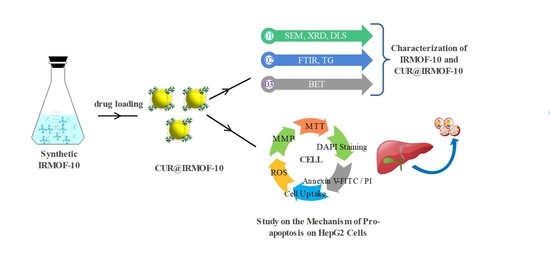
Preparation, Characterization, and In Vitro Release of Curcumin-Loaded IRMOF-10 Nanoparticles and Investigation of Their Pro-Apoptotic Effects on Human Hepatoma HepG2 Cells
Abstract
:1. Introduction
2. Results
2.1. Synthesis and Characterization
2.2. Drug Loading
2.3. In Vitro Release
2.4. In Vitro Study on the Safety of IRMOF-10
2.5. CUR@IRMOF-10 Induces Apoptosis of HepG2 Cells
2.5.1. Apoptosis
2.5.2. Cellular Uptake
2.5.3. ROS
2.5.4. Mitochondrial Membrane Potential
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Preparation of IRMOF-10 and Drug-Loading
4.2.1. Preparation of IRMOF-10
4.2.2. Drug Loading
4.3. In Vitro Release Study
4.4. Characterization
4.5. Cell Cultures and Treatments
4.6. Cell Viability Assay
4.7. Nuclear Morphology Assay Using DAPI Staining
4.8. Analysis of Apoptosis by Flow Cytometry
4.9. CLSM for Cell Uptake
4.10. Intracellular ROS Detection
4.11. Mitochondrial Membrane Potential (MMP)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Li, S.; Zhang, L.; Zhao, H.; Chen, Y. Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis. Open Life Sci. 2021, 16, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Shafei, L.; Mohamed Ibrahim, M.I.; Billa, N. Is Curcumin at the Threshold of Therapeutic Effectiveness on Patients with Colon Cancer?—A Systematic. Review. Front. Pharmacol. 2021, 12, 707231. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, M.; Sahebkar, A.; Askari, G.; Johnston, T.P.; Alikiaii, B.; Bagherniya, M. The clinical use of curcumin on neu-rological disorders: An updated systematic review of clinical trials. Phytother. Res. 2021, 35, 6862–6882. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Qin, L.; Pang, L.; Ma, B.; Bai, J.; Liu, J. Amino-functionalized Zn metal organic frameworks as antitumor drug curcumin carriers. New J. Chem. 2020, 44, 17693–17704. [Google Scholar] [CrossRef]
- Zhao, G.; Shi, Y.; Gong, C.; Liu, T.; Nan, W.; Ma, L.; Wu, Z.; Da, C.; Zhou, K.; Zhang, H. Curcumin Exerts Antino-ciceptive Effects in Cancer-Induced Bone Pain via an Endogenous Opioid Mechanism. Front. Neurosci. 2021, 15, 696861. [Google Scholar] [CrossRef]
- Wong, K.E.; Ngai, S.C.; Chan, K.G.; Lee, L.H.; Goh, B.H.; Chuah, L.H. Curcumin Nanoformulations for Colorectal Cancer: A Review. Front. Pharmacol. 2019, 10, 152. [Google Scholar] [CrossRef]
- Balogh, J.; Victor, D.; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Xian, L.; Ghobrial, R.M.; Monsour, H. Hepatocellular carcinoma: A review. J. Hepatocell Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Xie, P.; Liu, R.; Huang, H.; Qin, X.; Huang, R.; Zhao, F. Research Progress of Curcumin Intervention in Hepatocellular Carcinoma Cells. Sci. Consult. (Technol. Manag.) 2021, 10, 60–62. [Google Scholar]
- Tønnesena, H.H.; Loftsson, T. Studies of curcumin and curcuminoids. XXVII. Cyclodextrin complexation: Solubility, chemical and photochemical stability. Int. J. Pharm. 2002, 244, 127–135. [Google Scholar] [CrossRef]
- Paul, S.; Sa, G. Curcumin as an Adjuvant to Cancer Immunotherapy. Front. Oncol. 2021, 11, 675923. [Google Scholar] [CrossRef]
- Roth Stefaniak, K.; Epley, C.C.; Novak, J.J.; McAndrew, M.L.; Cornell, H.D.; Zhu, J.; McDaniel, D.K.; Davis, J.L.; Allen, I.C.; Morris, A.J.; et al. Photo-triggered release of 5-fluorouracil from a MOF drug delivery vehicle. Chem. Commun. (Camb.) 2018, 54, 7617–7620. [Google Scholar] [CrossRef]
