Thorough Investigation of the Phenolic Profile of Reputable Greek Honey Varieties: Varietal Discrimination and Floral Markers Identification Using Liquid Chromatography–High-Resolution Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
2.1. Target Screening
2.2. Suspect Screening
2.3. Non-Target Screening
3. Materials and Methods
3.1. Chemicals
3.2. Standard Preparation
3.3. Honey Samples and Sample Preparation
3.4. Method Verification
3.5. UPLC-QToF-MS Analysis
3.6. Targeted, Suspect and Non-Targeted Screening Workflows
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Afrin, S.; Haneefa, S.M.; Fernandez-Cabezudo, M.J.; Giampieri, F.; Al-Ramadi, B.K.; Battino, M. Therapeutic and preventive properties of honey and its bioactive compounds in cancer: An evidence-based review. Nutr. Res. Rev. 2020, 33, 50–76. [Google Scholar] [CrossRef] [PubMed]
- Raptopoulou, K.G.; Pasias, I.N.; Proestos, C. Recent Advances in Analytical Techniques for the Determination of Authenticity and Adulteration of Honey and its Products. Recent Adv. Anal. Tech. 2020, 4, 125–145. [Google Scholar]
- Tsagkaris, A.S.; Koulis, G.A.; Danezis, G.P.; Martakos, I.; Dasenaki, M.; Georgiou, C.A.; Thomaidis, N.S. Honey authenticity: Analytical techniques, state of the art and challenges. RSC Adv. 2021, 11, 11273–11294. [Google Scholar] [CrossRef]
- Oymen, B.; Aşır, S.; Türkmen, D.; Denizli, A. Determination of multi-pesticide residues in honey with a modified QuEChERS procedure followed by LC-MS/MS and GC-MS/MS. J. Apic. Res. 2021, 1–13. [Google Scholar] [CrossRef]
- Soyseven, M.; Sezgin, B.; Arli, G. A novel, rapid and robust HPLC-ELSD method for simultaneous determination of fructose, glucose and sucrose in various food samples: Method development and validation. J. Food Compos. Anal. 2022, 107, 104400. [Google Scholar] [CrossRef]
