Cellulolytic and Xylanolytic Enzymes from Yeasts: Properties and Industrial Applications
Abstract
:1. Introduction
2. Source, Composition and Physico-Chemical Properties of Cellulose
3. Classification and Mechanism of Cellulolytic Enzymes
4. Cellulolytic Microorganisms
5. Cellulolytic Yeasts
| Origin | Source | Isolated From | Strains | Reference |
|---|---|---|---|---|
| Terrestrial | Bali National Park, Indonesia | Dendrobium flower | Isolates D.2.7 and W.3.8 | [2] |
| - | Decayed wood | Aureobasidium microstictum, Trichosporon pullulans | [75] | |
| - | Enriched soil sample | Candida tropicalis (MK-118) | [78] | |
| Laboratory of Bioprocesses and Sustainable Products (LBIOS–UNESP, Rio Claro, SP, Brazil) | - | Aureobasidium pullulans LB83 | [84] | |
| Mushroom farm in Yala Local Government Area of Cross River State, Nigeria | Soil samples | Saccharomyces cerevisiae SCPW 17 | [85] | |
| Southeast Sulawesi, Indonesia | Leaf and leaf litter samples | Candida sp. Sporodiobolus sp. Pichia sp. Pseudozyma sp. Sporobolomyces sp. | [86] | |
| Onam-ri, Gyeonggi Province, South Korea | Gut of Grasshopper | Moesziomyces sp. | [87] | |
| Toledo River, Parana (PR), Brazil | Water samples | Apiotrichum mycotoxinivorans | [88] | |
| decaying leaves, wood and ant nests from Brazil | Decaying leaves, wood and ant nest | Trichosporon laibachii MG270406-1A14 strain | [89] | |
| - | - | Trichosporon sp. | [90] | |
| Eastern Ghats region of Thandikudi, Tamil Nadu, India | Forest soil samples | Trichosporon asahii | [91] | |
| - | - | Saccharomyces cerevisiae | [92] | |
| Five different regions around the local area, Jodhpur, India | Soil rich in cellulosic waste | Cystobasidium oligophagum | [93] | |
| Batanta Island Raja Ampat, West Papua Province, Indonesia | Multiple soil cores (15 cm depth by 2 cm diameter) | Sporobolomyces poonsookiae, Rhodosporidium paludigenum, and Cryptococcus flavescens | [94] | |
| RongKho forest, Ubon Ratchathani University, Thailand | Soil, tree barks and insect frass | Candida sp. 05-7-186T, Candida easanensis and Candida sp. ST-390 | [95] | |
| Gunung Halimun National Park, Indonesia | Soil, and rhizosphere soil | Debaryomyces sp., Rhodotorula sp., Pichia sp. and Candida sp. | [96] | |
| King George Island, sub-Antarctic region | Soil samples | Leucosporidiella fragaria and Mrakia sp. | [97] | |
| Marine | Yellow Sea, China | Sea saltern | Aureobasidium pullulans 98 | [79] |
| - | - | Aureobasidium pullulans 98 | [98] |
6. Basic Properties of Cellulolytic Yeasts
| Species | Type of Enzyme | Some of the Biochemical Properties of Cellulase Isolated from Yeasts | References |
|---|---|---|---|
| Trichosporon sp. | Endoglucanase and FPase | CMCase worked optimally at 55 °C and pH 5, while for FPase activity, temperature 60 °C and pH 4 to 6 found optimal. Both the enzymes showed best activity when reacted with 100 mg/mL of their respective substrates. FPase required more time to catalyze the reaction, probably due to the time taken for adsorption to the insoluble substrate | [90] |
| Aureobasidium pullulans 98 | Endoglucanase | The strain produced a titer of 4.51 CMCase U mg−1 of protein. The enzyme has a molecular mass of 67.0 kDa, as determined by SDS PAGE. The enzyme exhibited its maximum catalytic efficiency at 40 °C and pH 5.6, but it did not lose its activity in the buffer of pH 5.0–6.0. Na+, Mg2+, Ca2+, K+ Fe2+ and Cu2+ were found to be activators of the enzyme, while Fe3+, Ba2+, Zn2+, Mn2+ and Ag+ inhibited its activity. Km and Vmax values of the purified enzyme were 4.7 mg mL−1 and 0.57 µmol L−1 min−1 (mg protein)−1, respectively. | [79] |
| Trichosporon asahii | Endoglucanase | Under optimum conditions, the strain produced 35.70 U of the cellulase, which was reduced to 23.87 U when CMC in the medium was replaced by Napier biomass. The enzyme preparation saccharified the crude substrate with 33.15% yield in 3 days. | [91] |
| Sporobolomyces poonsookiae, Rhodosporidium paludigenum, and Cryptococcus flavescens | Endoglucanase | The cellulolytic potential of the three strains Sporobolomyces poonsookiae Y08RA07, Rhodosporidium paludigenum Y08RA29 and Cryptococcus flavescens Y08RA33 was evaluated where R. paludigenum appeared promising with an index value of 2.60. The isolate S. poonsookiae produced CMCase optimally at pH 8 and 37 °C, while R. paludigenum and C. flavescens produced the highest CMCase levels at pH 6 and 28 °C. Paper waste appeared a good substrate for S. poonsookiae, while bamboo leaf was also utilized by the same substrate by this strain and by C. flavescens. R. paludigenum, however, was cultivated on soluble CMC to produce optimal titers of cellulase. | [94] |
| Candida sp. 05-7-186T, Candida easanensis and Candida sp. ST-390 | Endoglucanase | The optimization studies showed that 1.0% CMC causes the most effective induction of the enzyme production by all three strains. An incubation period of 96 h was found suitable for cellulase production by Candida sp. 05-7-186T (0.224 UmL−1), while Candida easanensis (0.238 UmL−1 and Candida sp. ST-390 (0.26 UmL−1) produced highest titers in 120 h. | [95] |
| Candida tropicalis (MK-118) | Endoglucanase and β-glucosidase | This strain co-produced endoglucanase (EG) and β-glucosidase (BGL) at 40 °C but at different pH. Neutral pH favored the EG production, while enhanced BGL titers were obtained in acidic medium. For both the enzyme productions, an inoculum size of 2% was found appropriate, but in the presence of respective inducer i.e., CMC for EG and salicin for BGL. | [78] |
7. Application of Cellulolytic Yeasts
8. What Is Xylan?
9. Xylan Degradation
10. Xylanolytic Microorganisms
11. Xylanolytic Yeasts
12. Basic Properties of Xylanolytic Enzymes from Yeasts
| Species | Type of Xylanase | Some of the Biochemical Properties of Xylanase Isolated from Yeasts | References |
|---|---|---|---|
| Candida tropicalis (MK-118) | The highest titers of xylanase were obtained at 25 °C under neutral pH and in the presence of 1% xylan. | [78] | |
| Candida tropicalis MK-160 | The strain produced 23.6 IU mL−1 of xylanase at 40 °C, in an acidic medium containing 2% xylan. It also produced 5.45% ethanol in glucose-supplemented medium. PO4 2−, Co+2, Cu+2 or Ca+2 activated the xylanase activity, while Na+, Mg+2, Mn+2 and K+ appeared as inhibitors | [172] | |
| Aureobasidium pullulans CBS 135684 | The enzyme was purified 17.3-fold with a recovery yield of 13.7%. Its molecular mass was determined through SDS PAGE as 72 kDa. Temperature 70 °C and pH 6.0 were found appropriate for this enzyme’s activity. However, the enzyme retained more than 50% of its activity for 3 h at 50 °C; the addition of 0.75 mM sorbitol improved thermostability up to 10-fold at 70 °C. The enzyme was activated in the presence of Ca2+, Co2+, and Mg2+ and inhibited by Fe2+ and Cu2+ | [176] | |
| Pichia membranifaciens | Pichia membranifaciens produced the highest titers of 38.8 U mL−1 of xylanase in 4 days in a culture medium with pH adjusted to 7.0. Xylanase activity showed maximum activity (42.6 U mL−1) at 35 °C in the presence of beechwood xylan. An increase in the Km values of xylanase was noted with a decrease in pH from 7.0 to 4.5. | [183] | |
| Candida pseudorhagii | The strain could utilize both xylan and D-xylose but produced more enzyme on xylan (1.73 U mL−1) than on D-xylose 0.98 U mL−1. The study also reported the identification of four novel strains with the ability to ferment D-xylose and to produce ethanol. Particularly, C. pseudorhagii SSA-1542 T yielded 0.31 g/g of ethanol, with a productivity of 0.31 g L−1 h−1 and fermentation efficiency of 60.7% in 48 h. | [181] | |