- Dolgopolova, E.A.; Rice, A.M.; Martin, C.R.; Shustova, N.B. Photochemistry and photophysics of MOFs: Steps towards MOF-based sensing enhancements. Chem Soc. Rev. 2018, 47, 4710–4728. [Google Scholar] [CrossRef]
- Zou, K.Y.; Li, Z.X. Controllable Syntheses of MOF-Derived Materials. Chemistry 2018, 24, 6506–6518. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Wu, Y.; Guan, C.; Cheetham, A.K.; Wang, J. MOF-derived nanohybrids for electrocatalysis and energy storage: Current status and perspectives. Chem. Commun. (Camb.) 2018, 54, 5268–5288. [Google Scholar] [CrossRef]
- Eddaoudi, M.; Kim, J.; Rosi, N.; Vodak, D.; Wachter, J.; O’Keeffe, M.; Yaghi, O.M. Systematic Design of Pore Size and Functionality in Isoreticular MOFs and Their Application in Methane Storage. Science 2002, 25, 469–472. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Liu, D. Structure—Activity Relationship and Design of Metal-Organic Framework Materials; Science Press: Beijing, China, 2014; pp. 5–8. [Google Scholar]
- Rao, D.; Lu, R.; Xiao, C.; Kan, E.; Deng, K. Lithium-doped MOF impregnated with lithium-coated fullerenes: A hydrogen storage route for high gravimetric and volumetric uptakes at ambient temperatures. Chem. Commun. (Camb.) 2011, 47, 7698–7700. [Google Scholar] [CrossRef]
- Hicks, J.M.; Desgranges, C.; Delhommelle, J. Characterization and Comparison of the Performance of IRMOF-1, IRMOF-8, and IRMOF-10 for CO2 Adsorption in the Subcritical and Supercritical Regimes. J. Phys. Chem. C 2012, 116, 22938–22946. [Google Scholar] [CrossRef]
- Borycz, J.; Tiana, D.; Haldoupis, E.; Sung, J.C.; Farha, O.K.; Siepmann, J.I.; Gagliardi, L. CO2 Adsorption in M-IRMOF-10 (M = Mg, Ca, Fe, Cu, Zn, Ge, Sr, Cd, Sn, Ba). J. Phys. Chem. C 2016, 120, 12819–12830. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, K.; Wu, Y.; Xi, H. New functionalized IRMOF-10 with strong affinity for Methanol: A simulation study. Appl. Surf. Sci. 2018, 440, 351–358. [Google Scholar] [CrossRef]
- Lee, J.S.; Kapustin, E.A.; Pei, X.; Llopis, S.; Yaghi, O.M.; Toste, F.D. Architectural Stabilization of a Gold(III) Catalyst in Metal-Organic Frameworks. Chem 2020, 6, 142–152. [Google Scholar] [CrossRef]
- Li, X.Y.; Guan, Q.X.; Shang, Y.Z.; Wang, Y.H.; Lv, S.W.; Yang, Z.X.; Wang, R.; Feng, Y.F.; Li, W.N.; Li, Y.J. Metal-organic framework IRMOFs coated with a temperature-sensitive gel delivering norcantharidin to treat liver cancer. World J. Gastroenterol. 2021, 27, 4208–4220. [Google Scholar] [CrossRef]
- Chen, G.; Luo, J.; Cai, M.; Qin, L.; Wang, Y.; Gao, L.; Huang, P.; Yu, Y.; Ding, Y.; Dong, X.; et al. Investigation of Metal-Organic Framework-5 (MOF-5) as an Antitumor Drug Oridonin Sustained Release Carrier. Molecules 2019, 24, 3369. [Google Scholar] [CrossRef] [Green Version]
- Kotzabasaki, M.; Galdadas, I.; Tylianakis, E.; Klontzas, E.; Cournia, Z.; Froudakis, G.E. Multiscale simulations reveal IRMOF-74-III as a potent drug carrier for gemcitabine delivery. J. Mater. Chem. B 2017, 5, 3277–3282. [Google Scholar] [CrossRef]
- Chen, Z.; Xia, Y.; Liao, S.; Huang, Y.; Li, Y.; He, Y.; Tong, Z.; Li, B. Thermal degradation kinetics study of curcumin with nonlinear methods. Food Chem. 2014, 155, 81–86. [Google Scholar] [CrossRef]
