A New Alternative Tool to Analyse Glycosylation in Monoclonal Antibodies Based on Drop-Coating Deposition Raman imaging: A Proof of Concept
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Proteins for the Spectral Characterization of Commercial mAbs Solutions
2.2. Chemicals, Reagents, and Proteins Used to Study the Specificity of SVR Models
2.3. Solution Preparation
2.3.1. Detection Limits
2.3.2. Removal of Excipients in mAbs Formulations
2.4. Raman Analysis
2.4.1. Raman Spectral Measurements
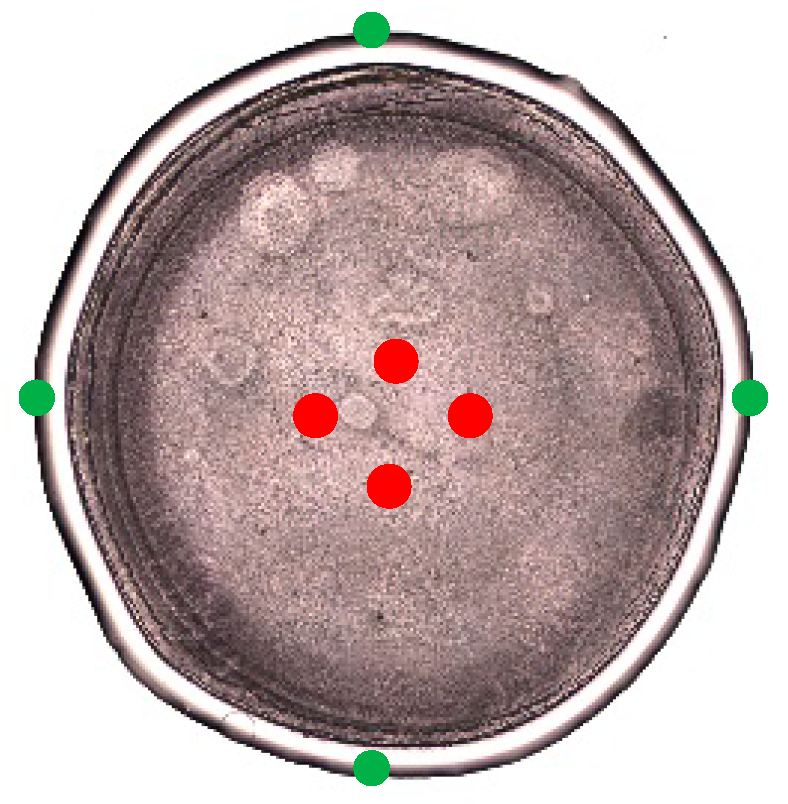
2.4.2. Raman Hyperspectral Imaging
2.5. Reference Analysis: LC-FLR-MS N-Glycans Characterization
2.5.1. Glycoworks RapiFluor-MS N-Glycan
2.5.2. UPLC-FLR-MS Analysis
2.6. Multivariate Data Analysis
2.6.1. Data Preprocessing
2.6.2. Multivariate Curve Resolution Alternating Least Squares (MCR-ALS)
- -
- corresponds to the input original data.
- -
- corresponds to the residual calculated from the difference between the input original data and the MCR-ALS reproduction.
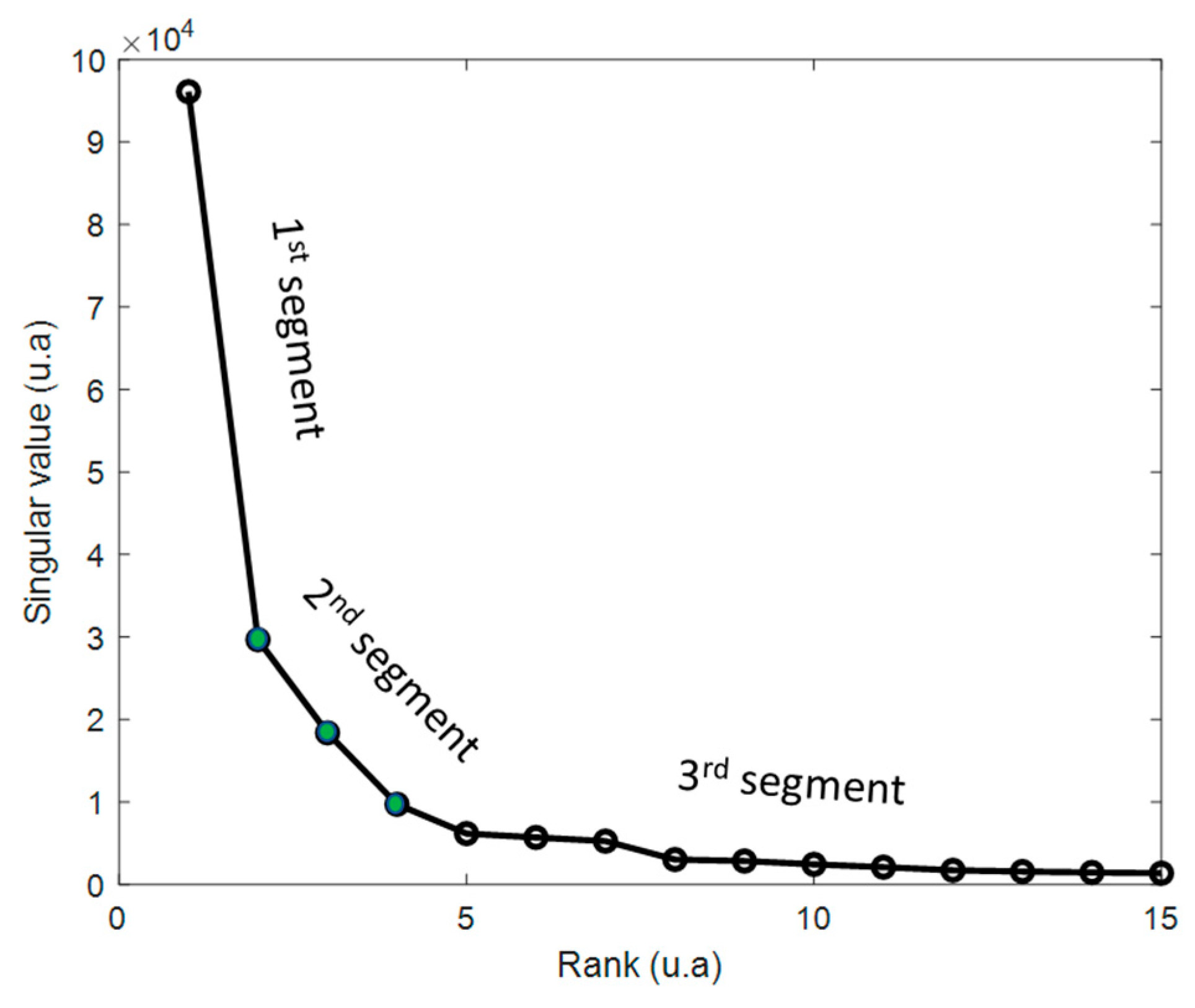
2.6.3. Singular Value Decomposition (SVD Truncation Strategy)
- -
- The first step of the SVD is a factorization of the data matrix aswhere:
- and are left and right singular vectors matrices, respectively.