| Pichia stipitis | The strain was cultivated under the solid-state fermentation of a corncob and wheat bran mixture and released 5536 U of xylanase per g of substrate. The enzyme was purified, and the molecular weight was determined as 31.6 kDa using SDS PAGE. The optimum temperature and pH were 50 °C and 6.0, respectively. Kinetics parameters were determined as Km 4.52 mg mL−1 and Vmax 9.17 μmol min−1 mL−1. Xylanase did not lose half of its activity by exposure to 50 °C for 80 min and to pH 5–8 for 60 min. The enzyme remained unaffected in the presence of Cu2+ and K+. | [184] | |
| Cryptococcus adeliae | A statistical approach was used to optimize xylanase production from this strain and an increase by 4.3-fold in the enzyme titers was achieved. The optimized conditions include 12.1 g L−1 xylan, 5.1 g L−1 yeast extract initial pH of 7.5, temperature 4 °C and incubation time 168 h. A titer of 400nkat of xylanase was obtained in the medium. Although the maximum xylanase titers achieved in presence of xylan, the strain also utilized lignocellulosics. Amongst nitrogenous sources, the organism effectively utilized yeast extract, soymeal, pharmamedia (cotton seed protein) and alburex (potato protein). The crude xylanase remained stable at pH 4–9 for 21 h (at 4°C) but exhibited optimal activity at pH 5.0–5.5. The optimum temperature for the activity was found to be 45–50 °C, but it appeared very thermolabile, with a half-life of 78 min at 35 °C, and at 40–50 °C, it lost 71–95% activity within 5 min. | [175] | |
| Cryptococcus adeliae | The study characterized this cold-adaptive enzyme with an apparent molecular weight of 43 kDa as estimated through SDS PAGE. The enzyme had a half-life of 60 min at 30 °C and a melting temperature of 48 °C. The activation energy, Ea, of the psychrophilic enzyme was 47.7 kJ mol−1. | [174] | |
| Pseudozyma hubeiensis, Hannaella pagnoccae, Papiliotrema mangalensis, Kodamaea ohmeri TS21 | The thermotolerance of the strains Pseudozyma hubeiensis STAG 1.7 and Hannaella pagnoccae STAG 1.14 was evident at 45 °C with the concomitant production of xylanase activities of 1.31 and 1.17 IU, respectively. The other two strains yielded less than 1 IU of xylanases. | [179] |
13. Application of Xylanolytic Yeasts
14. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Butinar, L.; Santos, S.; Spencer-Martins, I.; Oren, A.; Gunde-Cimerman, N. Yeast diversity in hypersaline habitats. FEMS Microbiol. Lett. 2005, 244, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Arman, Z.; Sondana, G.; Fikriyyah, N.; Afifah, Z.; Balqis, M.; Hasanah, R.; Risandi, A.; Sofiana, I.; Nisa, H. Ridawati. Screening of Amylolytic and Cellulolytic Yeast from Dendrobium Spathilingue in Bali Botanical Garden, Indonesia. AIP Conf. Proc. 2020, 2242, 050013. [Google Scholar]
- Souza, P.M.D.; Magalhães, P.D.O. Application of microbial α-amylase in industry—A review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Adelabu, B.A.; Kareem, S.O.; Oluwafemi, F.; Adeogun, I.A. Bioconversion of corn straw to ethanol by cellulolytic yeasts immobilized in Mucuna urens matrix. J. King Saud Univ. Sci. 2019, 31, 136–141. [Google Scholar] [CrossRef]
- Kasana, R.C.; Gulati, A. Cellulases from psychrophilic microorganisms: A review. J. Basic Microbiol. 2011, 51, 572–579. [Google Scholar] [CrossRef]
- Arora, R.; Sharma, N.K.; Kumar, S.; Sani, R.K. Lignocellulosic ethanol: Feedstocks and bioprocessing. In Bioethanol Production from Food Crops; Elsevier: Amsterdam, The Netherlands, 2019; pp. 165–185. [Google Scholar]
- Howard, R.; Abotsi, E.; Van Rensburg, E.J.; Howard, S. Lignocellulose biotechnology: Issues of bioconversion and enzyme production. Afr. J. Biotechnol. 2003, 2, 602–619. [Google Scholar] [CrossRef]
- Dellanerra, D.; Risandi, A.; Sunari, A.; Sukmawati, D.; Husna, S.N.A.; El-Enshasy, H.A. Screening and Characterization of amylolitic mold originated from ghost crab (Ocypode sp.) in Cidaon, Ujung Kulon National Park, Indonesia. AIP Conf. Proceedings 2019, 2120, 070008. [Google Scholar]
- Biely, P. Microbial xylanolytic systems. Trends Biotechnol. 1985, 3, 286–290. [Google Scholar] [CrossRef]
- Katahira, S.; Fujita, Y.; Mizuike, A.; Fukuda, H.; Kondo, A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 2004, 70, 5407–5414. [Google Scholar] [CrossRef]
- Du, L.; Cui, X.; Li, H.; Wang, Y.; Fan, L.; He, R.; Jiang, F.; Yu, A.; Xiao, D.; Ma, L. Enhancing the enzymatic hydrolysis efficiency of lignocellulose assisted by artificial fusion enzyme of swollenin-xylanase. Ind. Crop. Prod. 2021, 173, 114106. [Google Scholar] [CrossRef]
- Kaushal, J.; Khatri, M.; Singh, G.; Arya, S.K. A multifaceted enzyme conspicuous in fruit juice clarification: An elaborate review on xylanase. Int. J. Biol. Macromol. 2021, 193, 1350–1361. [Google Scholar] [CrossRef] [PubMed]
- Kuhad, R.C.; Singh, A. Lignocellulose biotechnology: Current and future prospects. Crit. Rev. Biotechnol. 1993, 13, 151–172. [Google Scholar] [CrossRef]
- Jeffries, T.W. Utilization of xylose by bacteria, yeasts, and fungi. In Pentoses Lignin; Elsevier: Amsterdam, The Netherlands, 1983; pp. 1–32. [Google Scholar]
- Holtzapple, M. Cellulose. In Encyclopedia of Food Science and Nutrition, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Khandelwal, M.; Windle, A.H. Hierarchical organisation in the most abundant biopolymer–cellulose. In MRS Online Proceedings Library (OPL); Cambridge University Press: Cambridge, UK, 2013; p. 1504. [Google Scholar]
- Ramachandran, G.; Ramakrishnan, C.; Venkatachalam, C. Structure of polyglycine II with direct and inverted chains. In Conformation of Biopolymers; Elsevier: Amsterdam, The Netherlands, 1967; pp. 429–438. [Google Scholar]
- Krässig, H.A. Cellulose: Structure, Accessibility and Reactivity; Gordon and Breach Science Publishers: Lodon, UK, 1993. [Google Scholar]
- Dumitriu, S. Polysaccharides: Structural Diversity and Functional Versatility; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Valishina, Z.T.; Starostina, I.A.; Pavlovets, G.Y.; Deberdeev, R.Y. Structure and properties of synthetic cellulose. Chem. Phys. Mesoscopy 2021, 2, 176–189. [Google Scholar] [CrossRef]
- Perez, S.; Samain, D. Structure and engineering of celluloses. Adv. Carbohydr. Chem. Biochem. 2010, 64, 25–116. [Google Scholar] [PubMed]
- Ilham, Z. Biomass classification and characterization for conversion to biofuels. In Value-Chain of Biofuels; Elsevier: Amsterdam, The Netherlands, 2022; pp. 69–87. [Google Scholar]
- Rongpipi, S.; Ye, D.; Gomez, E.D.; Gomez, E.W. Progress and opportunities in the characterization of cellulose—An important regulator of cell wall growth and mechanics. Front. Plant. Sci. 2019, 9, 1894. [Google Scholar] [CrossRef] [PubMed]
- Momzyakova, K.; Deberdeev, T.; Valishina, Z.; Deberdeev, R.Y.; Ibragimov, A.; Alexandrov, A. Materials Science Forum. In Research of Physical and Chemical Properties of Powder Cellulose from Various Type of Raw Materials; Trans Tech Publications Ltd.: Kapellweg, Switzerland, 2020; pp. 791–795. [Google Scholar]
- Valishina, Z.T.; Matukhin, E.L.; Khakimzyanova, R.I.; Kostochko, A.V. Comprehensive System of Analytical Monitoring of Raw Materials to Promptly Manage Producing Cellulose Ni-trates. Bull. Kazan Technol. Univ. 2018, 12, 46–51. [Google Scholar]
- Gupta, P.K.; Raghunath, S.S.; Prasanna, D.V.; Venkat, P.; Shree, V.; Chithananthan, C.; Choudhary, S.; Surender, K.; Geetha, K. An update on overview of cellulose, its structure and applications. In Cellulose; Pasculal, A., Martin, M., Eds.; Intechopen: London, UK, 2019; pp. 846–1297. [Google Scholar]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Fernandes, A.N.; Thomas, L.H.; Altaner, C.M.; Callow, P.; Forsyth, V.T.; Apperley, D.C.; Kennedy, C.J.; Jarvis, M.C. Nanostructure of cellulose microfibrils in spruce wood. Proc. Natl. Acad. Sci. USA 2011, 108, E1195–E1203. [Google Scholar] [CrossRef]
- Ramawat, K.G.; Mérillon, J.-M. (Eds.) Polysaccharides; Springer Cham: Berlin/Heidelberg, Germany, 2014; pp. 219–246. Available online: https://link.springer.com/referencework/10.1007/978-3-319-16298-0?noAccess=true#book-header (accessed on 8 June 2022).