- Sun, X.Z.; Williams, G.R.; Hou, X.X.; Zhu, L.M. Electrospun curcumin-loaded fibers with potential biomedical applications. Carbohydr. Polym. 2013, 94, 147–153. [Google Scholar] [CrossRef]
- Jiao, Y.; Wilkinson, J.; Pletsch, E.C.; Buss, J.L.; Wang, W.; Planalp, R.; Torti, F.M.; Torti, S.V. Iron chelation in the biological activity of curcumin. Free Radic. Biol. Med. 2006, 40, 1152–1160. [Google Scholar] [CrossRef]
- Yang, C.G.; Ma, X.Y.; Wang, Z.H.; Zeng, X.; Hu, Z.Q.; Ye, Z.Q.; Shen, G.X. Curcumin induces apoptosis and protective autophagy in castration-resistant prostate cancer cells through iron chelation. Drug Des. Dev. Ther. 2017, 11, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Chen, C.M.; Shi, H.F.; Yang, M.Y.; Liu, Y.; Ji, P.; Chen, H.J.; Tan, R.X.; Li, E.G. Curcumin is a biologically active copper chelator with antitumor activity. Phytomedicine 2016, 23, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsini, K.I. The Chemistry of Curcumin: From Extraction to Therapeutic Agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philip, L.; Ritger, N.; Peppas, A. A simple equation for description of solute release I. Fickian and non-fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs. J. Control. Release 1987, 5, 23–36. [Google Scholar]
- Hartojo, W.; Silvers, A.L.; Thomas, D.G.; Seder, C.W.; Lin, L.; Rao, H.; Wang, Z.; Greenson, J.K.; Giordano, T.J.; Orringer, M.B.; et al. Curcumin promotes apoptosis, increases chemosensitivity, and inhibits nuclear factor κB in esophageal adenocarcinoma. Transl. Oncol. 2010, 3, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Pan, H.; Li, L.; Yu, Y.; Liu, B. An insight into the in vivo imaging potential of curcumin analogues as fluorescence probes. Asian J. Pharm. Sci. 2021, 16, 419–431. [Google Scholar] [CrossRef]
- Priyadarsini, K.I. Photophysics, photochemistry and photobiology of curcumin:Studies from organic solutions, bio-mimetics and living cells. J. Photochem. Photobiol. C Photochem. Rev. 2009, 10, 81–95. [Google Scholar] [CrossRef]
- Green, D.R.; Llambi, F. Cell Death Signaling. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–24. [Google Scholar] [CrossRef]
- You, L.; Yang, C.; Du, Y.; Liu, Y.; Chen, G.; Sai, N.; Dong, X.; Yin, X.; Ni, J. Matrine Exerts Hepatotoxic Effects via the ROS Dependent Mitochondrial Apoptosis Pathway and Inhibition of Nrf2-Mediated Antioxidant Response. Oxidative Med. Cell. Longev. 2019, 2019, 1045345. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Xia, Y. A dual emission metal-organic framework for rapid ratiometric fluorescence detection of CO32− in seawater. RSC Adv. 2020, 10, 24764–24771. [Google Scholar] [CrossRef]
- Chung, J.Y.; Liao, C.W.; Chang, Y.W.; Chang, B.K.; Wang, H.; Li, J.; Wang, C.Y. Influence of Metal–Organic Framework Porosity on Hydrogen Generation from Nanoconfined Ammonia Borane. J. Phys. Chem. C 2017, 121, 27369–27378. [Google Scholar] [CrossRef]
- Ooms, K.J.; Wasylishen, R.E. 129Xe NMR study of xenon in iso-reticular metal–organic frameworks. Microporous Mesoporous Mater. 2007, 103, 341–351. [Google Scholar]
- Gutiérrez, I.; Díaz, E.; Ordóñez, S. Consequences of cavity size and palladium addition on the selective hydrogen adsorption in isoreticular metal-organic frameworks. Thermochim. Acta 2013, 567, 79–84. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, R.; Sun, D.; Sun, X.; Geng, Z.; Liu, H.; Wang, S.L. Intracellular Uptake of Curcumin-Loaded Solid Lipid Nano-particles Exhibit Anti-Inflammatory Activities Superior to Those of Curcumin Through the NF-κB Signaling Pathway. J. Biomed. Nanotechnol. 2015, 11, 403–415. [Google Scholar] [CrossRef]