- is the diagonal matrix of the singular values for with being the rank of the matrix .
- -
- The second step consists of plotting the singular values according to their value. Thus, the first, second, and third segment are identified. Only the second (between two breaks) is considered since it represents a good compromise between the relevant information carried by the segment on the left and the noise carried by the segment on the right. Each of this segment is considered as a rank and then as a threshold.
- -
- The last step consists of a reconstruction of the different truncated matrices (different ranks ) like . In this study, 3 truncated matrices were considered.
2.6.4. Performance Evaluation of the SVR Models
2.6.5. Interpretation of SVR Models
3. Results and Discussion
3.1. Spectral Characterization of Commercial mAbs Solutions after Excipients Removal
- Analysis Filtrate: concentrated mAbs
- Analysis filter: excipients
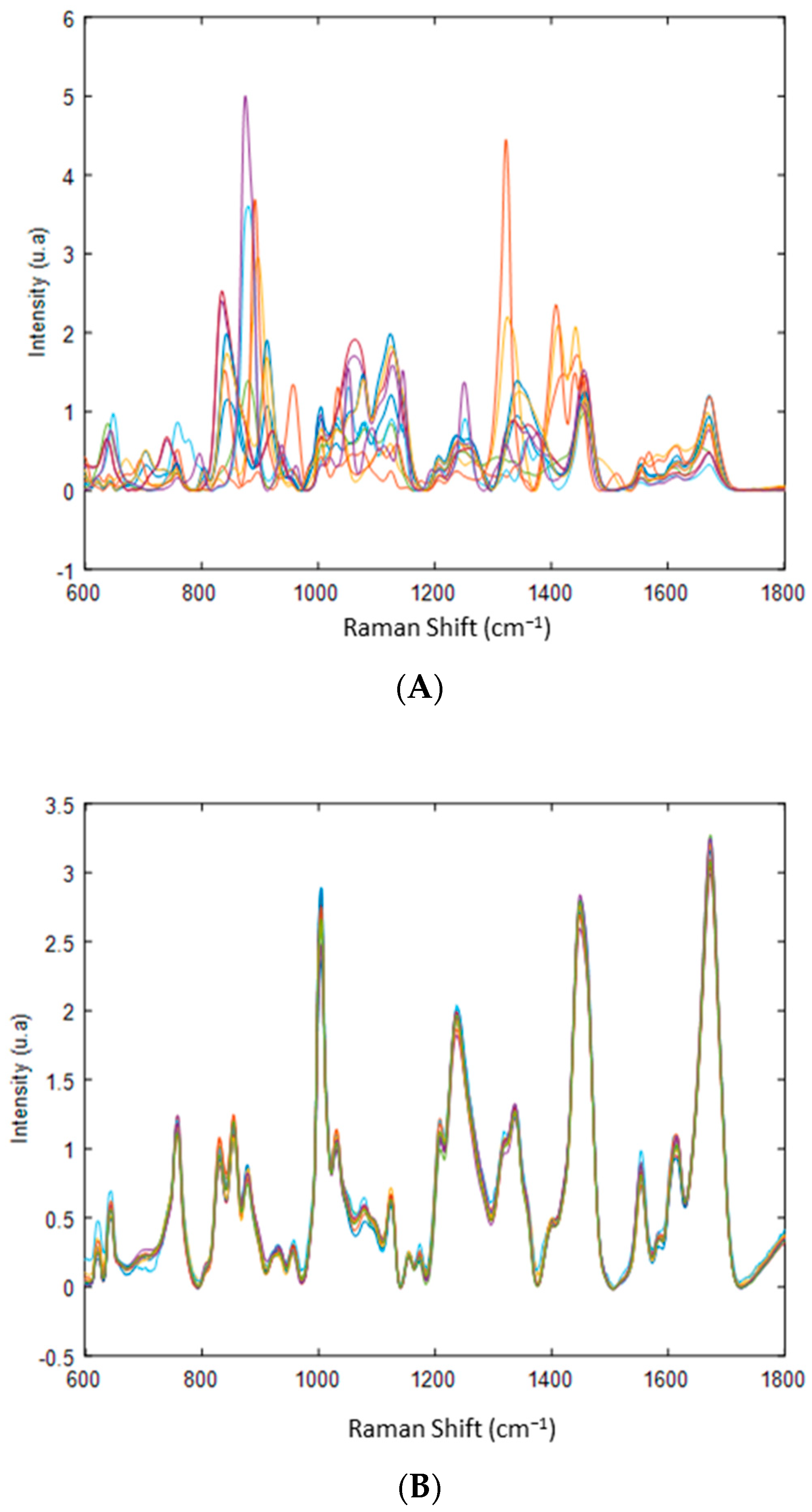
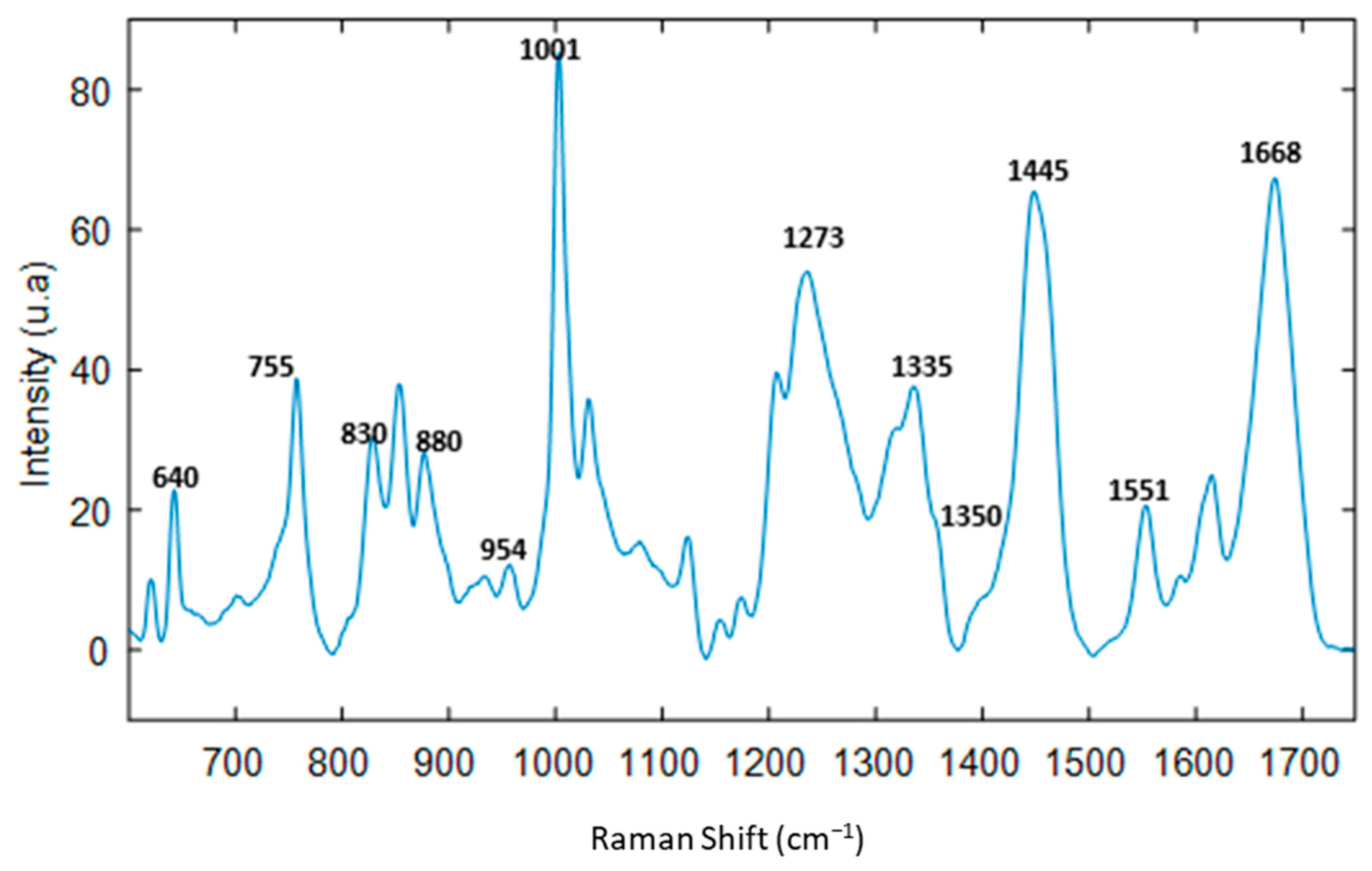
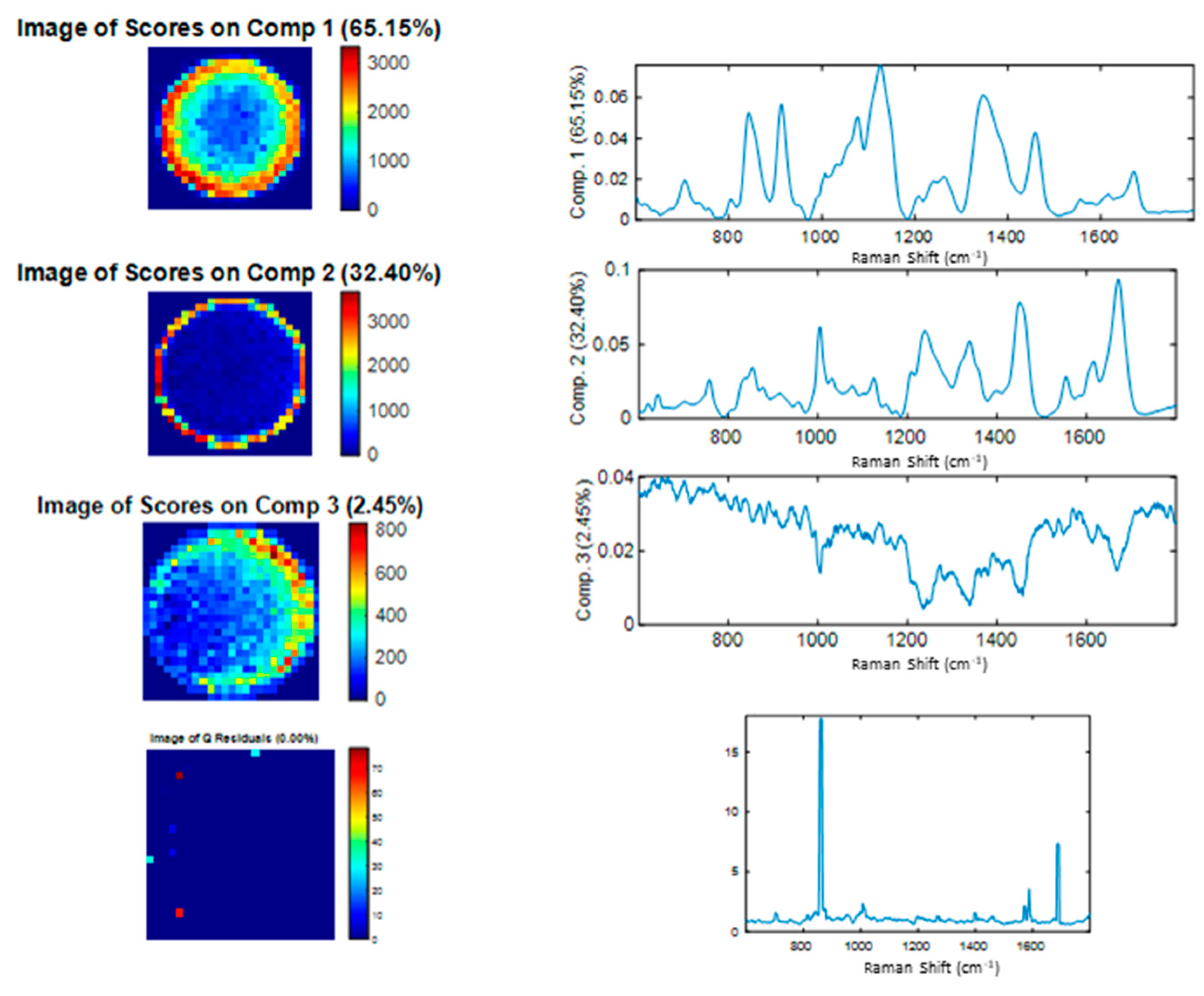
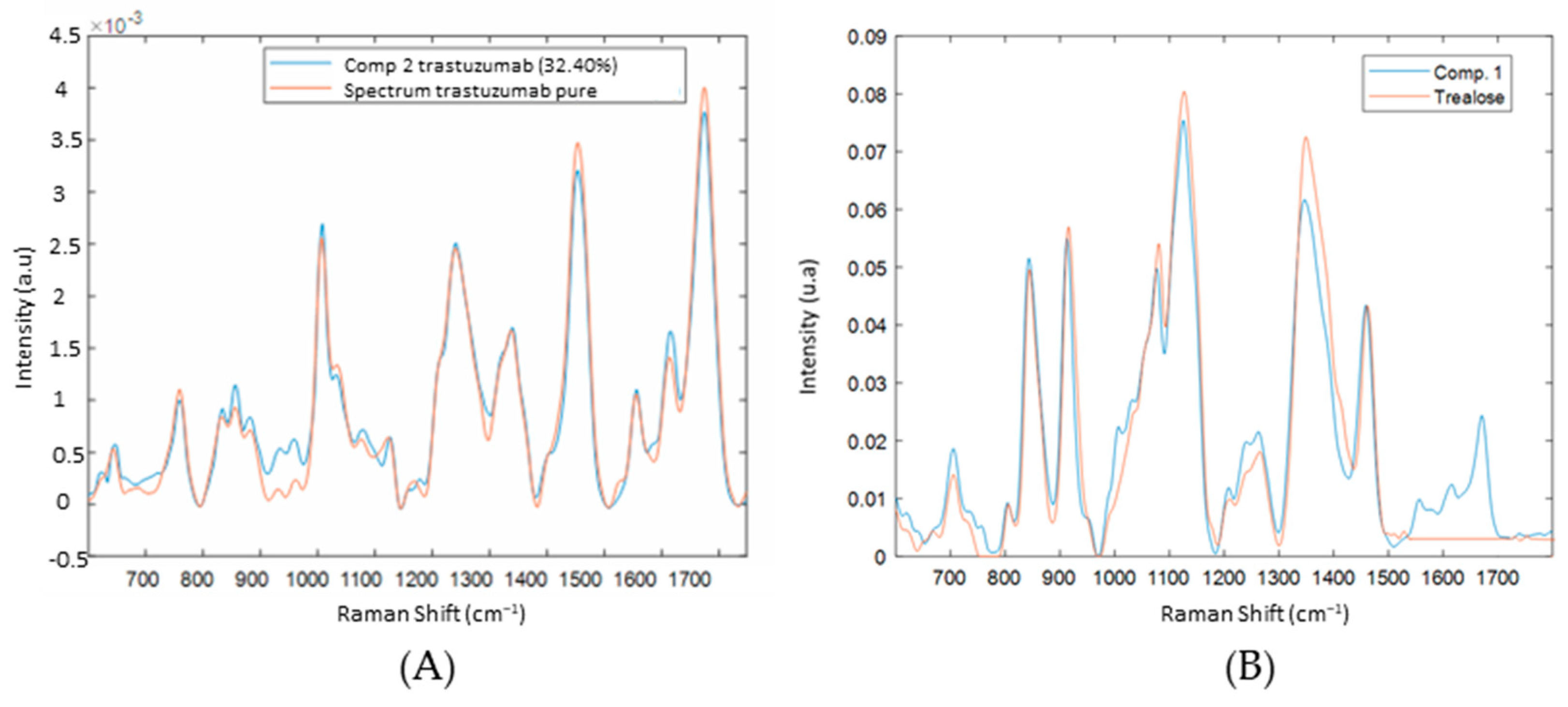
3.2. Spectral Characterization of Commercial mAbs Solutions by DCDR Imaging and MCR-ALS Analysis
3.3. Limit of Detection of DCDR Analysis
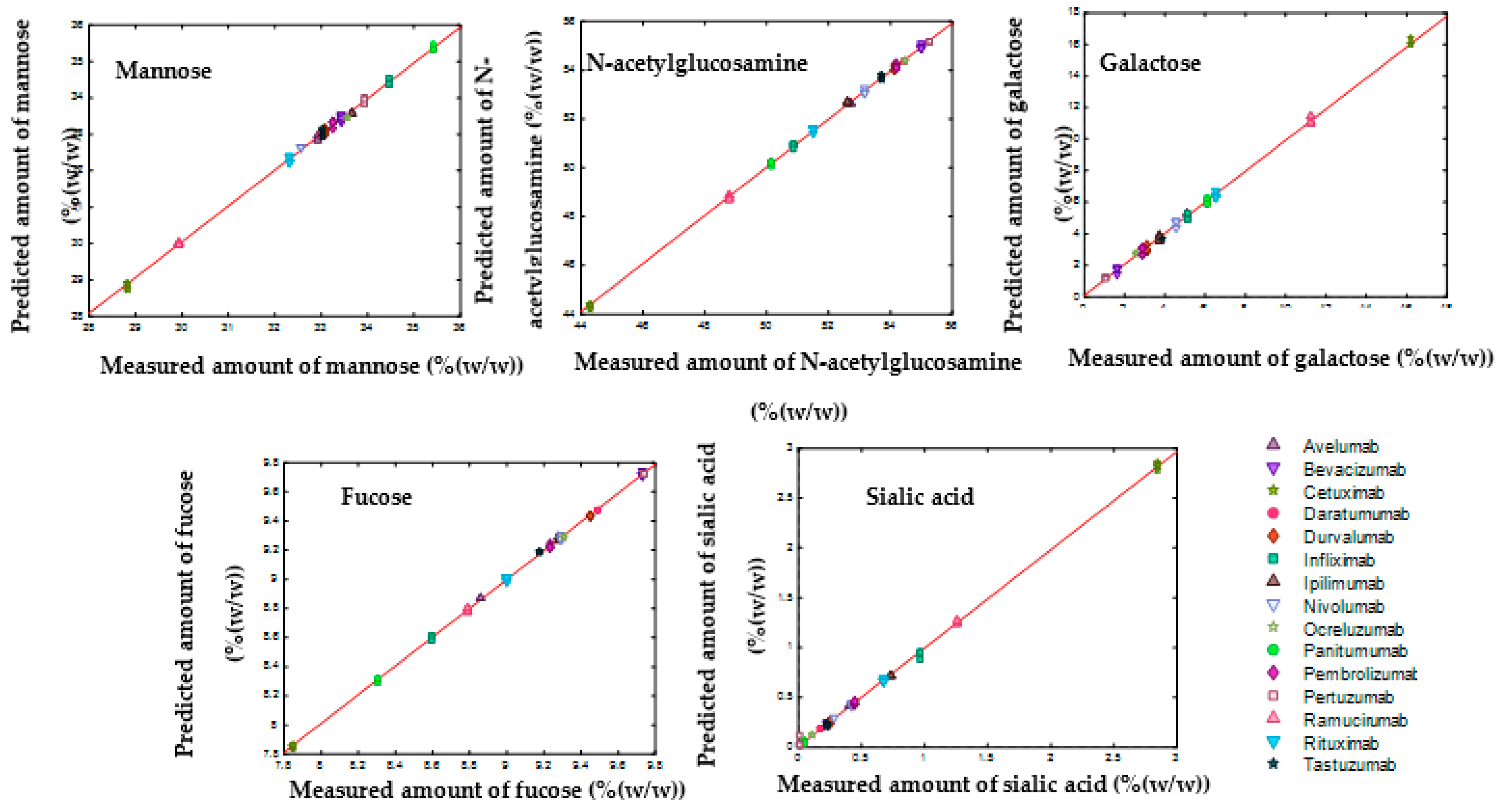
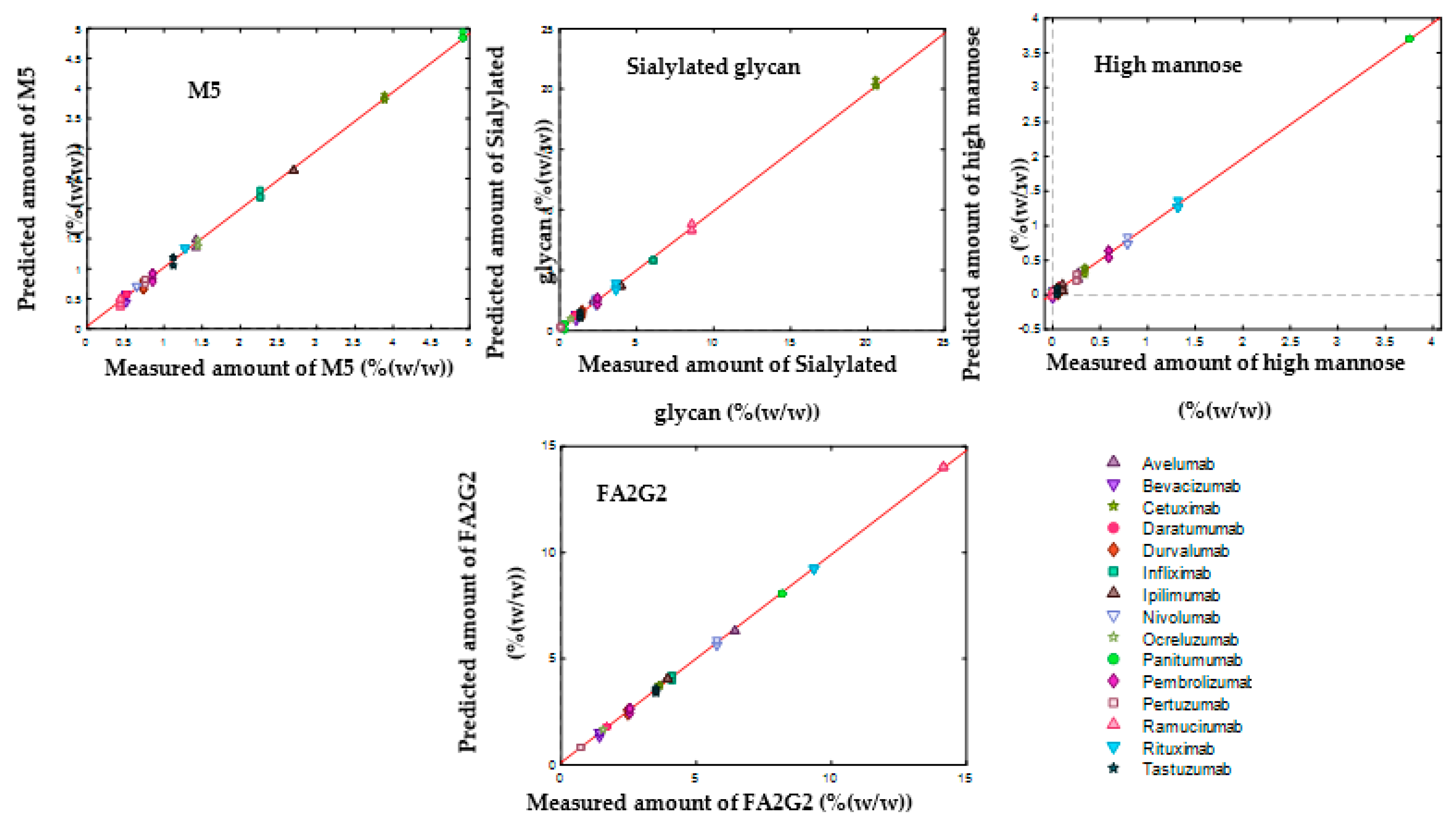
3.4. SVR Models to Determine the Composition of Monosaccharides and Glycans