- Vazquez, L.; Armada, D.; Celeiro, M.; Dagnac, T.; Llompart, M. Evaluating the Presence and Contents of Phytochemicals in Honey Samples: Phenolic Compounds as Indicators to Identify Their Botanical Origin. Foods 2021, 10, 2616. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Chen, L.; Zhou, J. Analytical Strategies for LC–MS-Based Untargeted and Targeted Metabolomics Approaches Reveal the Entomological Origins of Honey. J. Agric. Food Chem. 2022, 70, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Habib, H.M.; Kheadr, E.; Ibrahim, W.H. Inhibitory effects of honey from arid land on some enzymes and protein damage. Food Chem. 2021, 364, 130415. [Google Scholar] [CrossRef] [PubMed]
- Xagoraris, M.; Skouria, A.; Revelou, P.-K.; Alissandrakis, E.; Tarantilis, P.A.; Pappas, C.S. Response Surface Methodology to Optimize the Isolation of Dominant Volatile Compounds from Monofloral Greek Thyme Honey Using SPME-GC-MS. Molecules 2021, 26, 3612. [Google Scholar] [CrossRef]
- Tsavea, E.; Vardaka, F.-P.; Savvidaki, E.; Kellil, A.; Kanelis, D.; Bucekova, M.; Grigorakis, S.; Godocikova, J.; Gotsiou, P.; Dimou, M.; et al. Physicochemical Characterization and Biological Properties of Pine Honey Produced across Greece. Foods 2022, 11, 943. [Google Scholar] [CrossRef]
- Graikou, K.; Andreou, A.; Chinou, I. Chemical profile οf Greek Arbutus unedo honey: Biological properties. J. Apic. Res. 2022, 61, 100–106. [Google Scholar] [CrossRef]
- Koulis, G.A.; Tsagkaris, A.S.; Aalizadeh, R.; Dasenaki, M.E.; Panagopoulou, E.I.; Drivelos, S.; Halagarda, M.; Georgiou, C.A.; Proestos, C.; Thomaidis, N.S. Honey Phenolic Compound Profiling and Authenticity Assessment Using HRMS Targeted and Untargeted Metabolomics. Molecules 2021, 26, 2769. [Google Scholar] [CrossRef] [PubMed]
- Gašić, U.; Kečkeš, S.; Dabić, D.; Trifković, J.; Milojković-Opsenica, D.; Natić, M.; Tešić, Z. Phenolic profile and antioxidant activity of Serbian polyfloral honeys. Food Chem. 2014, 145, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.; Meenu, M.; Yu, X.; Xu, B. Phenolic acids and flavonoids profiles of commercial honey from different floral sources and geographic sources. Int. J. Food Prop. 2019, 22, 290–308. [Google Scholar] [CrossRef]