- Díez-Pascual, A.M. Synthesis and Applications of Biopolymer Composites; Multidisciplinary Digital Publishing Institute: Basel, Switzerland, 2019; Volume 20, p. 2321. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Szijártó, N.; Siika-Aho, M.; Tenkanen, M.; Alapuranen, M.; Vehmaanperä, J.; Réczey, K.; Viikari, L. Hydrolysis of amorphous and crystalline cellulose by heterologously produced cellulases of Melanocarpus albomyces. J. Biotechnol. 2008, 136, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current perspective on production and applications of microbial cellulases: A review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef]
- Harris, P.V.; Xu, F.; Kreel, N.E.; Kang, C.; Fukuyama, S. New enzyme insights drive advances in commercial ethanol production. Curr. Opin. Chem. Biol. 2014, 19, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Herpoël-Gimbert, I.; Margeot, A.; Dolla, A.; Jan, G.; Mollé, D.; Lignon, S.; Mathis, H.; Sigoillot, J.-C.; Monot, F.; Asther, M. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 2008, 1, 18. [Google Scholar] [CrossRef]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Factories 2016, 15, 106. [Google Scholar] [CrossRef]
- Chundawat, S.; Bellesia, G.; Uppugundla, N.; Sousa, L.; da Costa, G.D.; Cheh, A.M.; Agarwal, U.P.; Bianchetti, C.M.; Phillips, G.N.; Langan, P.; et al. Restructuring the Crystalline Cellulose Hydrogen Bond Network Enhances Its Depolymerization Rate. J. Am. Chem. Soc. 2011, 133, 11163. [Google Scholar] [CrossRef]
- Igarashi, K.; Wada, M.; Samejima, M. Activation of crystalline cellulose to cellulose IIII results in efficient hydrolysis by cellobiohydrolase. FEBS J. 2007, 274, 1785–1792. [Google Scholar] [CrossRef]
- Douglas, A. Endosymbionts and Iintracellular Parasites. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Ejaz, U.; Sohail, M.; Ghanemi, A. Cellulases: From bioactivity to a variety of industrial applications. Biomimetics 2021, 6, 44. [Google Scholar] [CrossRef]
- Qadir, F.; Shariq, M.; Ahmed, A.; Sohail, M. Evaluation of a yeast co-culture for cellulase and xylanase production under solid state fermentation of sugarcane bagasse using multivariate approach. Ind. Crop. Prod. 2018, 123, 407–415. [Google Scholar] [CrossRef]
- Doi, R.H.; Tamaru, Y. The Clostridium cellulovorans cellulosome: An enzyme complex with plant cell wall degrading activity. Chem. Rec. 2001, 1, 24–32. [Google Scholar] [CrossRef]
- Boudreau, B.P. A kinetic model for microbic organic-matter decomposition in marine sediments. FEMS Microbiol. Ecol. 1992, 11, 1–14. [Google Scholar] [CrossRef]
- Ichikawa, S.; Karita, S.; Kondo, M.; Goto, M. Cellulosomal carbohydrate-binding module from Clostridium josui binds to crystalline and non-crystalline cellulose, and soluble polysaccharides. FEBS Lett. 2014, 588, 3886–3890. [Google Scholar] [CrossRef]
- Bayer, E.A.; Belaich, J.-P.; Shoham, Y.; Lamed, R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.B.; Kostylev, M. Cellulase processivity. In Biomass Conversion; Springer: Berlin/Heidelberg, Germany, 2012; pp. 93–99. [Google Scholar]
- Brás, J.L.; Carvalho, A.L.; Viegas, A.; Najmudin, S.; Alves, V.D.; Prates, J.A.; Ferreira, L.M.; Romao, M.J.; Gilbert, H.J.; Fontes, C.M. Escherichia coli expression, purification, crystallization, and structure determination of bacterial cohesin–dockerin complexes. Methods Enzymol. 2012, 510, 395–415. [Google Scholar] [PubMed]
- Xu, Q.; Resch, M.G.; Podkaminer, K.; Yang, S.; Baker, J.O.; Donohoe, B.S.; Wilson, C.; Klingeman, D.M.; Olson, D.G.; Decker, S.R. Dramatic performance of Clostridium thermocellum explained by its wide range of cellulase modalities. Sci. Adv. 2016, 2, e1501254. [Google Scholar] [CrossRef] [PubMed]
- Willson, D.B. Cellulases and Biofuels. Curr. Opin. Biotechnol. 2009, 20, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.B. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 2008, 1125, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.B. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 2011, 14, 259–263. [Google Scholar] [CrossRef]
- Wilson, D.B. Evidence for a novel mechanism of microbial cellulose degradation. Cellulose 2009, 16, 723–727. [Google Scholar] [CrossRef]
- Zhu, Y.; Han, L.; Hefferon, K.L.; Silvaggi, N.R.; Wilson, D.B.; McBride, M.J. Periplasmic Cytophaga hutchinsonii endoglucanases are required for use of crystalline cellulose as the sole source of carbon and energy. Appl. Environ. Microbiol. 2016, 82, 4835–4845. [Google Scholar] [CrossRef]
- Suen, G.; Weimer, P.J.; Stevenson, D.M.; Aylward, F.O.; Boyum, J.; Deneke, J.; Drinkwater, C.; Ivanova, N.N.; Mikhailova, N.; Chertkov, O. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 2011, 6, e18814. [Google Scholar] [CrossRef] [PubMed]
- Sabathé, F.; Soucaille, P. Characterization of the CipA scaffolding protein and in vivo production of a minicellulosome in Clostridium acetobutylicum. J. Bacteriol. 2003, 185, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; Salyers, A.A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 1989, 171, 3192–3198. [Google Scholar] [CrossRef] [PubMed]
- Premalatha, N.; Gopal, N.O.; Jose, P.A.; Anandham, R.; Kwon, S.-W. Optimization of cellulase production by Enhydrobacter sp. ACCA2 and its application in biomass saccharification. Front. Microbiol. 2015, 6, 1046. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, M.N.; Falkoski, D.L.; Guimarães, V.M.; de Rezende, S.T. Study of gamba grass as carbon source for cellulase production by Fusarium verticillioides and its application on sugarcane bagasse saccharification. Ind. Crop. Prod. 2019, 133, 33–43. [Google Scholar] [CrossRef]
- Crognale, S.; Liuzzi, F.; D’Annibale, A.; de Bari, I.; Petruccioli, M. Cynara cardunculus a novel substrate for solid-state production of Aspergillus tubingensis cellulases and sugar hydrolysates. Biomass Bioenergy 2019, 127, 105276. [Google Scholar] [CrossRef]
- Goldbeck, R.; Ramos, M.M.; Pereira, G.A.; Maugeri-Filho, F. Cellulase production from a new strain Acremonium strictum isolated from the Brazilian Biome using different substrates. Bioresour. Technol. 2013, 128, 797–803. [Google Scholar] [CrossRef]
- Picart, P.; Diaz, P.; Pastor, F. Cellulases from two Penicillium sp. strains isolated from subtropical forest soil: Production and characterization. Lett. Appl. Microbiol. 2007, 45, 108–113. [Google Scholar] [CrossRef]
- Annamalai, N.; Rajeswari, M.V.; Balasubramanian, T. Enzymatic saccharification of pretreated rice straw by cellulase produced from Bacillus carboniphilus CAS 3 utilizing lignocellulosic wastes through statistical optimization. Biomass Bioenergy 2014, 68, 151–160. [Google Scholar] [CrossRef]
- Gutierrez-Correa, M.; Tengerdy, R.P. Production of cellulase on sugar cane bagasse by fungal mixed culture solid substrate fermentation. Biotechnol. Lett. 1997, 19, 665–667. [Google Scholar] [CrossRef]