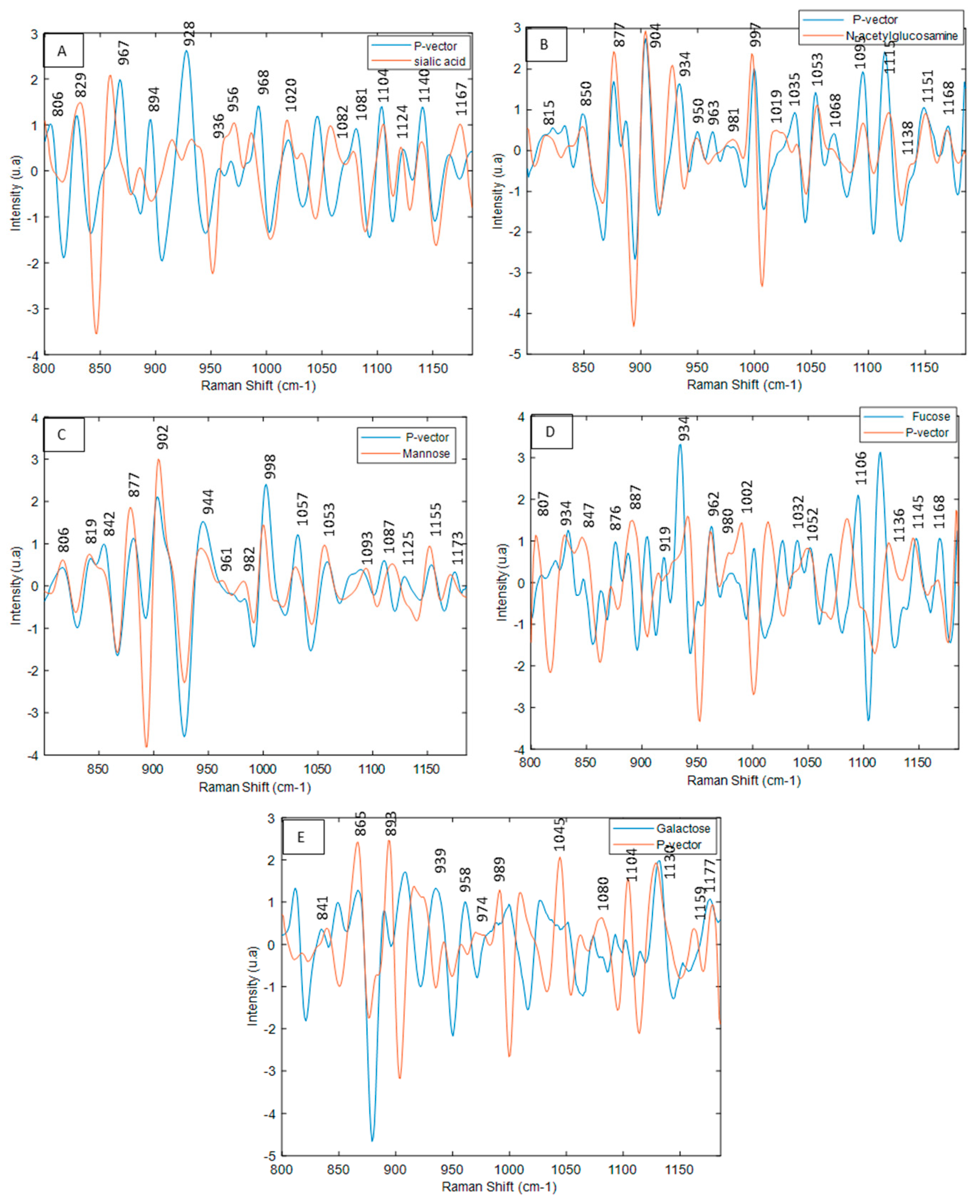
3.5. Interpretation SVR Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hamla, S.; Sacré, P.-Y.; Derenne, A.; Derfoufi, K.-M.; Cowper, B.; Butré, C.I.; Delobel, A.; Goormaghtigh, E.; Hubert, P.; Ziemons, E. A new alternative tool to analyse glycosylation in pharmaceutical proteins based on infrared spectroscopy combined with nonlinear support vector regression. Analyst 2022, 147, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Wuhrer, M.; Deelder, A.M.; Hokke, C.H. Protein glycosylation analysis by liquid chromatography-mass spectrometry. J. Chromatogr. B 2005, 825, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Luo, S.; Zhang, B. Glycan analysis of therapeutic glycoproteins. MAbs 2016, 8, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Lalonde, M.E.; Durocher, Y. Therapeutic glycoprotein production in mammalian cells. J. Biotechnol. 2017, 251, 128–140. [Google Scholar] [CrossRef]
- Hajba, L.; Csanky, E.; Guttman, A. Liquid phase separation methods for N-glycosylation analysis of glycoproteins of biomedical and biopharmaceutical interest. A critical review. Anal. Chim. Acta 2016, 943, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Derenne, A.; Derfoufi, K.M.; Cowper, B.; Delporte, C.; Goormaghtigh, E. FTIR spectroscopy as an analytical tool to compare glycosylation in therapeutic monoclonal antibodies. Anal. Chim. Acta 2020, 1112, 62–71. [Google Scholar] [CrossRef]