- Escriche, I.; Kadar, M.; Juan-Borrás, M.; Domenech, E. Using flavonoids, phenolic compounds and headspace volatile profile for botanical authentication of lemon and orange honeys. Food Res. Int. 2011, 44, 1504–1513. [Google Scholar] [CrossRef]
- Can, Z.; Yildiz, O.; Sahin, H.; Akyuz Turumtay, E.; Silici, S.; Kolayli, S. An investigation of Turkish honeys: Their physico-chemical properties, antioxidant capacities and phenolic profiles. Food Chem. 2015, 180, 133–141. [Google Scholar] [CrossRef]
- Sahin, H. Honey as an apitherapic product: Its inhibitory effect on urease and xanthine oxidase. J. Enzyme Inhib. Med. Chem. 2016, 31, 490–494. [Google Scholar] [CrossRef] [Green Version]
- Kolayli, S.; Sahin, H.; Can, Z.; Yildiz, O.; Sahin, K. Honey shows potent inhibitory activity against the bovine testes hyaluronidase. J. Enzyme Inhib. Med. Chem. 2016, 31, 599–602. [Google Scholar] [CrossRef]
- Recklies, K.; Peukert, C.; Kölling-Speer, I.; Speer, K. Differentiation of Honeydew Honeys from Blossom Honeys and According to Their Botanical Origin by Electrical Conductivity and Phenolic and Sugar Spectra. J. Agric. Food Chem. 2021, 69, 1329–1347. [Google Scholar] [CrossRef]
- Bertoncelj, J.; Polak, T.; Kropf, U.; Korošec, M.; Golob, T. LC-DAD-ESI/MS analysis of flavonoids and abscisic acid with chemometric approach for the classification of Slovenian honey. Food Chem. 2011, 127, 296–302. [Google Scholar] [CrossRef]
- Kato, Y.; Fujinaka, R.; Ishisaka, A.; Nitta, Y.; Kitamoto, N.; Takimoto, Y. Plausible authentication of manuka honey and related products by measuring leptosperin with methyl syringate. J. Agric. Food Chem. 2014, 62, 6400–6407. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; Sobolev, A.P.; Di Lorenzo, A.; Vista, S.; Tenore, G.C.; Daglia, M. Chemical Composition of Different Botanical Origin Honeys Produced by Sicilian Black Honeybees (Apis mellifera ssp. sicula). J. Agric. Food Chem. 2015, 63, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.T.; Dillon, A.; Warren, J.; Walker, M.J. A Critical Review of the Factors Available for the Identification and Determination of Mānuka Honey. Food Anal. Methods 2018, 11, 1561–1567. [Google Scholar] [CrossRef] [Green Version]
- Jonathan Chessum, K.; Chen, T.; Hamid, N.; Kam, R. A comprehensive chemical analysis of New Zealand honeydew honey. Food Res. Int. 2022, 111436. [Google Scholar] [CrossRef]
- Stompor, M. A Review on Sources and Pharmacological Aspects of Sakuranetin. Nutrients 2020, 12, 513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciucure, C.T.; Geană, E. Phenolic compounds profile and biochemical properties of honeys in relationship to the honey floral sources. Phytochem. Anal. 2019, 30, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Tuberoso, C.I.G.; Jerković, I.; Bifulco, E.; Marijanovic, Z.; Congiu, F.; Bubalo, D. Riboflavin and lumichrome in Dalmatian sage honey and other unifloral honeys determined by LC–DAD technique. Food Chem. 2012, 135, 1985–1990. [Google Scholar] [CrossRef]
- Escuredo, O.; Silva, L.R.; Valentão, P.; Seijo, M.C.; Andrade, P.B. Assessing Rubus honey value: Pollen and phenolic compounds content and antibacterial capacity. Food Chem. 2012, 130, 671–678. [Google Scholar] [CrossRef]
- Ouchemoukh, S.; Amessis-Ouchemoukh, N.; Gómez-Romero, M.; Aboud, F.; Giuseppe, A.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Characterisation of phenolic compounds in Algerian honeys by RP-HPLC coupled to electrospray time-of-flight mass spectrometry. LWT-Food Sci. Technol. 2017, 85, 460–469. [Google Scholar] [CrossRef]