- Yu, Q.; Meng, Z.; Liu, Y.; Li, Z.; Sun, X.; Zhao, Z. Photocuring Hyaluronic Acid/Silk Fibroin Hydrogel Containing Curcumin Loaded CHITOSAN Nanoparticles for the Treatment of MG-63 Cells and ME3T3-E1 Cells. Polymers 2021, 13, 2302. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, W.; Sadeghi-Soureh, S.; Amirsaadat, S.; Pourpirali, R.; Alijani, S. Dual drug release mechanisms through mesoporous silica nanoparticle/electrospun nanofiber for enhanced anticancer efficiency of curcumin. J. Biomed. Mater. Res. A 2022, 110, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Liang, J.; Zhang, M.; Li, Z.; Wang, Z.; Dang, B.; Feng, N. Mucosal transfer of wheat germ agglutinin modified li-pid-polymer hybrid nanoparticles for oral delivery of oridonin. Nanomedicine 2017, 13, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Zhang, Z.; Wu, H.; Jia, X.; Wang, Y. Optimization and evaluation of Oridonin-loaded Soluplus((R))-Pluronic P105 mixed micelles for oral administration. Int. J. Pharm. 2017, 518, 193–202. [Google Scholar] [CrossRef]
- Ding, Y.; Ding, C.; Ye, N.; Liu, Z.; Wold, E.A.; Chen, H.; Wild, C.; Shen, Q.; Zhou, J. Discovery and development of natural product oridonin-inspired anticancer agents. Eur. J. Med. Chem. 2016, 122, 102–117. [Google Scholar] [CrossRef] [Green Version]
- Xing, Z.H.; Wei, J.H.; Cheang, T.Y.; Wang, Z.R.; Zhou, X.; Wang, S.S.; Chen, W.; Wang, S.M.; Luo, J.H.; Xu, A.W. Bifunctional pH-sensitive Zn(ii)-curcumin nanoparticles/siRNA effectively inhibit growth of human bladder cancer cells in vitro and in vivo. J. Mater. Chem. B 2014, 2, 2714–2724. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, T.; Wang, L.; Yang, H.; Zhang, S.; Zhou, P. Interaction of curcumin with Zn(II) and Cu(II) ions based on experiment and theoretical calculation. J. Mol. Struct. 2010, 984, 316–325. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Jameei, A.; Karande, A.A.; Chakravarty, A.R. BODIPY-attached zinc(II) complexes of curcumin drug for visible light assisted photo-sensitization, cellular imaging and targeted PDT. Eur. J. Med. Chem. 2021, 220, 113438. [Google Scholar] [CrossRef]
- Laha, D.; Pal, K.; Ray Chowdhuri, A.; Parida, P.K.; Sahu, S.K.; Jana, K.; Karmakar, P. Fabrication of curcumin-loaded folic acid-tagged metal organic framework for triple negative breast cancer therapy in in vitro and in vivo systems. New J. Chem. 2019, 43, 217–229. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.; Li, Y.; Li, N.; Shi, X.; Ding, H.; Zhang, Y.; Li, X.; Liu, G.; Wang, Z. Non-esterified fatty acids activate the ROS-p38-p53/Nrf2 signaling pathway to induce bovine hepatocyte apoptosis in vitro. Apoptosis 2014, 19, 984–997. [Google Scholar] [CrossRef]
- Zeng, Y.; Du, Q.; Zhang, Z.; Ma, J.; Han, L.; Wang, Y.; Yang, L.; Tao, N.; Qin, Z. Curcumin promotes cancer-associated fibroblasts apoptosis via ROS-mediated endoplasmic reticulum stress. Arch. Biochem. Biophys. 2020, 694, 108613. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, J.; Yuan, L.; Xiao, C.; Wang, Y.; Liu, X. Chicoric acid induces apoptosis in 3T3-L1 preadipocytes through ROS-mediated PI3K/Akt and MAPK signaling pathways. J. Agric. Food Chem. 2013, 61, 1509–1520. [Google Scholar] [CrossRef]