- Chrimes, A.F.; Khoshmanesh, K.; Stoddart, P.R.; Mitchell, A.; Kalantar-Zadeh, K. Microfluidics and raman microscopy: Current applications and future challenges. Chem. Soc. Rev. 2013, 42, 5880–5906. [Google Scholar] [CrossRef]
- Zhang, D.; Mrozek, M.F.; Xie, Y.; Ben-Amotz, D. Chemical segregation and reduction of Raman background interference using drop-coating deposition. Appl. Spectrosc. 2004, 58, 929–933. [Google Scholar] [CrossRef]
- Barman, I.; Dingari, N.C.; Kang, J.W.; Horowitz, G.L.; Dasari, R.R.; Feld, M.S. Raman spectroscopy-based sensitive and specific detection of glycated hemoglobin. Anal. Chem. 2012, 84, 2474–2482. [Google Scholar] [CrossRef] [Green Version]
- Magistrale, L. In Scienze Chimiche per la Conservazione Prova Finale di Laurea Chemometric Analysis of the Photooxidative Decolorization of the azo dye Acid Red 97; Università Ca’ Foscari: Venezia, Italy, 2012. [Google Scholar]
- Haouchine, M.; Biache, C.; Lorgeoux, C.; Faure, P.; Marc, O. Handle Matrix Rank Deficiency, Noise, and Interferences in 3D Emission—Excitation Matrices: Effective Truncated Singular-Value Decomposition in Chemometrics Applied to the Analysis of Polycyclic Aromatic Compounds. ACS Omega 2022. [Google Scholar] [CrossRef]
- Üstün, B.; Melssen, W.J.; Buydens, L.M.C. Visualisation and interpretation of Support Vector Regression models. Anal. Chim. Acta 2007, 595, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Walls, D.; Walker, J.M. Protein Chromatography. Protein Chromatogr. 1485, 423, 2017. [Google Scholar] [CrossRef]