- Gautam, S.; Bundela, P.; Pandey, A.; Khan, S.; Awasthi, M.; Sarsaiya, S. Optimization for the production of cellulase enzyme from municipal solid waste residue by two novel cellulolytic fungi. Biotechnol. Res. Int. 2011, 2011, 810425. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, R.; Yang, X.; Wu, H.; Xu, D.; Tang, Z.; Shen, Q. Thermostable cellulase production of Aspergillus fumigatus Z5 under solid-state fermentation and its application in degradation of agricultural wastes. Int. Biodeterior. Biodegrad. 2011, 65, 717–725. [Google Scholar] [CrossRef]
- Jo, K.-I.; Lee, Y.-J.; Kim, B.-K.; Lee, B.-H.; Chung, C.-H.; Nam, S.-W.; Kim, S.-K.; Lee, J.-W. Pilot-scale production of carboxymethylcellulase from rice hull by Bacillus amyloliquefaciens DL-3. Biotechnol. BioProcess Eng. 2008, 13, 182–188. [Google Scholar] [CrossRef]
- Waghmare, P.; Kshirsagar, S.; Saratale, R.; Govindwar, S.; Saratale, G. Production and characterization of cellulolytic enzymes by isolated Klebsiella sp. PRW-1 using agricultural waste biomass. Emir. J. Food Agric. 2014, 26, 44–59. [Google Scholar] [CrossRef]
- Zehra, M.; Syed, M.N.; Sohail, M. Banana peels: A promising substrate for the coproduction of pectinase and xylanase from Aspergillus fumigatus MS16. Pol. J. Microbiol. 2020, 69, 19–26. [Google Scholar] [CrossRef]
- Shah, M.P.; Reddy, G.; Banerjee, R.; Babu, P.R.; Kothari, I. Microbial degradation of banana waste under solid state bioprocessing using two lignocellulolytic fungi (Phylosticta spp. MPS-001 and Aspergillus spp. MPS-002). Process Biochem. 2005, 40, 445–451. [Google Scholar] [CrossRef]
- Olajuyigbe, F.M.; Nlekerem, C.M.; Ogunyewo, O.A. Production and characterization of highly thermostable β-glucosidase during the biodegradation of methyl cellulose by Fusarium oxysporum. Biochem. Res. Int. 2016, 2016, 2016. [Google Scholar] [CrossRef]
- Alam, M.Z.; Muyibi, S.A.; Wahid, R. Statistical optimization of process conditions for cellulase production by liquid state bioconversion of domestic wastewater sludge. Bioresour. Technol. 2008, 99, 4709–4716. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Bai, F.; Zhao, X. Overproduction of cellulase by Trichoderma reesei RUT C30 through batch-feeding of synthesized low-cost sugar mixture. Bioresour. Technol. 2016, 216, 503–510. [Google Scholar] [CrossRef]
- Annamalai, N.; Rajeswari, M.V.; Elayaraja, S.; Balasubramanian, T. Thermostable, haloalkaline cellulase from Bacillus halodurans CAS 1 by conversion of lignocellulosic wastes. Carbohydr. Polym. 2013, 94, 409–415. [Google Scholar] [CrossRef]
- Jimenez, M.; Gonzalez, A.; Martinez, M.; Martinez, A.; Dale, B. Screening of yeasts isolated from decayed wood for lignocellulose-degrading enzyme activities. Mycol. Res. 1991, 95, 1299–1302. [Google Scholar] [CrossRef]
- Oikawa, T.; Tsukagawa, Y.; Soda, K. Endo-β-glucanase secreted by a psychrotrophic yeast: Purification and characterization. Biosci. Biotechnol. Biochem. 1998, 62, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fu, X. Industrial technologies for bioethanol production from lignocellulosic biomass. Renew. Sustain. Energy Rev. 2016, 57, 468–478. [Google Scholar] [CrossRef]
- Shariq, M.; Sohail, M. Production of cellulase and xylanase from Candida tropicalis (MK-118) on purified and crude substrates. Pak. J. Bot. 2020, 52, 323–328. [Google Scholar] [CrossRef]
- Rong, Y.; Zhang, L.; Chi, Z.; Wang, X. A carboxymethyl cellulase from a marine yeast (Aureobasidium pullulans 98): Its purification, characterization, gene cloning and carboxymethyl cellulose digestion. J. Ocean Univ. China 2015, 14, 913–921. [Google Scholar] [CrossRef]
- Kudanga, T.; Mwenje, E. Extracellular cellulase production by tropical isolates of Aureobasidium pullulans. Can. J. Microbiol. 2005, 51, 773–776. [Google Scholar] [CrossRef]
- Barzkar, N.; Sohail, M. An overview on marine cellulolytic enzymes and their potential applications. Appl. Microbiol. Biotechnol. 2020, 104, 6873–6892. [Google Scholar] [CrossRef]
- Barzkar, N.; Sohail, M.; Jahromi, S.T.; Nahavandi, R.; Khodadadi, M. Marine microbial L-glutaminase: From pharmaceutical to food industry. Appl. Microbiol. Biotechnol. 2021, 105, 4453–4466. [Google Scholar] [CrossRef]
- Barzkar, N. Marine microbial alkaline protease: An efficient and essential tool for various industrial applications. Int. J. Biol. Macromol. 2020, 161, 1216–1229. [Google Scholar] [CrossRef]
- Vieira, M.M.; Kadoguchi, E.; Segato, F.; da Silva, S.S.; Chandel, A.K. Production of cellulases by Aureobasidium pullulans LB83: Optimization, characterization, and hydrolytic potential for the production of cellulosic sugars. Prep. Biochem. Biotechnol. 2021, 51, 153–163. [Google Scholar] [CrossRef]
- Amadi, O.C.; Egong, E.J.; Nwagu, T.N.; Okpala, G.; Onwosi, C.O.; Chukwu, G.C.; Okolo, B.N.; Agu, R.C.; Moneke, A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon 2020, 6, e04566. [Google Scholar] [CrossRef]
- Kanti, A. Carboxymethyl cellulose hydrolyzing yeast isolated from South East Sulawesi, Indonesia. J. Biol. Indones. 2015, 11, 285–294. [Google Scholar]
- Kim, J.-Y.; Jung, H.-Y.; Park, J.-S.; Cho, S.-J.; Lee, H.B.; Sung, G.-H.; Subramani, G.; Kim, M.K. Isolation and characterization of cellulolytic yeast belonging to Moesziomyces sp. from the gut of Grasshopper. Korean, J. Microbiol. 2019, 55, 234–241. [Google Scholar]
- Carvalho, J.K.; Panatta, A.A.S.; Silveira, M.A.D.; Tav, C.; Johann, S.; Rodrigues, M.L.F.; Martins, C.V.B. Yeasts isolated from a lotic continental environment in Brazil show potential to produce amylase, cellulase and protease. Biotechnol. Rep. 2021, 30, e00630. [Google Scholar] [CrossRef]
- Giese, E.C.; Dussán, K.J.; Pierozzi, M.; Chandel, A.K.; Pagnocca, F.C.; Da Silva, S.S. Cellulase production by Trichosporon laibachii. Orbital 2017, 9, 271–278. [Google Scholar] [CrossRef]
- Touijer, H.; Benchemsi, N.; Ettayebi, M.; Janati Idrissi, A.; Chaouni, B.; Bekkari, H. Thermostable Cellulases from the Yeast Trichosporon sp. Enzym. Res. 2019, 2019, 2790414. [Google Scholar] [CrossRef]
- Valliammai, M.; Gopal, N.; Anandham, R. Probing cellulolytic yeast from forest ecosystem for the saccharification of Napier fodder biomass. J. Environ. Biol. 2021, 42, 1152–1161. [Google Scholar] [CrossRef]
- Liu, Z.; Ho, S.-H.; Sasaki, K.; Den Haan, R.; Inokuma, K.; Ogino, C.; Van Zyl, W.H.; Hasunuma, T.; Kondo, A. Engineering of a novel cellulose-adherent cellulolytic Saccharomyces cerevisiae for cellulosic biofuel production. Sci. Rep. 2016, 6, 24550. [Google Scholar] [CrossRef]
- Vyas, S.; Chhabra, M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour. Technol. 2017, 223, 250–258. [Google Scholar] [CrossRef]
- Kanti, A.; Sukarno, N.; Sukara, E.; Darusman, L.K. Cellulolytic Yeast Isolated From Raja Ampat Indonesia. Sci. J. Life Sci. 2012, 16, 27–34. [Google Scholar]
