Tree-Code Based Improvement of Computational Performance of the X-ray-Matter-Interaction Simulation Tool XMDYN
Abstract
:1. Introduction
2. Materials and Methods
2.1. XMDYN Code
- 1.
- Recombination (RE) Block. Within this block, all classical electron–ion pairs are analyzed in a search for configurations when an electron stays in the vicinity of an ion and satisfies the conditions for a recombination event to occur.
- 2.
- Secondary Ionization (SI) Block. Within this block, all classical electron–ion pairs are analyzed in a search for configurations when an electron stays in the vicinity of an atom or ion and satisfies the conditions for a secondary ionization event to occur.
- 3.
- Monte Carlo (MC) Block. Tracking of atomic processes, i.e., photoionization, Auger decay and fluorescence, depends on the probabilities of such events during a single time step. These probabilities are derived from atomic cross section and rates calculated by the ab initio XATOM code [18,22]. For each atom or ion, a random number is generated, which determines which event occurs during a single time step.
- 4.
- Molecular Dynamics (MD) Block. Within this block, all classical particles, i.e., the atoms, ions and classical electron particles are propagated in real space, during a single time step. XMDYN uses for the propagation the well-known velocity Verlet algorithm [26].
2.2. Computational Bottlenecks in XMDYN
- 1.
- MD Block: Evaluation of long-range Coulomb interactions between all charged particle pairs, both for force and potential calculations. In case of the most straightforward implementation (i.e., with two nested loops, both running over the number of charged particles in the sample, ), the computational time scales as, . This computational strategy is also called the ‘brute-force’ method.
- 2.