- Smith, J.P.; Smith, F.C.; Booksh, K.S. Multivariate Curve Resolution–Alternating Least Squares (MCR-ALS) with Raman Imaging Applied to Lunar Meteorites. Appl. Spectrosc. 2018, 72, 404–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.P.; Holahan, E.C.; Smith, F.C.; Marrero, V.; Booksh, K.S. A novel multivariate curve resolution-alternating least squares (MCR-ALS) methodology for application in hyperspectral Raman imaging analysis. Analyst 2019, 144, 5425–5438. [Google Scholar] [CrossRef] [PubMed]
- Gourvénec, S.; Massart, D.L.; Rutledge, D.N. Determination of the number of components during mixture analysis using the Durbin-Watson criterion in the Orthogonal Projection Approach and in the SIMPLe-to-use Interactive Self-modelling Mixture Analysis approach. Chemom. Intell. Lab. Syst. 2002, 61, 51–61. [Google Scholar] [CrossRef]
- Brewster, V.L.; Ashton, L.; Goodacre, R. Monitoring the glycosylation status of proteins using raman spectroscopy. Anal. Chem. 2011, 83, 6074–6081. [Google Scholar] [CrossRef]
- Derenne, A.; Vandersleyen, O.; Goormaghtigh, E. Lipid quantification method using FTIR spectroscopy applied on cancer cell extracts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 1200–1209. [Google Scholar] [CrossRef]