- Thongekkaew, J.; Kongsanthia, J. Screening and identification of cellulase producing yeast from rongkho forest, ubon ratchathani university. Bioeng. Biosci 2016, 4, 29–33. [Google Scholar] [CrossRef]
- Kanti, A.; Sudiana, I.M. Cellulolytic Yeast Isolated From Soil Gunung Halimun National Park*[Khamir Selulotik Yang Diisolasi Dari Tanah Taman Nasional Gunung Halimun]. Ber. Biol. 2002, 6, 85–90. [Google Scholar]
- Carrasco, M.; Villarreal, P.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Screening and characterization of amylase and cellulase activities in psychrotolerant yeasts. BMC Microbiol. 2016, 16, 21–29. [Google Scholar] [CrossRef]
- Zhang, L.; Chi, Z. Screening and identification of a cellulase producing marine yeast and optimization of medium and cultivation conditions for cellulase production. J. Ocean Univ China 2007, 37, 101–108. [Google Scholar]
- Otero, D.; Cadaval, C.; Teixeira, L.; Rosa, C.; Sanzo, A.; Kalil, S. Screening of yeasts capable of producing cellulase-free xylanase. Afr. J. Biotechnol. 2015, 14, 1961–1969. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Gupta, R.; Khasa, Y.P.; Singh, A.; Zhang, Y.-H.P. Bioethanol production from pentose sugars: Current status and future prospects. Renew. Sustain. Energy Rev. 2011, 15, 4950–4962. [Google Scholar] [CrossRef]
- Bhat, M. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 2000, 18, 355–383. [Google Scholar] [CrossRef]
- Choudhary, J.; Singh, S.; Nain, L. Thermotolerant fermenting yeasts for simultaneous saccharification fermentation of lignocellulosic biomass. Electron. J. Biotechnol. 2016, 21, 82–92. [Google Scholar] [CrossRef]
- Anandharaj, M.; Lin, Y.-J.; Rani, R.P.; Nadendla, E.K.; Ho, M.-C.; Huang, C.-C.; Cheng, J.-F.; Chang, J.-J.; Li, W.-H. Constructing a yeast to express the largest cellulosome complex on the cell surface. Proc. Natl. Acad. Sci. USA 2020, 117, 2385–2394. [Google Scholar] [CrossRef]
- Khah, E.; Kakava, E.; Mavromatis, A.; Chachalis, D.; Goulas, C. Effect of grafting on growth and yield of tomato (Lycopersicon esculentum Mill.) in greenhouse and open-field. J. Appl. Hortic. 2006, 8, 3–7. [Google Scholar] [CrossRef]
- Oh, E.J.; Jin, Y.-S. Engineering of Saccharomyces cerevisiae for efficient fermentation of cellulose. FEMS Yeast Res. 2020, 20, foz089. [Google Scholar] [CrossRef] [PubMed]
- Schulze, E. Information regarding chemical composition of plant cell membrane. Ber. Dtsch. Chem. Ges. 1891, 24, 2277–2287. [Google Scholar] [CrossRef]
- Verma, D.; Satyanarayana, T. Molecular approaches for ameliorating microbial xylanases. Bioresour. Technol. 2012, 117, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Shallom, D.; Shoham, Y. Microbial hemicellulases. Curr. Opin. Microbiol. 2003, 6, 219–228. [Google Scholar] [CrossRef]
- Beg, Q.; Kapoor, M.; Mahajan, L.; Hoondal, G. Microbial xylanases and their industrial applications: A review. Appl. Microbiol. Biotechnol. 2001, 56, 326–338. [Google Scholar] [CrossRef]
- Wierzbicki, M.P.; Maloney, V.; Mizrachi, E.; Myburg, A.A. Xylan in the middle: Understanding xylan biosynthesis and its metabolic dependencies toward improving wood fiber for industrial processing. Front. Plant Sci. 2019, 10, 176. [Google Scholar] [CrossRef]
- Peralta, A.G.; Venkatachalam, S.; Stone, S.C.; Pattathil, S. Xylan epitope profiling: An enhanced approach to study organ development-dependent changes in xylan structure, biosynthesis, and deposition in plant cell walls. Biotechnol. Biofuels 2017, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Mert, M.J.; La Grange, D.C.; Rose, S.H.; van Zyl, W.H. Engineering of Saccharomyces cerevisiae to utilize xylan as a sole carbohydrate source by co-expression of an endoxylanase, xylosidase and a bacterial xylose isomerase. J. Ind. Microbiol. Biotechnol. 2016, 43, 431–440. [Google Scholar] [CrossRef]
- Pinto, P.; Evtuguin, D.; Neto, C.P. Structure of hardwood glucuronoxylans: Modifications and impact on pulp retention during wood kraft pulping. Carbohydr. Polym. 2005, 60, 489–497. [Google Scholar] [CrossRef]
- Bajpai, P. Chapter 2-Xylan: Occurrence and structure. In Xylanolytic Enzym; Elsevier: Amsterdam, The Netherlands, 2014; pp. 9–18. [Google Scholar]
- Bouveng, H. Phenylisocyanate derivatives of carbohydrates. 2. Location of o-acetyl groups in birch xylan. Acta Chem. Scand. 1961, 15, 87–95. [Google Scholar] [CrossRef]
- Puls, J. Chemistry and biochemistry of hemicelluloses: Relationship between hemicellulose structure and enzymes required for hydrolysis. Macromol. Symp. 1997, 120, 183–196. [Google Scholar] [CrossRef]
- Sunna, A.; Antranikian, G. Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol. 1997, 17, 39–67. [Google Scholar] [CrossRef] [PubMed]
- Schendel, R.R.; Becker, A.; Tyl, C.E.; Bunzel, M. Isolation and characterization of feruloylated arabinoxylan oligosaccharides from the perennial cereal grain intermediate wheat grass (Thinopyrum intermedium). Carbohydr. Res. 2015, 407, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Juturu, V.; Wu, J.C. Microbial xylanases: Engineering, production and industrial applications. Biotechnol. Adv. 2012, 30, 1219–1227. [Google Scholar] [CrossRef]
- Joseleau, J.; Comtat, J.; Ruel, K. Chemical Structure of Xylans and Their Interaction in the Plant Cell Walls; Elsevier: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Hao, Z.; Mohnen, D. A review of xylan and lignin biosynthesis: Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 212–241. [Google Scholar] [CrossRef]
- Moreira, L. Insights into the mechanism of enzymatic hydrolysis of xylan. Appl. Microbiol. Biotechnol. 2016, 100, 5205–5214. [Google Scholar] [CrossRef]
- Chanda, S.; Hirst, E.; Jones, J.; Percival, E. 262. The constitution of xylan from esparto grass (stipa tenacissima, L.). J. Chem. Soc. (Resumed) 1950, 1289–1297. [Google Scholar] [CrossRef]
- Eda, S.; Ohnishi, A.; KatŌ, K. Xylan isolated from the stalk of Nicotiana tabacum. Agric. Biol. Chem. 1976, 40, 359–364. [Google Scholar]
- Montgomery, R.; Smith, F.; Srivastava, H. Structure of Corn Hull Hemicellulose. I. Partial Hydrolysis and Identification of 2-O-(α-D-Glucopyranosyluronic Acid)-D-xylopyranose1, 2. J. Am. Chem. Soc. 1956, 78, 2837–2839. [Google Scholar] [CrossRef]
- Dekker, R.F.; Richards, G.N. Hemicellulases: Their occurrence, purification, properties, and mode of action. Adv. Carbohydr. Chem. Biochem. 1976, 32, 277–352. [Google Scholar] [PubMed]
- Barry, V.C.; Dillon, T. Occurrence of xylans in marine algae. Nature 1940, 146, 620. [Google Scholar] [CrossRef]