- SI block: In XMDYN, the occurrence rate of a secondary ionization event for a free electron and an atom/ion depends on the relative distance and velocities of these particles [18]. The decision regarding whether SI takes place or not requires their detailed analysis. In general, all electron–atom/ion pairs have to be analyzed within a time step. Therefore, in case of a straightforward (i.e., ‘brute-force’) implementation of secondary ionization, the computational cost, t, scales with the product of the number of free electrons and the number of atoms and ions: .
- 3.
- RE block: A decision whether an electron recombines with an ion is also based on their relative distance and the velocities of the electron–ion pair [18]. Therefore, in the brute-force implementation the computational cost scales with the product of the number of electrons and number of ions: , similarly as within the SI Block.
2.3. Incorporating the PEPC Tree-Based Coulomb Solver into XMDYN
2.4. Tree Algorithm Developed to Speed-Up Secondary Ionization Calculation
3. Results
3.1. Improved Coulomb Force Calculations
3.2. Improvement of Secondary Ionization Calculation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| XFEL | X-ray free-electron laser |
| SPI | Single-particle imaging |
| LCLS | Linac Coherent Light Source |
| SIMEX | Simulation of Experiments at Advanced Laser Light Sources |
| XMDYN | Molecular-dynamics- and Monte-Carlo-based code for modelling X-ray driven dynamics in complex systems |
| XATOM | Atomic structure calculation tool |
| RE | Recombination |
| SI | Secondary ionization |
| MC | Monte Carlo |
| MD | Molecular dynamics |
| PEPC | Pretty Efficient Parallel Coulomb-solver |
| FWHM | Full width at half maximum |
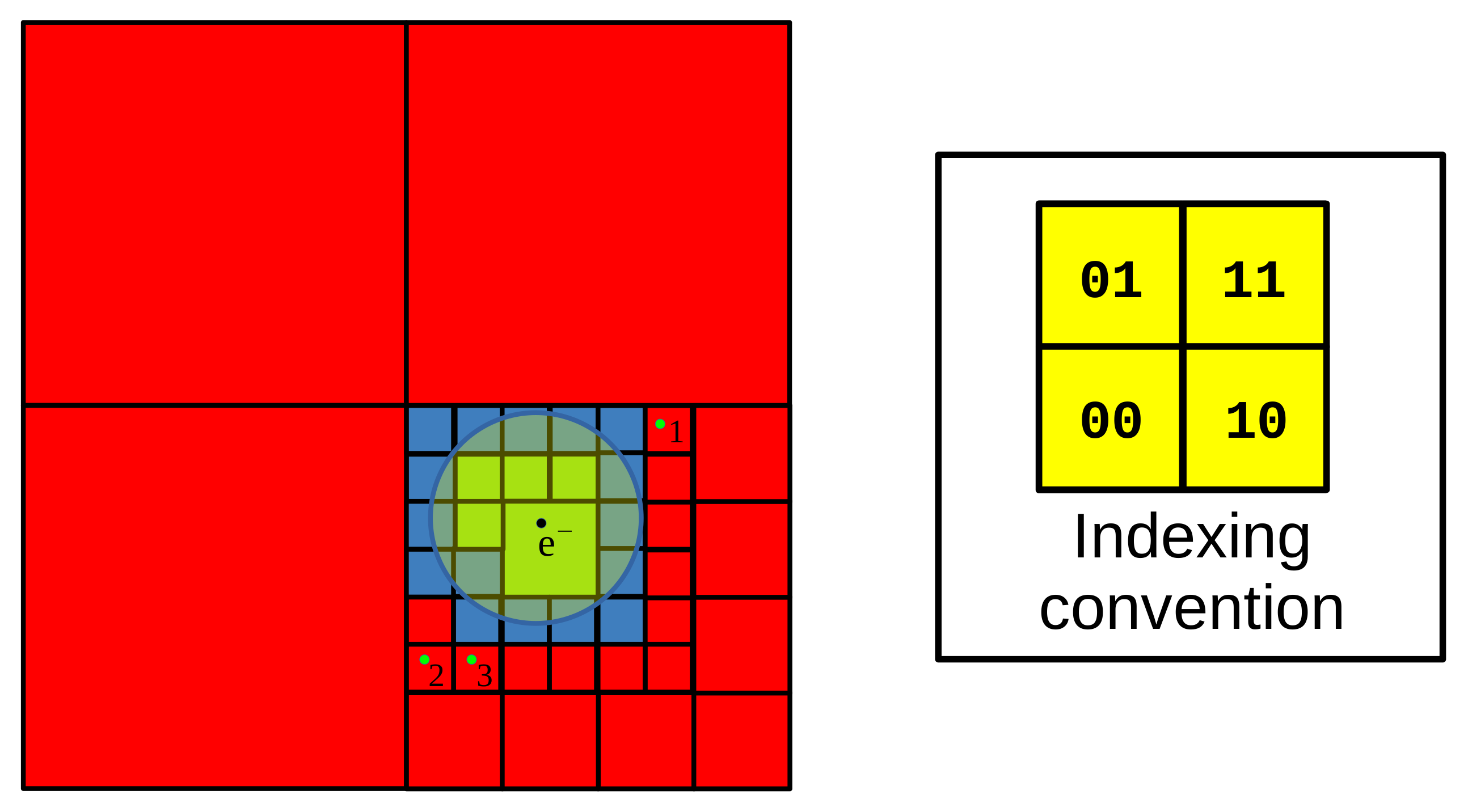
Appendix A
- (i)
- skip all boxes without intersection with the sphere (illustrated by red squares),
- (ii)
- select all atoms and ions from boxes contained fully by the sphere (green squares) as candidates,
- (iii)
- if a box has an intersection with the sphere but the sphere does not contain it entirely, then the box is subdivided and searched recursively. If this occurs after the final division (blue squares), the atoms and ions are searched one after another, in order to check if they lie within the sphere.