- Makki, A.A.; Massot, V.; Byrne, H.J.; Respaud, R.; Bertrand, D.; Mohammed, E.; Chourpa, I.; Bonnier, F. Understanding the discrimination and quantification of monoclonal antibodies preparations using Raman spectroscopy. J. Pharm. Biomed. Anal. 2021, 194, 113734. [Google Scholar] [CrossRef]
- Strickley, R.G.; Lambert, W.J. A review of Formulations of Commercially Available Antibodies. J. Pharm. Sci. 2021, 110, 2590–2608.e56. [Google Scholar] [CrossRef]
- Oberg, K.A.; Fink, A.L. A new attenuated total reflectance fourier transform infrared spectroscopy method for the study of proteins in solution. Anal. Biochem. 1998, 256, 92–106. [Google Scholar] [CrossRef]
- Smith, J.; Arnolds, H. Polytetrafluoroethylene tape as a low-cost hydrophobic substrate for drop-coating deposition Raman spectroscopy of proteins. J. Raman Spectrosc. 2018, 49, 1236–1239. [Google Scholar] [CrossRef] [Green Version]
- Offroy, M. Développement de la Super-Résolution Appliquée à L’imagerie des Spectroscopies Vibrationnelles; University of Lille: Villeneuve-d’Ascq, France, 2012. [Google Scholar]
- Kumar, A.K.S.; Zhang, Y.; Li, D.; Compton, R.G. A mini-review: How reliable is the drop casting technique? Electrochem. Commun. 2020, 121, 106867. [Google Scholar] [CrossRef]
- Hu, P.; Zheng, X.-S.; Zong, C.; Li, M.-H.; Zhang, L.-Y.; Li, W.; Ren, B. Drop-coating deposition and surface-enhanced Raman spectroscopies (DCDRS and SERS) provide complementary information of whole human tears. J. Raman Spectrosc. 2014, 45, 565–573. [Google Scholar] [CrossRef]