- Hv, S.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Arfi, Y.; Shamshoum, M.; Rogachev, I.; Peleg, Y.; Bayer, E.A. Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc. Natl. Acad. Sci. USA 2014, 111, 9109–9114. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.N.; Schmitz, E.; Linares-Pastén, J.A.; Adlercreutz, P. Endo-xylanases as tools for production of substituted xylooligosaccharides with prebiotic properties. Appl. Microbiol. Biotechnol. 2018, 102, 9081–9088. [Google Scholar] [CrossRef] [PubMed]
- Mendis, M.; Simsek, S. Production of structurally diverse wheat arabinoxylan hydrolyzates using combinations of xylanase and arabinofuranosidase. Carbohydr. Polym. 2015, 132, 452–459. [Google Scholar] [CrossRef]
- Biely, P.; Malovíková, A.; Hirsch, J.; Krogh, K.M.; Ebringerová, A. The role of the glucuronoxylan carboxyl groups in the action of endoxylanases of three glycoside hydrolase families: A study with two substrate mutants. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 2246–2255. [Google Scholar] [CrossRef]
- Malgas, S.; Mafa, M.S.; Mkabayi, L.; Pletschke, B.I. A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J. Microbiol. Biotechnol. 2019, 35, 187. [Google Scholar] [CrossRef]
- Valenzuela, S.V.; Lopez, S.; Biely, P.; Sanz-Aparicio, J.; Pastor, F.J. The glycoside hydrolase family 8 reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 is active on branched xylooligosaccharides. Appl. Environ. Microbiol. 2016, 82, 5116–5124. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, L.; Jiang, Z.; Yang, S.; Zhu, H.; Li, L. A xylose-tolerant β-xylosidase from Paecilomyces thermophila: Characterization and its co-action with the endogenous xylanase. Bioresour. Technol. 2008, 99, 5402–5410. [Google Scholar] [CrossRef]
- Huy, N.D.; Le Nguyen, C.; Seo, J.-W.; Kim, D.-H.; Park, S.-M. Putative endoglucanase PcGH5 from Phanerochaete chrysosporium is a β-xylosidase that cleaves xylans in synergistic action with endo-xylanase. J. Biosci. Bioeng. 2015, 119, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Lagaert, S.; Pollet, A.; Courtin, C.M.; Volckaert, G. β-Xylosidases and α-l-arabinofuranosidases: Accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014, 32, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.; Emami, K.; Fontes, C.M.; Ferreira, L.M.; Humphry, D.R.; Gilbert, H.J. The membrane-bound α-glucuronidase from Pseudomonas cellulosa hydrolyzes 4-O-methyl-d-glucuronoxylooligosaccharides but not 4-O-methyl-d-glucuronoxylan. J. Bacteriol. 2002, 184, 4925–4929. [Google Scholar] [CrossRef]
- Nurizzo, D.; Nagy, T.; Gilbert, H.J.; Davies, G.J. The structural basis for catalysis and specificity of the Pseudomonas cellulosa α-glucuronidase, GlcA67A. Structure 2002, 10, 547–556. [Google Scholar] [CrossRef]
- Golan, G.; Shallom, D.; Teplitsky, A.; Zaide, G.; Shulami, S.; Baasov, T.; Stojanoff, V.; Thompson, A.; Shoham, Y.; Shoham, G. Crystal structures of Geobacillus stearothermophilus α-glucuronidase complexed with its substrate and products: Mechanistic implications. J. Biol. Chem. 2004, 279, 3014–3024. [Google Scholar] [CrossRef] [PubMed]
- Tenkanen, M.; Siika-aho, M. An α-glucuronidase of Schizophyllum commune acting on polymeric xylan. J. Biotechnol. 2000, 78, 149–161. [Google Scholar] [CrossRef]
- McKee, L.S.; Sunner, H.; Anasontzis, G.E.; Toriz, G.; Gatenholm, P.; Bulone, V.; Vilaplana, F.; Olsson, L. A GH115 α-glucuronidase from Schizophyllum commune contributes to the synergistic enzymatic deconstruction of softwood glucuronoarabinoxylan. Biotechnol. Biofuels 2016, 9, 2. [Google Scholar] [CrossRef]
- Arnling Bååth, J.; Giummarella, N.; Klaubauf, S.; Lawoko, M.; Olsson, L. A glucuronoyl esterase from Acremonium alcalophilum cleaves native lignin-carbohydrate ester bonds. FEBS Lett. 2016, 590, 2611–2618. [Google Scholar] [CrossRef]
- Driss, D.; Bhiri, F.; Elleuch, L.; Bouly, N.; Stals, I.; Miled, N.; Blibech, M.; Ghorbel, R.; Chaabouni, S.E. Purification and properties of an extracellular acidophilic endo-1, 4-β-xylanase, naturally deleted in the “thumb”, from Penicillium occitanis Pol6. Process Biochem. 2011, 46, 1299–1306. [Google Scholar] [CrossRef]
- Walia, A.; Mehta, P.; Chauhan, A.; Shirkot, C.K. Optimization of cellulase-free xylanase production by alkalophilic Cellulosimicrobium sp. CKMX1 in solid-state fermentation of apple pomace using central composite design and response surface methodology. Ann. Microbiol. 2013, 63, 187–198. [Google Scholar] [CrossRef]
- Singh, S.; Madlala, A.M.; Prior, B.A. Thermomyces lanuginosus: Properties of strains and their hemicellulases. FEMS Microbiol. Rev. 2003, 27, 3–16. [Google Scholar] [CrossRef]
- Basu, M.; Kumar, V.; Shukla, P. Recombinant approaches for microbial xylanases: Recent advances and perspectives. Curr. Protein Pept. Sci. 2018, 19, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Chhabra, D.; Shukla, P. Xylanase production from Thermomyces lanuginosus VAPS-24 using low cost agro-industrial residues via hybrid optimization tools and its potential use for saccharification. Bioresour. Technol. 2017, 243, 1009–1019. [Google Scholar] [CrossRef]
- Kumar, V.; Marín-Navarro, J.; Shukla, P. Thermostable microbial xylanases for pulp and paper industries: Trends, applications and further perspectives. World J. Microbiol. Biotechnol. 2016, 32, 34. [Google Scholar] [CrossRef]
- Leggio, L.L.; Kalogiannis, S.; Bhat, M.; Pickersgill, R. High resolution structure and sequence of T. aurantiacus Xylanase I: Implications for the evolution of thermostability in family 10 xylanases and enzymes with βα-barrel architecture. Proteins Struct. Funct. Bioinform. 1999, 36, 295–306. [Google Scholar] [CrossRef]
- Zverlov, V.; Piotukh, K.; Dakhova, O.; Velikodvorskaya, G.; Borriss, R. The multidomain xylanase A of the hyperthermophilic bacterium Thermotoga neapolitana is extremely thermoresistant. Appl. Microbiol. Biotechnol. 1996, 45, 245–247. [Google Scholar] [CrossRef]
- Abou-Hachem, M.; Olsson, F.; Karlsson, E.N. Probing the stability of the modular family 10 xylanase from Rhodothermus marinus. Extremophiles 2003, 7, 483–491. [Google Scholar] [CrossRef]
- Lüthi, E.; Jasmat, N.B.; Bergquist, P.L. Xylanase from the extremely thermophilic bacterium” Caldocellum saccharolyticum”: Overexpression of the gene in Escherichia coli and characterization of the gene product. Appl. Environ. Microbiol. 1990, 56, 2677–2683. [Google Scholar] [CrossRef]
- Khasin, A.; Alchanati, I.; Shoham, Y. Purification and characterization of a thermostable xylanase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 1993, 59, 1725–1730. [Google Scholar] [CrossRef]