References
- Neutze, R.; Wouts, R.; van der Spoel, D.; Weckert, E.; Hajdu, J. Potential for Biomolecular Imaging with Femtosecond X-ray Pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Seibert, M.M.; Ekeberg, T.; Maia, F.R.N.C.; Svenda, M.; Andreasson, J.; Jönsson, O.; Odić, D.; Iwan, B.; Rocker, A.; Westphal, D.; et al. Single Mimivirus Particles Intercepted and Imaged with an X-ray Laser. Nature 2011, 470, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Ekeberg, T.; Svenda, M.; Abergel, C.; Maia, F.R.; Seltzer, V.; Claverie, J.M.; Hantke, M.; Jönsson, O.; Nettelblad, C.; Van Der Schot, G. Three-Dimensional Reconstruction of the Giant Mimivirus Particle with an X-ray Free-Electron Laser. Phys. Rev. Lett. 2015, 114, 098102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munke, A.; Andreasson, J.; Aquila, A.; Awel, S.; Ayyer, K.; Barty, A.; Bean, R.J.; Berntsen, P.; Bielecki, J.; Boutet, S.; et al. Coherent Diffraction of Single Rice Dwarf Virus Particles Using Hard X-rays at the Linac Coherent Light Source. Sci. Data 2016, 3, 160064. [Google Scholar] [CrossRef] [Green Version]
- Ayyer, K.; Lan, T.Y.; Elser, V.; Loh, N.D. Dragonfly: An Implementation of the Expand–Maximize–Compress Algorithm for Single-Particle Imaging. J. Appl. Cryst. 2016, 49, 1320–1335. [Google Scholar] [CrossRef] [Green Version]
- Loh, N.T.D.; Elser, V. Reconstruction Algorithm for Single-Particle Diffraction Imaging Experiments. Phys. Rev. E 2009, 80, 026705. [Google Scholar] [CrossRef] [Green Version]
- Fienup, J.R. Phase Retrieval Algorithms: A Comparison. Appl. Opt. 1982, 21, 2758–2769. [Google Scholar] [CrossRef] [Green Version]
- Fortmann-Grote, C.; Buzmakov, A.; Jurek, Z.; Loh, N.T.D.; Samoylova, L.; Santra, R.; Schneidmiller, E.A.; Tschentscher, T.; Yakubov, S.; Yoon, C.H.; et al. Start-to-End Simulation of Single-Particle Imaging Using Ultra-Short Pulses at the European X-ray Free-Electron Laser. IUCrJ 2017, 4, 560–568. [Google Scholar] [CrossRef] [Green Version]
- Bielecki, J.; Maia, F.R.N.C.; Mancuso, A.P. Perspectives on Single Particle Imaging with x Rays at the Advent of High Repetition Rate X-ray Free Electron Laser Sources. Struct. Dyn. 2020, 7, 040901. [Google Scholar] [CrossRef]
- Decking, W.; Abeghyan, S.; Abramian, P.; Abramsky, A.; Aguirre, A.; Albrecht, C.; Alou, P.; Altarelli, M.; Altmannet, P.; Amyan, K.; et al. A MHz-repetition-rate hard X-ray free-electron laser driven by a superconducting linear accelerator. Nat. Photonics 2020, 14, 391. [Google Scholar] [CrossRef]
- LCLS-II Project Team. LCLS-II Project Team. LCLS-II Final Design Report. In LCLSII-1.1-DR-0251-R0; SLAC: Menlo Park, CA, USA, 2015; Volume 14, p. 391. [Google Scholar]
- Zhu, Z.Y.; Zhao, Z.T.; Wang, D.; Liu, Z.; Li, R.X.; Yin, L.X.; Yang, Z.H. SCLF: An 8-GeV CW SCRF Linac-Based X-ray FEL Facility in Shanghai. In Proceedings of the FEL2017, Santa Fe, NM, USA, 20–25 August 2017; pp. 20–25. [Google Scholar]
- Rose, M.; Bobkov, S.; Ayyer, K.; Kurta, R.P.; Dzhigaev, D.; Kim, Y.Y.; Morgan, A.J.; Yoon, C.H.; Westphal, D.; Bielecki, J.; et al. Single-Particle Imaging without Symmetry Constraints at an X-ray Free-Electron Laser. IUCrJ 2018, 5, 727–736. [Google Scholar] [CrossRef] [Green Version]
- Sobolev, E.; Zolotarev, S.; Giewekemeyer, K.; Bielecki, J.; Okamoto, K.; Reddy, H.K.N.; Andreasson, J.; Ayyer, K.; Barak, I.; Bari, S.; et al. Megahertz single-particle imaging at the European XFEL. Communications Physics 2020, 3, 97. [Google Scholar] [CrossRef]
- Fortmann-Grote, C.; E, J.C. SimEx. 2020. Available online: https://github.com/PaNOSC-ViNYL/SimEx (accessed on 13 May 2022).