- Zeid, W.B.; Brutin, D. Influence of relative humidity on spreading, pattern formation and adhesion of a drying drop of whole blood. Colloids Surfaces A Physicochem. Eng. Asp. 2013, 430, 1–7. [Google Scholar] [CrossRef]
- Ortiz, C.; Zhang, D.; Xie, Y.; Ribbe, A.E.; Ben-Amotz, D. Validation of the drop-coating deposition Raman method for protein analysis. Anal. Biochem. 2006, 353, 157–166. [Google Scholar] [CrossRef] [PubMed]



| Initial Concentration (mg mL−1) | Dilution Range (mg mL−1) |
|---|---|
| 5 | 4, 3, 2, 1, 0.5, 0.1, 0.01 |
| 10 | 7, 5, 3, 1, 0.5, 0.1, 0.01 |
| 20 | 10, 5, 3, 1, 0.5, 0.1, 0.01 |
| 21 | 10, 5, 3, 1, 0.5, 0.1, 0.01 |
| 25 | 12.5, 5, 3, 1, 0.5, 0.1, 0.01 |
| 30 | 15, 5, 3, 1, 0.5, 0.1, 0.01 |
| 50 | 25, 5, 3, 1, 0.5, 0.1, 0.01 |
| mAbs | Excipient Detected |
|---|---|
| Pembrolizumab Infliximab Pertuzumab | Sucrose |
| Cetuximab Ramucirumab | Glycine |
| Ocreluzumab Durvalumab Trastuzumab Bevacizumab | Trehalose |
| Avelumab Daratumumab Nivolumab Ipilimumab | Mannitol |
| Panitumumab | Sodium acetate |
| Rituximab | Sodium citrate |
| Mannose | N-acetylglucosamine | Galactose | Fucose | Sialic Acid | |
|---|---|---|---|---|---|
| Number of support vectors | 60 | 70 | 72 | 71 | 66 |
| R2Cal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| R2CV | 1.00 | 0.95 | 0.96 | 0.84 | 0.86 |