- Zhang, Q.; Du, Z.; Zhang, Y.; Zheng, Z.; Li, Q.; Wang, K. Apoptosis induction activity of polysaccharide from Lentinus edodes in H22-bearing mice through ROS-mediated mitochondrial pathway and inhibition of tubulin polymerization. Food Nutr. Res. 2020, 64, 1554. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, X.; Guo, T.; Wang, H.; Fan, H.; Fang, L. Study of Cinobufagin as a Promising Anticancer Agent in Uveal Melanoma Through Intrinsic Apoptosis Pathway. Front. Oncol. 2020, 10, 325. [Google Scholar] [CrossRef]
- Chung, Y.M.; Bae, Y.S.; Lee, S.Y. Molecular ordering of ROS production, mitochondrial changes, and caspase activation during sodium salicylate-induced apoptosis. Free Radic. Biol. Med. 2003, 34, 434–442. [Google Scholar] [CrossRef]
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef]
- Merino, D.; Kelly, G.L.; Lessene, G.; Wei, A.H.; Roberts, A.W.; Strasser, A. BH3-Mimetic Drugs: Blazing the Trail for New Cancer Medicines. Cancer Cell 2018, 34, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.W.; Davids, M.S.; Pagel, J.M.; Kahl, B.S.; Puvvada, S.D.; Gerecitano, J.F.; Kipps, T.J.; Anderson, M.A.; Brown, J.R.; Gressick, L.; et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2016, 374, 311–322. [Google Scholar] [CrossRef]
- Yue, R.; Hu, H.; Yiu, K.H.; Luo, T.; Zhou, Z.; Xu, L.; Zhang, S.; Li, K.; Yu, Z. Lycopene protects against hypoxia/reoxygenation-induced apoptosis by preventing mitochondrial dysfunction in primary neonatal mouse cardiomyocytes. PLoS ONE 2012, 7, e50778. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Morselli, E.; Kepp, O.; Kroemer, G. Targeting post-mitochondrial effectors of apoptosis for neuroprotection. Biochim. Biophys. Acta 2009, 1787, 402–413. [Google Scholar] [CrossRef] [Green Version]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, D.; Hu, X.; Cai, M.; Wang, K.; Peng, H.; Bai, J.; Xv, Y.; Fu, T.; Dong, X.; Ni, J.; et al. Preparation, Characterization, and In Vitro Release of Curcumin-Loaded IRMOF-10 Nanoparticles and Investigation of Their Pro-Apoptotic Effects on Human Hepatoma HepG2 Cells. Molecules 2022, 27, 3940. https://doi.org/10.3390/molecules27123940
Yin D, Hu X, Cai M, Wang K, Peng H, Bai J, Xv Y, Fu T, Dong X, Ni J, et al. Preparation, Characterization, and In Vitro Release of Curcumin-Loaded IRMOF-10 Nanoparticles and Investigation of Their Pro-Apoptotic Effects on Human Hepatoma HepG2 Cells. Molecules. 2022; 27(12):3940. https://doi.org/10.3390/molecules27123940
Chicago/Turabian StyleYin, Dongge, Xueling Hu, Mengru Cai, Kaixin Wang, Hulinyue Peng, Jie Bai, Yvchen Xv, Tingting Fu, Xiaoxv Dong, Jian Ni, and et al. 2022. "Preparation, Characterization, and In Vitro Release of Curcumin-Loaded IRMOF-10 Nanoparticles and Investigation of Their Pro-Apoptotic Effects on Human Hepatoma HepG2 Cells" Molecules 27, no. 12: 3940. https://doi.org/10.3390/molecules27123940
APA StyleYin, D., Hu, X., Cai, M., Wang, K., Peng, H., Bai, J., Xv, Y., Fu, T., Dong, X., Ni, J., & Yin, X. (2022). Preparation, Characterization, and In Vitro Release of Curcumin-Loaded IRMOF-10 Nanoparticles and Investigation of Their Pro-Apoptotic Effects on Human Hepatoma HepG2 Cells. Molecules, 27(12), 3940. https://doi.org/10.3390/molecules27123940