- Yoon, C.H.; Yurkov, M.V.; Schneidmiller, E.A.; Samoylova, L.; Buzmakov, A.; Jurek, Z.; Ziaja, B.; Santra, R.; Loh, N.D.; Tschentscher, T.; et al. A Comprehensive Simulation Framework for Imaging Single Particles and Biomolecules at the European X-ray Free-Electron Laser. Sci. Rep. 2016, 6, 24791. [Google Scholar] [CrossRef] [Green Version]
- E, J.; Stransky, M.; Jurek, Z.; Fortmann-Grote, C.; Juha, L.; Santra, R.; Ziaja, B.; Mancuso, A.P. Effects of radiation damage and inelastic scattering on single-particle imaging of hydrated proteins with an X-ray Free-Electron Laser. Sci. Rep. 2021, 11, 17976. [Google Scholar] [CrossRef]
- Jurek, Z.; Son, S.K.; Ziaja, B.; Santra, R. XMDYN and XATOM: Versatile Simulation Tools for Quantitative Modeling of X-ray Free-Electron Laser Induced Dynamics of Matter. J. Appl. Cryst. 2016, 49, 1048–1056. [Google Scholar] [CrossRef]
- Murphy, B.F.; Osipov, T.; Jurek, Z.; Fang, L.; Son, S.K.; Mucke, M.; Eland, J.H.D.; Zhaunerchyk, V.; Feifel, R.; Avaldi, L.; et al. Femtosecond X-ray-Induced Explosion of C 60 at Extreme Intensity. Nat. Commun. 2014, 5, 4281. [Google Scholar] [CrossRef] [Green Version]
- Tachibana, T.; Jurek, Z.; Fukuzawa, H.; Motomura, K.; Nagaya, K.; Wada, S.; Johnsson, P.; Siano, M.; Mondal, S.; Ito, Y.; et al. Nanoplasma Formation by High Intensity Hard X-rays. Sci. Rep. 2015, 5, 10977. [Google Scholar] [CrossRef] [Green Version]
- Gibbon, P. PEPC:Pretty Efficient Parallel Coulomb-Solver; Technical Report; Forschungszentrum Jülich GmbH Zentralinstitut für Angewandte Mathematik: Jülich, Germany, 2003; FZJ-ZAM-IB-2003-05. [Google Scholar]
- Son, S.K.; Young, L.; Santra, R. Impact of hollow-atom formation on coherent X-ray scattering at high intensity. Phys. Rev. A 2011, 83, 033402. [Google Scholar] [CrossRef] [Green Version]
- Son, S.-K.; Santra, R. Monte Carlo calculation of ion, electron, and photon spectra of xenon atoms in X-ray free-electron laser pulses. Phys. Rev. A 2012, 85, 063415. [Google Scholar] [CrossRef] [Green Version]
- Kumagai, Y.; Jurek, Z.; Xu, W.; Fukuzawa, H.; Motomura, K.; Iablonskyi, D.; Nagaya, K.; Wada, S.I.; Mondal, S.; Tachibana, T.; et al. Radiation-Induced Chemical Dynamics in Ar Clusters Exposed to Strong X-ray Pulses. Phys. Rev. Lett. 2018, 120, 223201. [Google Scholar] [CrossRef]
- Kumagai, Y.; Jurek, Z.; Xu, W.; Saxena, V.; Fukuzawa, H.; Motomura, K.; Iablonskyi, D.; Nagaya, K.; Wada, S.I.; Ito, Y.; et al. Suppression of thermal nanoplasma emission in clusters strongly ionized by hard X-rays. J. Phys. B 2021, 54, 044001. [Google Scholar] [CrossRef]
