 20.40 ± 3.95 | 18.65 ± 3.50 | 14.92 ± 4.67 | 19.60 ± 1.40 |
| Gene | Upstream Primers (5′ to 3′) | Downstream Primers (5′ to 3′) |
|---|---|---|
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATCATTCTCATGG |
| Actin | CCGTCTTCCCCTCCATCGT | ATCGTCCCAGTTGGTTACAATGC |
| MT-MMP-1 | GGTGGAGGTTGTAGGTGTGA | CAACCCTCAGAGAGCAAAGC |
| CDK2 | CCAAGTGAGACTGAGGGTGT | CCAAGTGAGACTGAGGGTGT |
| CDK4 | CTGCAGGCTCATACCATCCT | ACTCTTGAGGGCCACAAAGT |
| Cyclin D1 | CTCCTTTCTCCACCCACCTC | TCCTCTGCTGGACACCCC |
| Cyclin E1 | CAGCGGTTGTAATGTGACCC | AAAGCTCTTCCCCACCCAAT |
| CXCL8 | TGTCCTATTGAGAACCACGGT | GCAAGCTAAGACTCTCCAGC |
| p21 | CCCAAGCTCTACCTTCCCAC | CTGAGAGTCTCCAGGTCCAC |
| IL6 | TCCACTGGAATTTGCTTGCC | AGTGCCTCTTTGCTGCTTTC |
| FASN | TGTGGTGTGTGGGTTGGTAT | GGACGAAATGGGGATAGCCT |
| Snail | CTACCTGTTTGCACACTCGG | ATTTGGTCTTGGCAAAAGCC |
| Slug | TTCTACGTTCTCTGGGCTGG | ACCCAGGCTCACATATTCCT |
| E-cadherin | AACGCATTGCCACATACACTC | GACCTCCATCACAGAGGTTCC |
| Vimentin | TCAATGTTAAGATGGCCCTTG | TGAGTGGGTATCAACCAGAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, L.; Peng, C.; Zhu, X.; Liang, Y.; Xu, T.; Xu, P.; Wu, S. Dihydrotanshinone I Enhances Cell Adhesion and Inhibits Cell Migration in Osteosarcoma U−2 OS Cells through CD44 and Chemokine Signaling. Molecules 2022, 27, 3714. https://doi.org/10.3390/molecules27123714
Fan L, Peng C, Zhu X, Liang Y, Xu T, Xu P, Wu S. Dihydrotanshinone I Enhances Cell Adhesion and Inhibits Cell Migration in Osteosarcoma U−2 OS Cells through CD44 and Chemokine Signaling. Molecules. 2022; 27(12):3714. https://doi.org/10.3390/molecules27123714
Chicago/Turabian StyleFan, Lanyan, Chen Peng, Xiaoping Zhu, Yawen Liang, Tianyi Xu, Peng Xu, and Shihua Wu. 2022. "Dihydrotanshinone I Enhances Cell Adhesion and Inhibits Cell Migration in Osteosarcoma U−2 OS Cells through CD44 and Chemokine Signaling" Molecules 27, no. 12: 3714. https://doi.org/10.3390/molecules27123714