- Tolba, M.F.; Azab, S.S.; Khalifa, A.E.; Abdel-Rahman, S.Z.; Abdel-Naim, A.B. Caffeic acid phenethyl ester, a promising component of propolis with a plethora of biological activities: A review on its anti-inflammatory, neuroprotective, hepatoprotective, and cardioprotective effects. IUBMB Life 2013, 65, 699–709. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. A comprehensive review on the main honey authentication issues: Production and origin. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1072–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gašić, U.M.; Milojković-Opsenica, D.M.; Tešić, Ž.L. Polyphenols as possible markers of botanical origin of honey. J. AOAC Int. 2017, 100, 852–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridi, R.; Montenegro, G. The value of chilean honey: Floral origin related to their antioxidant and antibacterial activities. Honey Anal. 2017, 63–78. [Google Scholar]
- Kaškoniene, V.; Venskutonis, P.R. Floral Markers in Honey of Various Botanical and Geographic Origins: A Review. Compr. Rev. Food Sci. Food Saf. 2010, 9, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Aalizadeh, R.; Nika, M.C.; Thomaidis, N.S. Development and application of retention time prediction models in the suspect and non-target screening of emerging contaminants. J. Hazard. Mater. 2019, 363, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Seraglio, S.K.T.; Schulz, M.; Brugnerotto, P.; Silva, B.; Gonzaga, L.V.; Fett, R.; Costa, A.C.O. Quality, composition and health-protective properties of citrus honey: A review. Food Res. Int. 2021, 143, 110268. [Google Scholar] [CrossRef]
- de Melo, F.H.C.; Menezes, F.N.D.D.; de Sousa, J.M.B.; dos Santos Lima, M.; da Silva Campelo Borges, G.; de Souza, E.L.; Magnani, M. Prebiotic activity of monofloral honeys produced by stingless bees in the semi-arid region of Brazilian Northeastern toward Lactobacillus acidophilus LA-05 and Bifidobacterium lactis BB-12. Food Res. Int. 2020, 128, 108809. [Google Scholar] [CrossRef]