- Winterhalter, C.; Heinrich, P.; Candussio, A.; Wich, G.; Liebl, W. Identification of a novel cellulose-binding domain the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 1995, 15, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Simpson, H.D.; Haufler, U.R.; Daniel, R.M. An extremely thermostable xylanase from the thermophilic eubacterium Thermotoga. Biochem. J. 1991, 277, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Amel, B.-D.; Nawel, B.; Khelifa, B.; Mohammed, G.; Manon, J.; Salima, K.-G.; Farida, N.; Hocine, H.; Bernard, O.; Jean-Luc, C. Characterization of a purified thermostable xylanase from Caldicoprobacter algeriensis sp. nov. strain TH7C1T. Carbohydr. Res. 2016, 419, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Turunen, O.; Jänis, J.; Leisola, M.; Rouvinen, J. Three-dimensional structures of thermophilic β-1, 4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa: Comparison of twelve xylanases in relation to their thermal stability. Eur. J. Biochem. 2003, 270, 1399–1412. [Google Scholar] [CrossRef]
- Fan, G.; Yang, S.; Yan, Q.; Guo, Y.; Li, Y.; Jiang, Z. Characterization of a highly thermostable glycoside hydrolase family 10 xylanase from Malbranchea cinnamomea. Int. J. Biol. Macromol. 2014, 70, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.R.; Eswaramoorthy, S.; Vithayathil, P.J.; Viswamitra, M. The tertiary structure at 1.59 Å resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti Bainier. J. Mol. Biol. 2000, 295, 581–593. [Google Scholar] [CrossRef]
- Shrivastava, S.; Kumar, V.; Baweja, M.; Shukla, P. Enhanced xylanase production from Thermomyces lanuginosus NCIM 1374/DSM 28966 using statistical analysis. J. Pure Appl. Microbiol. 2016, 10, 2225–2231. [Google Scholar]
- Andrade, C.M.; Pereira Jr, N.; Antranikian, G. Extremely thermophilic microorganisms and their polymer-hidrolytic enzymes. Rev. Microbiol. 1999, 30, 287–298. [Google Scholar] [CrossRef]
- Cady, S.G.; Bauer, M.W.; Callen, W.; Snead, M.A.; Mathur, E.J.; Short, J.M.; Kelly, R.M. β-Endoglucanase from Pyrococcus furiosus. Methods Enzymol. 2001, 330, 346–354. [Google Scholar]
- Cannio, R.; Di Prizito, N.; Rossi, M.; Morana, A. A xylan-degrading strain of Sulfolobus solfataricus: Isolation and characterization of the xylanase activity. Extremophiles 2004, 8, 117–124. [Google Scholar] [CrossRef]
- Walia, A.; Guleria, S.; Mehta, P.; Chauhan, A.; Parkash, J. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech. 2017, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Lara, C.A.; Santos, R.O.; Cadete, R.M.; Ferreira, C.; Marques, S.; Girio, F.; Oliveira, E.S.; Rosa, C.A.; Fonseca, C. Identification and characterisation of xylanolytic yeasts isolated from decaying wood and sugarcane bagasse in Brazil. Antonie Van Leeuwenhoek 2014, 105, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.; Motta, F.; Andrade, C.; Rodrigues, M.; Maugeri-Filho, F. Thermo-stable xylanases from non conventional yeasts. J. Microbiol. Biochem. Technol 2011, 3, 36–42. [Google Scholar]
- Özcan, S.; Kötter, P.; Ciciary, M. Xylan-hydrolysing enzymes of the yeast Pichia stipitis. Appl. Microbiol. Biotechnol. 1991, 36, 190–195. [Google Scholar] [CrossRef]
- Gomes, F.C.; Safar, S.V.; Marques, A.R.; Medeiros, A.O.; Santos, A.R.O.; Carvalho, C.; Lachance, M.-A.; Sampaio, J.P.; Rosa, C.A. The diversity and extracellular enzymatic activities of yeasts isolated from water tanks of Vriesea minarum, an endangered bromeliad species in Brazil, and the description of Occultifur brasiliensis fa, sp. nov. Antonie Van Leeuwenhoek 2015, 107, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Shariq, M.; Sohail, M. Application of Candida tropicalis MK-160 for the production of xylanase and ethanol. J. King Saud Univ. Sci. 2019, 31, 1189–1194. [Google Scholar] [CrossRef]
- Handel, S.; Wang, T.; Yurkov, A.M.; König, H. Sugiyamaella mastotermitis sp. nov. and Papiliotrema odontotermitis fa, sp. nov. from the gut of the termites Mastotermes darwiniensis and Odontotermes obesus. Int. J. Syst. Evol. Microbiol. 2016, 66, 4600–4608. [Google Scholar] [CrossRef]
- Petrescu, I.; Lamotte-Brasseur, J.; Chessa, J.-P.; Ntarima, P.; Claeyssens, M.; Devreese, B.; Marino, G.; Gerday, C. Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles 2000, 4, 137–144. [Google Scholar] [CrossRef]
- Gomes, J.; Gomes, I.; Steiner, W. Thermolabile xylanase of the Antarctic yeast Cryptococcus adeliae: Production and properties. Extremophiles 2000, 4, 227–235. [Google Scholar] [CrossRef]
- Bankeeree, W.; Lotrakul, P.; Prasongsuk, S.; Chaiareekij, S.; Eveleigh, D.E.; Kim, S.W.; Punnapayak, H. Effect of polyols on thermostability of xylanase from a tropical isolate of Aureobasidium pullulans and its application in prebleaching of rice straw pulp. SpringerPlus 2014, 3, 37. [Google Scholar] [CrossRef]
- Morais, C.G.; Sena, L.M.; Lopes, M.R.; Santos, A.R.O.; Barros, K.O.; Alves, C.R.; Uetanabaro, A.P.T.; Lachance, M.-A.; Rosa, C.A. Production of ethanol and xylanolytic enzymes by yeasts inhabiting rotting wood isolated in sugarcane bagasse hydrolysate. Fungal Biol. 2020, 124, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Otero, D.M.; Braga, A.R.C.; Kalil, S.J. Diversification of nitrogen sources as a tool to improve endo-xylanase enzyme activity produced by Cryptococcus laurentii. Biocatal. Agric. Biotechnol. 2021, 32, 101941. [Google Scholar] [CrossRef]
- Tiwari, S.; Avchar, R.; Arora, R.; Lanjekar, V.; Dhakephalkar, P.K.; Dagar, S.S.; Baghela, A. Xylanolytic and ethanologenic potential of gut associated yeasts from different species of termites from India. Mycobiology 2020, 48, 501–511. [Google Scholar] [CrossRef]
- Duarte, A.; Dayo-Owoyemi, I.; Nobre, F.; Pagnocca, F.; Chaud, L.; Pessoa, A.; Felipe, M.; Sette, L. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Wu, J.; Xie, R.; Zhou, F.; Sun, J.; Huang, M. Screening and characterizing of xylanolytic and xylose-fermenting yeasts isolated from the wood-feeding termite, Reticulitermes chinensis. PLoS ONE 2017, 12, e0181141. [Google Scholar] [CrossRef] [PubMed]
- Morais, C.G.; Cadete, R.M.; Uetanabaro, A.P.T.; Rosa, L.H.; Lachance, M.-A.; Rosa, C.A. D-xylose-fermenting and xylanase-producing yeast species from rotting wood of two Atlantic Rainforest habitats in Brazil. Fungal Genet. Biol. 2013, 60, 19–28. [Google Scholar] [CrossRef]
- Salah, H.A.; Temerk, H.A.; Salah, N.A.; Alshehri, S.R.Z.; Al-Harbi, J.A.; Mawad, A.M.; Khaled, K.A.; Hesham, A.E.-L.; Amein, K.A. Production and Optimization of Xylanase and [alpha]-Amylase from Non-Saccharomyces Yeasts (Pichia membranifaciens). J. Pure Appl. Microbiol. 2021, 15, 452–462. [Google Scholar] [CrossRef]
- Ding, C.; Li, M.; Hu, Y. High-activity production of xylanase by Pichia stipitis: Purification, characterization, kinetic evaluation and xylooligosaccharides production. Int. J. Biol. Macromol. 2018, 117, 72–77. [Google Scholar] [CrossRef]
- Bankeeree, W.; Akada, R.; Lotrakul, P.; Punnapayak, H.; Prasongsuk, S. Enzymatic hydrolysis of black liquor xylan by a novel xylose-tolerant, thermostable β-xylosidase from a tropical strain of Aureobasidium pullulans CBS 135684. Appl. Biochem. Biotechnol. 2018, 184, 919–934. [Google Scholar] [CrossRef]