| RMSEC (%(w/w)) | 0.08 | 0.07 | 0.18 | 0.01 | 0.05 |
| RMSECV (%(w/w)) | 0.72 | 1.05 | 0.85 | 0.23 | 0.34 |
| M5 | Sialylated Glycan | High Mannose | FA2G2 | |
|---|---|---|---|---|
| Number of support vectors | 62 | 62 | 64 | 68 |
| R2Cal | 1.00 | 1.00 | 1.00 | 1.00 |
| R2CV | 0.52 | 0.95 | 0.83 | 0.88 |
| RMSEC (%(w/w)) | 0.06 | 0.25 | 0.04 | 0.11 |
| RMSECV (%(w/w)) | 0.92 | 1.80 | 0.58 | 1.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamla, S.; Sacré, P.-Y.; Derenne, A.; Cowper, B.; Goormaghtigh, E.; Hubert, P.; Ziemons, E. A New Alternative Tool to Analyse Glycosylation in Monoclonal Antibodies Based on Drop-Coating Deposition Raman imaging: A Proof of Concept. Molecules 2022, 27, 4405. https://doi.org/10.3390/molecules27144405
Hamla S, Sacré P-Y, Derenne A, Cowper B, Goormaghtigh E, Hubert P, Ziemons E. A New Alternative Tool to Analyse Glycosylation in Monoclonal Antibodies Based on Drop-Coating Deposition Raman imaging: A Proof of Concept. Molecules. 2022; 27(14):4405. https://doi.org/10.3390/molecules27144405
Chicago/Turabian StyleHamla, Sabrina, Pierre-Yves Sacré, Allison Derenne, Ben Cowper, Erik Goormaghtigh, Philippe Hubert, and Eric Ziemons. 2022. "A New Alternative Tool to Analyse Glycosylation in Monoclonal Antibodies Based on Drop-Coating Deposition Raman imaging: A Proof of Concept" Molecules 27, no. 14: 4405. https://doi.org/10.3390/molecules27144405