- Jerković, I.; Marijanović, Z.; Tuberoso, C.I.G.; Bubalo, D.; Kezić, N. Molecular diversity of volatile compounds in rare willow (Salix spp.) honeydew honey: Identification of chemical biomarkers. Mol. Divers. 2010, 14, 237–248. [Google Scholar] [CrossRef]
- Floris, I.; Pusceddu, M.; Satta, A. The Sardinian Bitter Honey: From Ancient Healing Use to Recent Findings. Antioxidants 2021, 10, 506. [Google Scholar] [CrossRef]
- Campillo, N.; Viñas, P.; Férez-Melgarejo, G.; Hernández-Córdoba, M. Dispersive liquid-liquid microextraction for the determination of flavonoid aglycone compounds in honey using liquid chromatography with diode array detection and time-of-flight mass spectrometry. Talanta 2015, 131, 185–191. [Google Scholar] [CrossRef]
- Gośliński, M.; Nowak, D.; Szwengiel, A. Multidimensional Comparative Analysis of Bioactive Phenolic Compounds of Honeys of Various Origin. Antioxidants 2021, 10, 530. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Schmidt, S.; Müller-Hannemann, M.; Neumann, S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinform. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malm, L.; Palm, E.; Souihi, A.; Plassmann, M.; Liigand, J.; Kruve, A. Guide to Semi-Quantitative Non-Targeted Screening Using LC/ESI/HRMS. Molecules 2021, 26, 3524. [Google Scholar] [CrossRef] [PubMed]
- Olmo-García, L.; Wendt, K.; Kessler, N.; Bajoub, A.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Exploring the Capability of LC-MS and GC-MS Multi-Class Methods to Discriminate Virgin Olive Oils from Different Geographical Indications and to Identify Potential Origin Markers. Eur. J. Lipid Sci. Technol. 2019, 121, 1800336. [Google Scholar] [CrossRef]
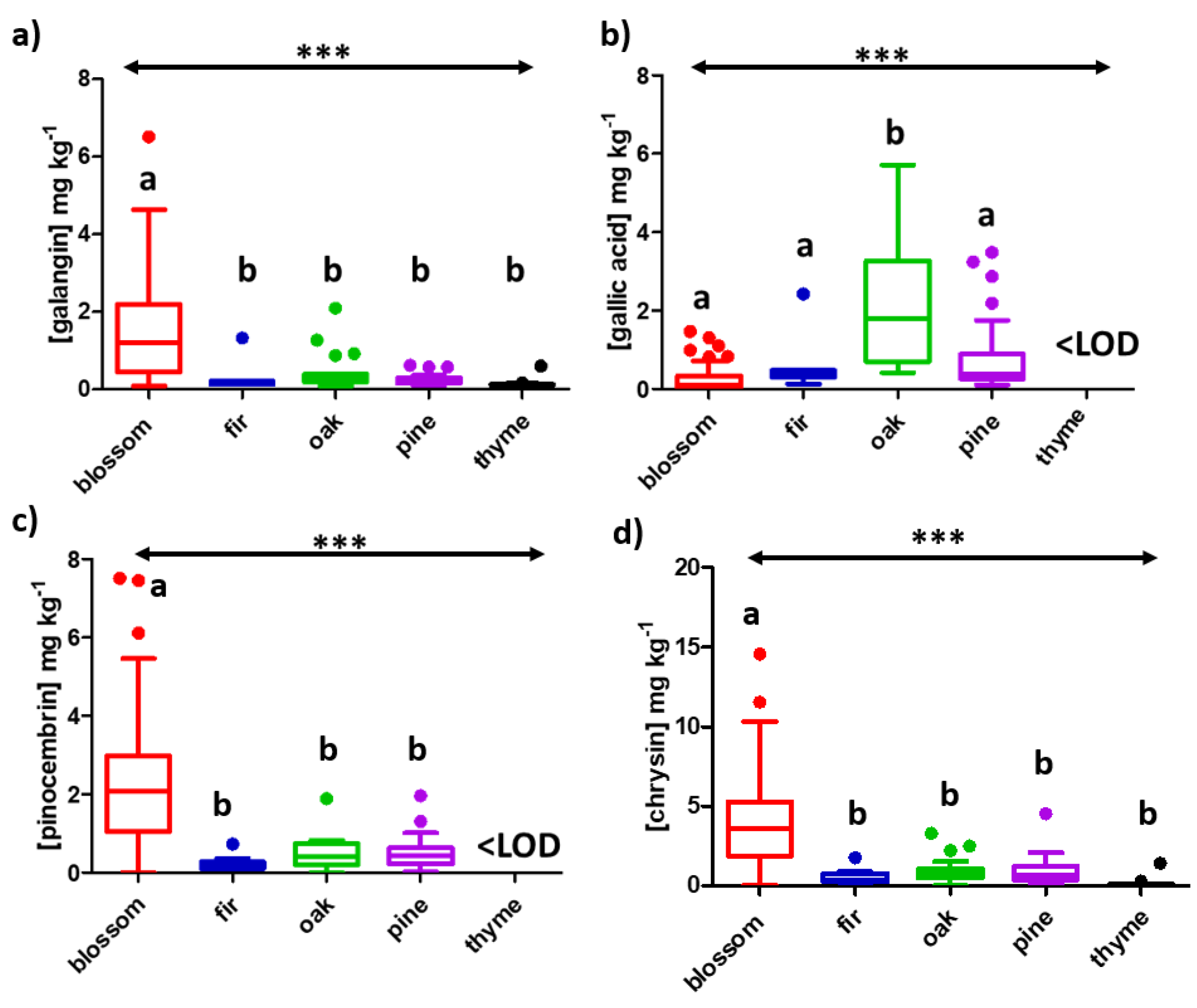
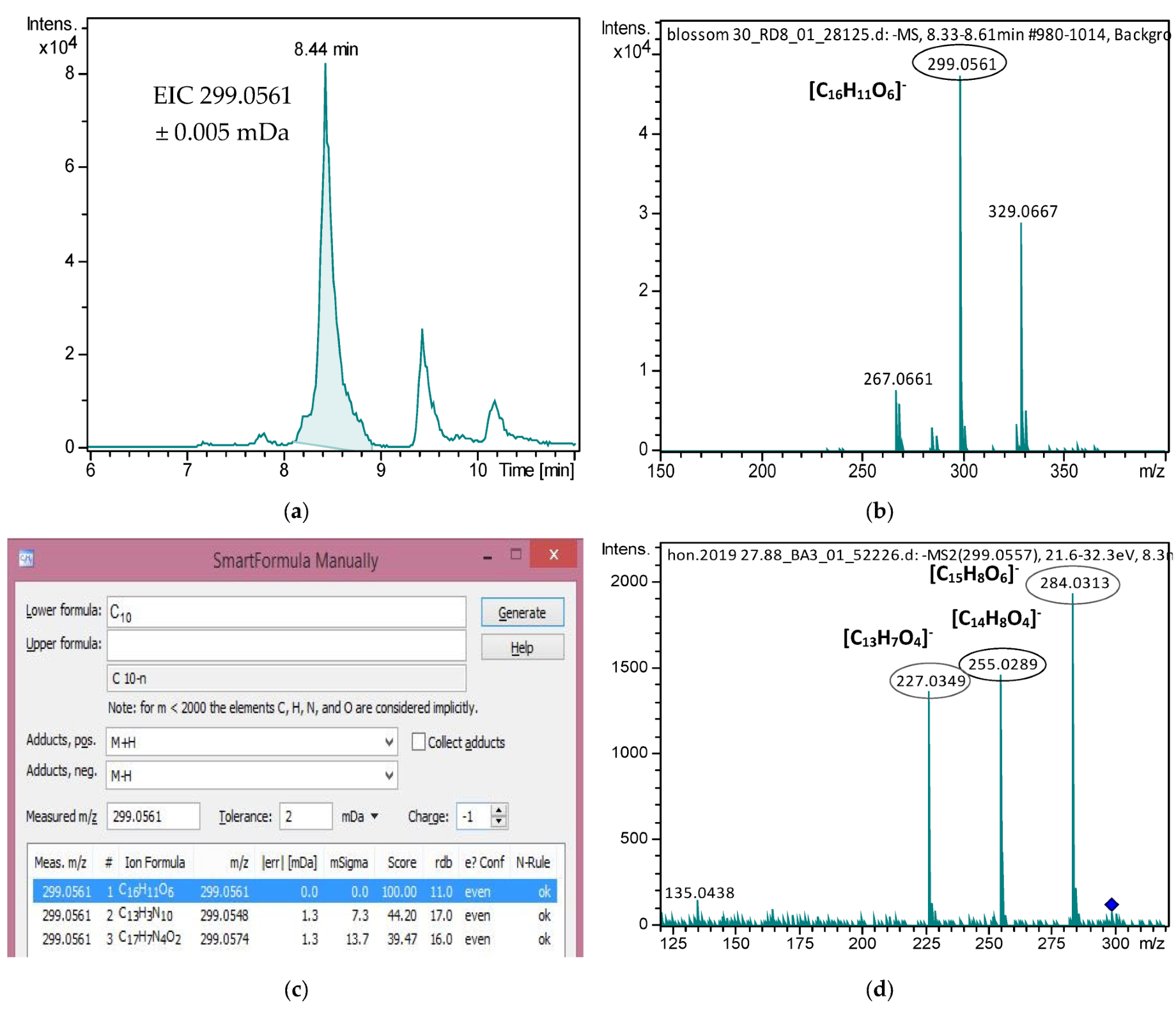
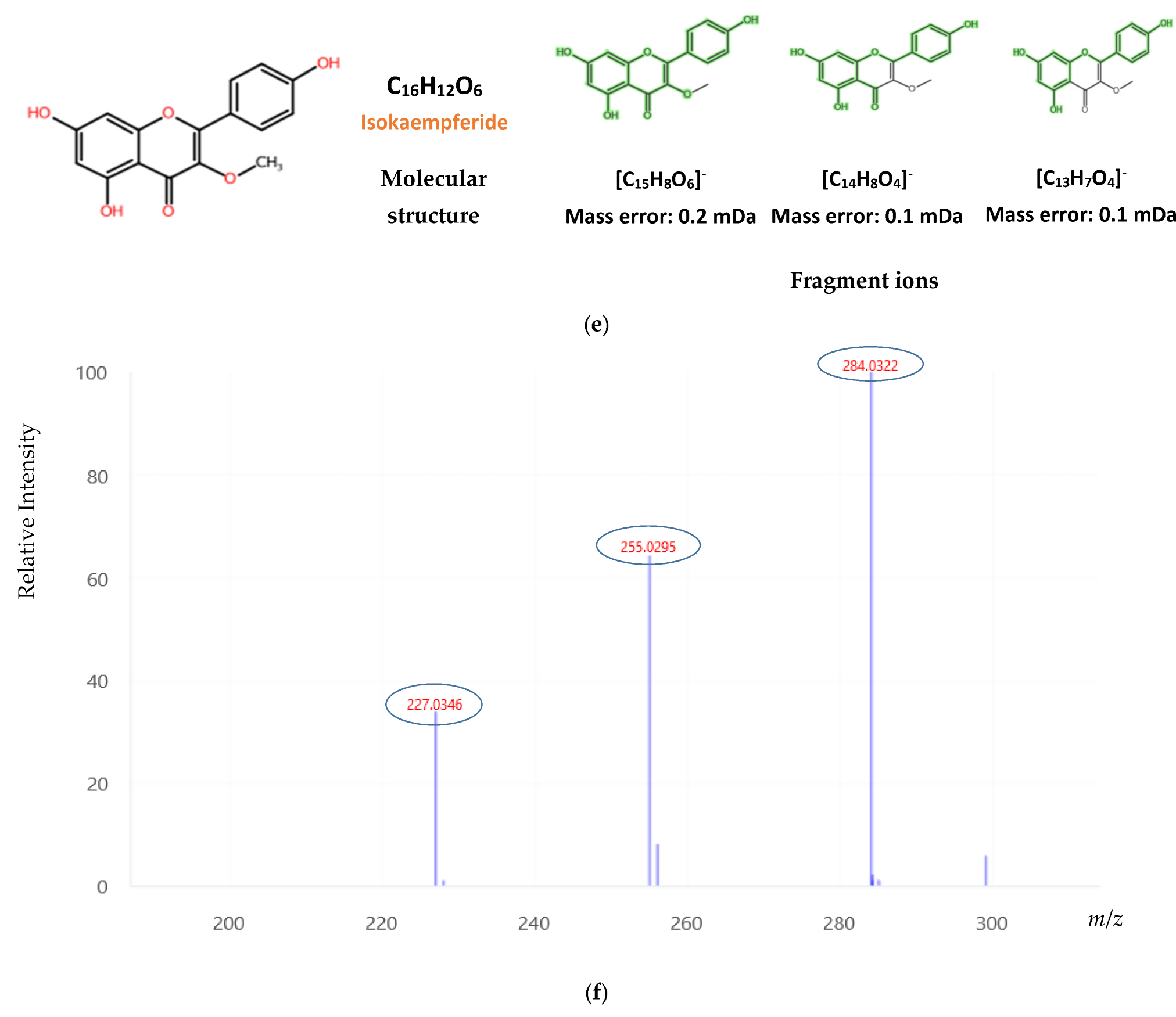
| Compound | LOD | Blossom, n = 62 | Fir, n = 10 | Oak, n = 24 | Pine, n = 39 | Thyme, n = 34 |
|---|---|---|---|---|---|---|
| 2,5 dihydroxybenzoic acid | 0.070 | 0.21 | 0.71 | 0.82 | 0.92 | 0.11 |