- Sena, L.M.; Morais, C.G.; Lopes, M.R.; Santos, R.O.; Uetanabaro, A.P.; Morais, P.B.; Vital, M.J.; de Morais, M.A.; Lachance, M.-A.; Rosa, C.A. d-Xylose fermentation, xylitol production and xylanase activities by seven new species of Sugiyamaella. Antonie Van Leeuwenhoek 2017, 110, 53–67. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Faria, N.T.; Marques, S.; Ferreira, F.C.; Fonseca, C. Production of xylanolytic enzymes by Moesziomyces spp. using xylose, xylan and brewery’s spent grain as substrates. New Biotechnol. 2019, 49, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Sreenath, H.K.; Shah, A.B.; Yang, V.W.; Gharia, M.M.; Jeffries, T.W. Enzymatic polishing of jute/cotton blended fabrics. J. Ferment. Bioeng. 1996, 81, 18–20. [Google Scholar] [CrossRef]
- Ahmad, Z.; Butt, M.S.; Hussain, R.; Ahmed, A.; Riaz, M. Effect of oral application of xylanase on some hematological and serum biochemical parameters in broilers. Pak. Vet. J. 2013, 33, 388–390. [Google Scholar]
- Patel, S.; Savanth, V. Review on fungal xylanases and their applications. Int. J. 2015, 3, 311–315. [Google Scholar]
- Ali, U.F.; Ibrahim, Z.M.; Isaac, G.S. Ethanol and xylitol production from xylanase broth of Thermomyces lanuginosus grown on some lignocellulosic wastes using Candida tropicalis EMCC2. Life Sci. J. 2013, 10, 968–978. [Google Scholar]
- Viikari, L.; Tenkanen, M.; Poutanen, K. Hemicellulases, Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation; Drew, S.W., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1999. [Google Scholar]
- Viikari, L.; Alapuranen, M.; Puranen, T.; Vehmaanperä, J.; Siika-Aho, M. Thermostable enzymes in lignocellulose hydrolysis. Biofuels 2007, 108, 121–145. [Google Scholar]
- Ohta, K.; Fujimoto, H.; Fujii, S.; Wakiyama, M. Cell-associated β-xylosidase from Aureobasidium pullulans ATCC 20524: Purification, properties, and characterization of the encoding gene. J. Biosci. Bioeng. 2010, 110, 152–157. [Google Scholar] [CrossRef]
- Manitchotpisit, P.; Leathers, T.D.; Peterson, S.W.; Kurtzman, C.P.; Li, X.-L.; Eveleigh, D.E.; Lotrakul, P.; Prasongsuk, S.; Dunlap, C.A.; Vermillion, K.E. Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aureobasidium pullulans. Mycol. Res. 2009, 113, 1107–1120. [Google Scholar] [CrossRef]
- Cui, L.; Du, G.; Zhang, D.; Chen, J. Thermal stability and conformational changes of transglutaminase from a newly isolated Streptomyces hygroscopicus. Bioresour. Technol. 2008, 99, 3794–3800. [Google Scholar] [CrossRef]
- George, S.P.; Ahmad, A.; Rao, M.B. A novel thermostable xylanase from Thermomonospora sp.: Influence of additives on thermostability. Bioresour. Technol. 2001, 78, 221–224. [Google Scholar] [CrossRef]
- Cadete, R.M.; Melo, M.A.; Dussan, K.J.; Rodrigues, R.C.; Silva, S.S.; Zilli, J.E.; Vital, M.J.; Gomes, F.C.; Lachance, M.-A.; Rosa, C.A. Diversity and physiological characterization of D-xylose-fermenting yeasts isolated from the Brazilian Amazonian Forest. PLoS ONE 2012, 7, e43135. [Google Scholar] [CrossRef] [PubMed]
- Cadete, R.M.; Rosa, C.A. The yeasts of the genus Spathaspora: Potential candidates for second-generation biofuel production. Yeast 2018, 35, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, L.V.; de Barros Grassi, M.C.; Gallardo, J.C.M.; Pirolla, R.A.S.; Calderón, L.L.; de Carvalho-Netto, O.V.; Parreiras, L.S.; Camargo, E.L.O.; Drezza, A.L.; Missawa, S.K. Second-generation ethanol: The need is becoming a reality. Ind. Biotechnol. 2016, 12, 40–57. [Google Scholar] [CrossRef]
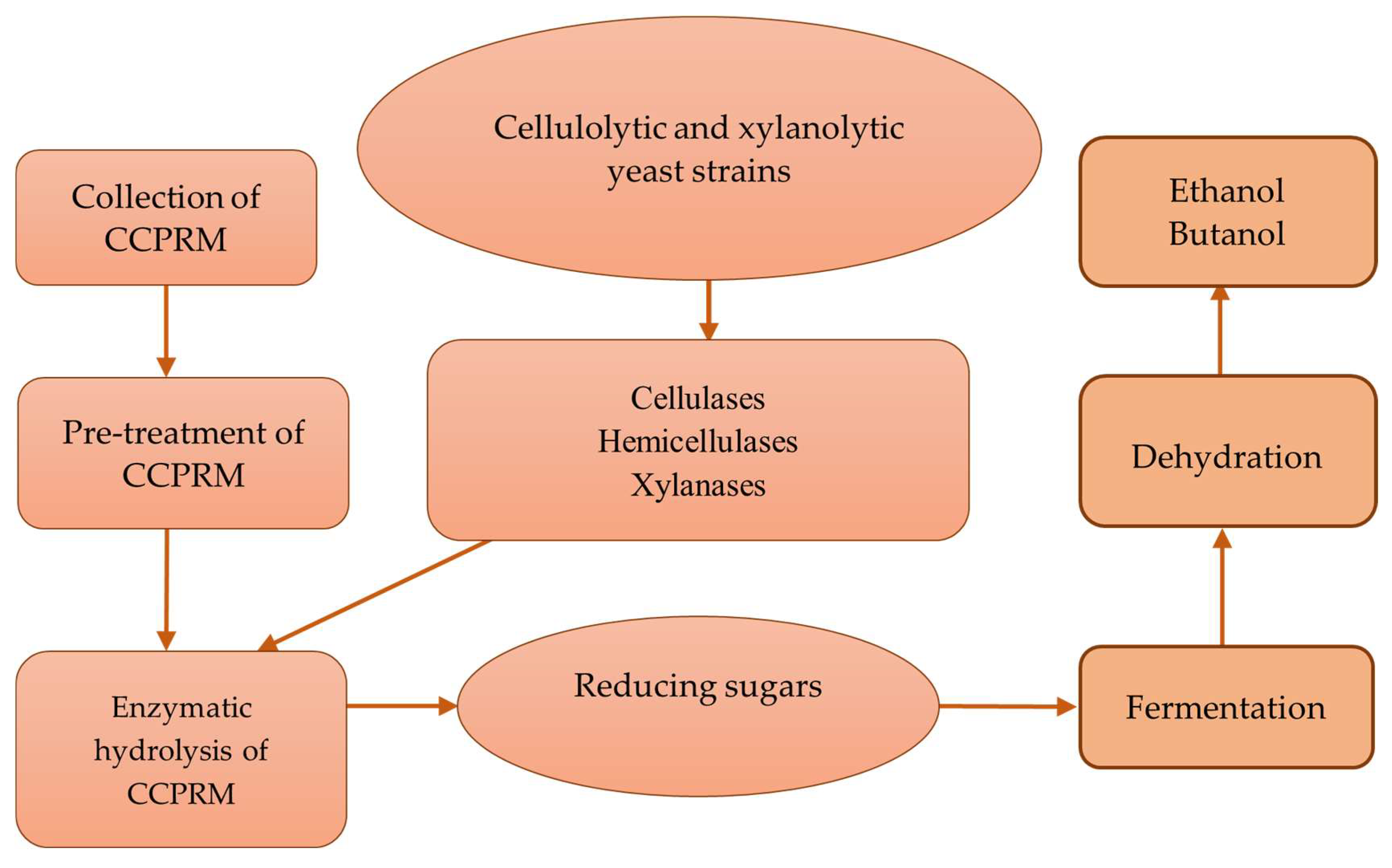
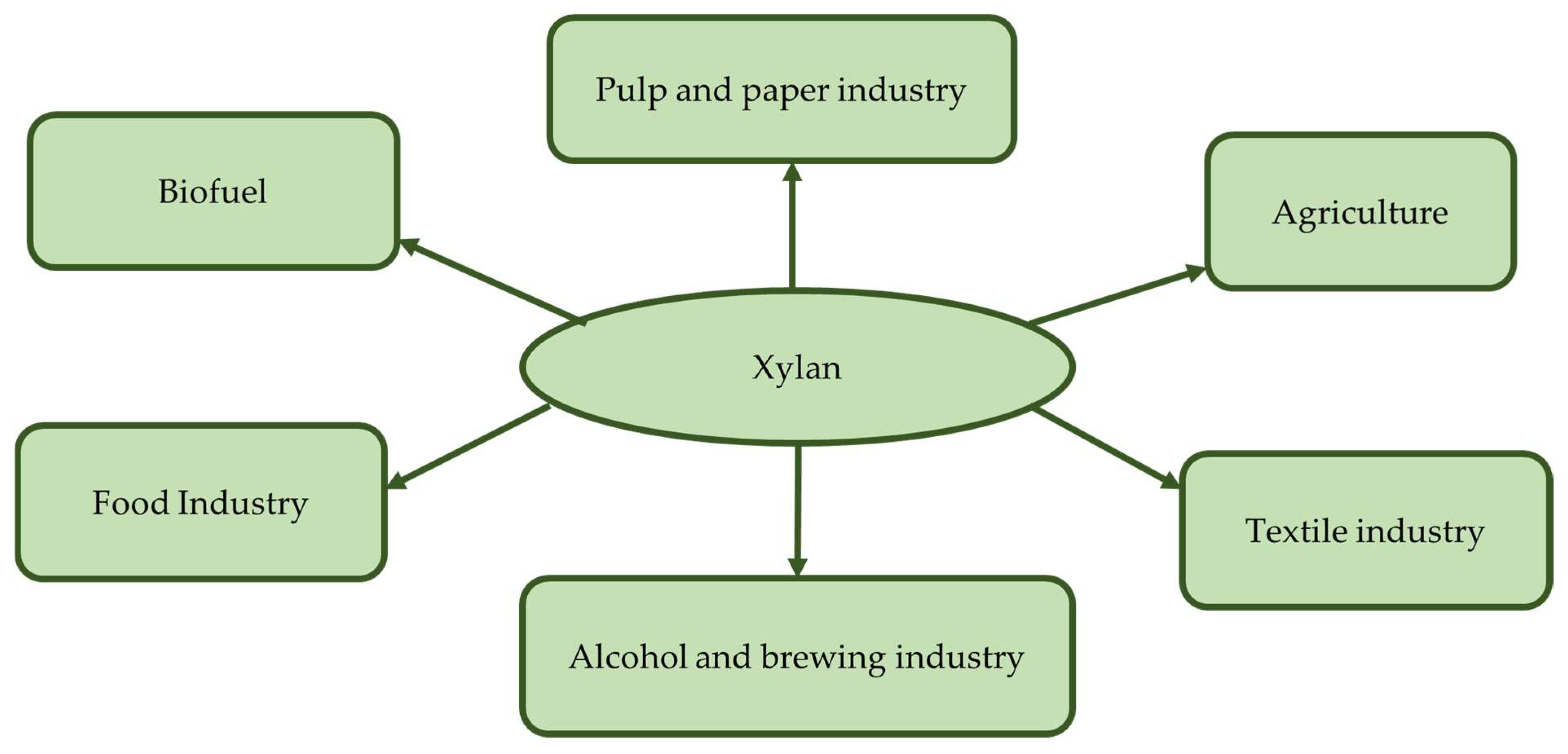
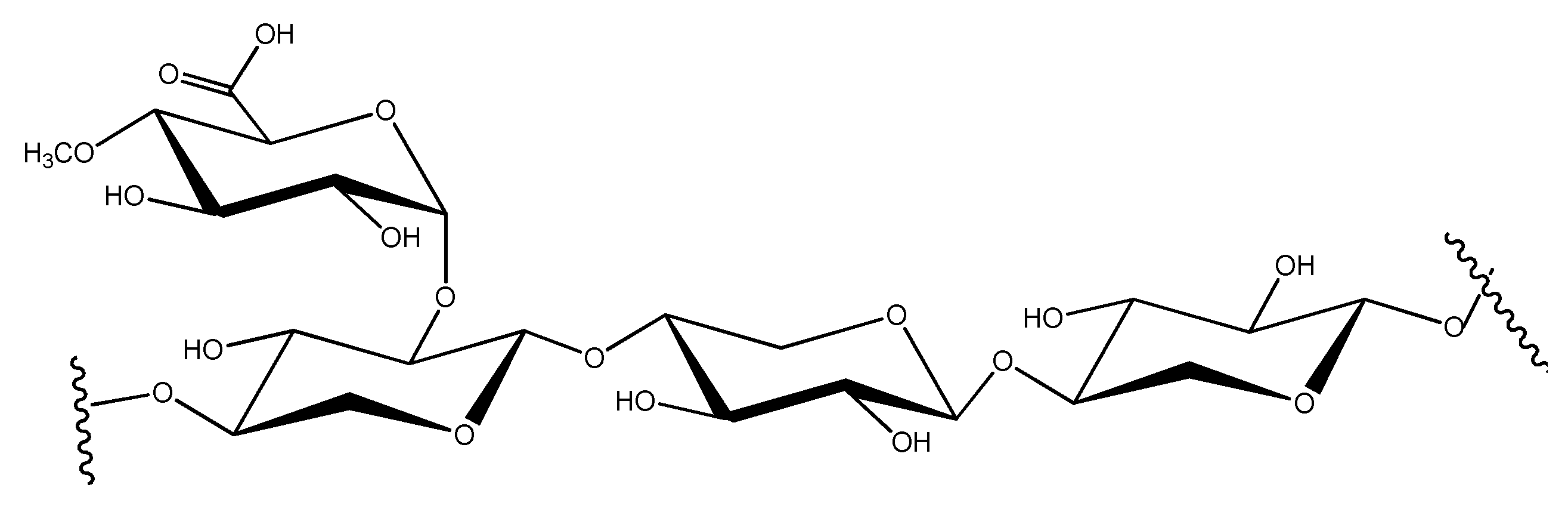
| Indicator, % | Types of Cellulose | Source | ||
|---|---|---|---|---|
| SC | CC | HC | ||
| α-cellulose | 99.0 | 98.9 | 95.4 | [20] |
| residual lignin | 0.51 | -- | 0.17 | [20] |
| pentosans | 0.17 | -- | -- | [25] |
| ash | 0.13 | 0.13 | 0.16 | [26] |
| Origin | Source | Isolated from | Strains | Reference |
|---|---|---|---|---|
| Terrestrial | Assiut region, Egypt | - | Saccharomyces cerevisiae | [10] |
| Mushroom farm in Yala Local Government Area of Cross River State, Nigeria | Soil samples | Saccharomyces cerevisiae SCPW 17 | [85] | |
| Brazil | Decaying wood and sugarcane bagasse | Meyerozyma guilliermondii, Scheffersomyces shehatae, Sugiyamaella smithiae | [168] | |
| Two Atlantic Rainforest habitats in Brazil | Environmental and food samples including garden soil, plant parts, grapes, lemon, green chili and orange juice | Candida tropicalis MK-160 | [172] | |
| Icepack at the Antarctic station Dumont d’Urville (60°409 S; 40°019 E) | Decayed algae | Cryptococcus adeliae | [174] | |
| Four Atlantic Rainforest sites in Brazil | Samples of rotting wood | Sugiyamaella xylanicola and Saitozyma podzolica, Apioctrichum sporotrichoides, Aureobasidium pullulans | [177] | |
| Different cities in southern Rio Grande do Sul | Chicory | Cryptococcus laurentii | [178] | |
| Agharkar Research Institute, Pune, Maharashtra (18.5207451° N, 73.8315643° E), and Singalandhapuram, Namakkal District, Tamil Nadu (11.420428° N, 78.220487° E) | Wood-feeding termites | Pseudozyma hubeiensis, Hannaella pagnoccae, Papiliotrema mangalensis, Kodamaea ohmeri TS21 | [179] | |
| Rotting wood trees at Huazhong Agricultural University, Wuhan, China | Wood-feeding termite Reticulitermes chinensis | Barnettozyma californica, Candida sp., Cyberlindnera sp., Sterigmatomyces halophilus, Sugiyamaella smithiae, Vanrija humicola and Wickerhamomyces sp. | [181] | |
| Two Atlantic Rainforest habitats in Brazil | Rotting wood | Spencermartinsiella sp. 1, Sugiyamaella xylanicola, Tremella sp. | [182] | |
| Assiut region, Egypt | Soil samples | Pichia membranifaciens | [183] | |
| - | Soil samples | Pichia stipitis | [184] | |
| Thailand | Leaves, painted wall surfaces and wood surfaces | Aureobasidium pullulans CBS 135684 | [185] | |
| Brazil | Rotting sugarcane bagasse and wood samples | Sugiyamaella sp. | [186] | |
| Marine | Antarctic | Sea squirt | Candidadavisiana | [180] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohail, M.; Barzkar, N.; Michaud, P.; Tamadoni Jahromi, S.; Babich, O.; Sukhikh, S.; Das, R.; Nahavandi, R. Cellulolytic and Xylanolytic Enzymes from Yeasts: Properties and Industrial Applications. Molecules 2022, 27, 3783. https://doi.org/10.3390/molecules27123783
Sohail M, Barzkar N, Michaud P, Tamadoni Jahromi S, Babich O, Sukhikh S, Das R, Nahavandi R. Cellulolytic and Xylanolytic Enzymes from Yeasts: Properties and Industrial Applications. Molecules. 2022; 27(12):3783. https://doi.org/10.3390/molecules27123783
Chicago/Turabian StyleSohail, Muhammad, Noora Barzkar, Philippe Michaud, Saeid Tamadoni Jahromi, Olga Babich, Stanislav Sukhikh, Rakesh Das, and Reza Nahavandi. 2022. "Cellulolytic and Xylanolytic Enzymes from Yeasts: Properties and Industrial Applications" Molecules 27, no. 12: 3783. https://doi.org/10.3390/molecules27123783