- Swope, W.C.; Andersen, H.C.; Berens, P.H.; Wilson, K.R. A computer simulation method for the calculation of equilibrium constants for the formation of physical clusters of molecules: Application to small water clusters. J. Chem. Phys. 1982, 76, 637–649. [Google Scholar] [CrossRef]
- Jurek, Z.; Santra, R.; Son, S.K.; Ziaja, B. XRAYPAC—A Software Package for Modeling X-ray-Induced Dynamics of Matter, v. 1.0.0; CFEL, DESY: Hamburg, Germany, 2016. [Google Scholar]
- Barnes, J.; Hut, P. A hierarchical O(N log N) force-calculation algorithm. Nature 1986, 324, 446. [Google Scholar] [CrossRef]
- Hamada, T.; Nitadori, K.; Benkrid, K.; Ohno, Y.; Morimoto, G.; Masada, T.; Shibata, Y.; Oguri, K.; Taiji, M. A novel multiple-walk parallel algorithm for the Barnes-Hut treecode on GPUs - towards cost effective, high performance N-body simulation. Comp. Sci. Res. Dev. 2009, 24, 21. [Google Scholar] [CrossRef]
- Bolten, M.; Fahrenberger, F.; Halver, R.; Heber, F.; Hofmann, M.; Kabadshow, I.; Lenz, O.; Pippig, M.; Sutmann, G. ScaFaCoS, C Subroutine Library. Available online: http://scafacos.github.com/ (accessed on 13 May 2022).
| Sample | Atoms & Ions | Ions | Electrons (Free) |
|---|---|---|---|
| 47 Å | 9722 | 2102 | 2806 |
| 60 Å | 20,534 | 4422 | 5881 |
| 75 Å | 40,358 | 8717 | 11,603 |
| 100 Å | 97,655 | 21,087 | 28,084 |
| 150 Å | 328,475 | 67,828 | 87,369 |
| Sample | Brute [s] | Tree [s] | Tree (4 Threads) [s] |
|---|---|---|---|
| 47 Å | 0.444 | 0.956 | 0.300 |
| 60 Å | 2.00 | 2.82 | 0.940 |
| 75 Å | 7.82 | 6.26 | 2.44 |
| 100 Å | 44.9 | 23.2 | 11.5 |
| 150 Å | 464.1 | 123.4 | 94.1 |
| Sample | Brute [s] | Tree [s] | Tree (4 Threads) [s] |
|---|---|---|---|
| 47 Å | 0.411 | 0.911 | 0.289 |
| 60 Å | 1.80 | 2.56 | 0.760 |
| 75 Å | 6.96 | 5.36 | 1.64 |
| 100 Å | 39.5 | 17.7 | 6.00 |
| 150 Å | 410.0 | 62.5 | 34.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stransky, M.; Jurek, Z.; Santra, R.; Mancuso, A.P.; Ziaja, B. Tree-Code Based Improvement of Computational Performance of the X-ray-Matter-Interaction Simulation Tool XMDYN. Molecules 2022, 27, 4206. https://doi.org/10.3390/molecules27134206
Stransky M, Jurek Z, Santra R, Mancuso AP, Ziaja B. Tree-Code Based Improvement of Computational Performance of the X-ray-Matter-Interaction Simulation Tool XMDYN. Molecules. 2022; 27(13):4206. https://doi.org/10.3390/molecules27134206
Chicago/Turabian StyleStransky, Michal, Zoltan Jurek, Robin Santra, Adrian P. Mancuso, and Beata Ziaja. 2022. "Tree-Code Based Improvement of Computational Performance of the X-ray-Matter-Interaction Simulation Tool XMDYN" Molecules 27, no. 13: 4206. https://doi.org/10.3390/molecules27134206