| 3,4 dihydroxybenzoic acid | 0.083 | 1.0 | 3.4 | 10 | 4.7 | 0.50 |
| 4 hydroxybenzoic acid | 0.098 | 1.5 | 1.3 | 1.0 | 1.8 | 0.35 |
| apigenin | 0.082 | 0.15 | 0.10 | 0.12 | 0.11 | 0.17 |
| caffeic acid | 0.065 | 0.77 | 0.21 | 0.38 | 0.49 | 0.080 |
| chrysin | 0.032 | 3.6 | 0.35 | 0.72 | 0.65 | 0.061 |
| cinnamic acid | 0.043 | 0.26 | 0.17 | 0.24 | 0.13 | 0.022 |
| eriodictyol | 0.048 | 0.098 | <LOD | 0.10 | 0.28 | 0.10 |
| ferulic acid | 0.030 | 0.58 | 0.21 | 0.27 | 0.37 | 0.045 |
| galangin | 0.070 | 1.2 | 0.15 | 0.27 | 0.21 | 0.12 |
| gallic acid | 0.067 | 0.092 | 0.37 | 1.8 | 0.37 | <LOD |
| genistein | 0.081 | 0.089 | <LOD | 0.098 | 0.10 | 0.11 |
| luteolin | 0.079 | 0.14 | 0.34 | 0.15 | 0.16 | 0.14 |
| naringenin | 0.050 | 1.4 | 0.25 | 0.59 | 0.60 | 0.073 |
| p-coumaric acid | 0.16 | 0.60 | 0.59 | 0.72 | 0.94 | 0.27 |
| pinobanksin | 0.055 | 1.4 | 0.25 | 0.57 | 0.59 | 0.069 |
| pinocembrin | 0.076 | 2.1 | 0.15 | 0.40 | 0.44 | <LOD |
| quercetin | 0.067 | 0.15 | 0.49 | 0.20 | 0.18 | 0.15 |
| rosmarinic acid | 0.084 | <LOD | 0.14 | <LOD | <LOD | 0.12 |
| salicylic acid | 0.33 | 0.76 | 0.81 | 1.6 | 1.3 | 0.34 |
| syringic acid | 0.081 | <LOD | <LOD | 0.25 | 0.36 | <LOD |
| taxifolin | 0.084 | 0.18 | 0.32 | 0.20 | 0.41 | 0.23 |
| vanillic acid | 0.12 | 0.12 | 0.15 | <LOD | 0.12 | <LOD |
| vanillin | 0.037 | 0.50 | 0.16 | 0.27 | 0.59 | 0.098 |
| Analytes | Honey Matrix | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Blossom | Fir | Oak | Pine | Thyme | ||||||
| Mean (mg/Kg) | SD | Mean (mg/Kg) | SD | Mean (mg/Kg) | SD | Mean (mg/Kg) | SD | Mean (mg/Kg) | SD | |
| Acacetin | 0.092 | 0.13 | ND | - | 0.053 | 0.032 | 0.062 | 0.043 | ND | - |
| 2-trans,4-trans-abscisic acid | 0.21 | 0.37 | 0.31 | 0.58 | 0.92 | 1.4 | 0.17 | 0.19 | 0.12 | 0.052 |
| 2-cis,4-trans-abscisic acid | 0.37 | 0.58 | 0.46 | 0.88 | 1.4 | 1.8 | 0.33 | 0.40 | 0.19 | 0.12 |
| Sakuranetin | 0.024 | 0.030 | 0.010 | 0.010 | 0.022 | 0.011 | ND | - | ND | - |
| Homogentisic acid | 0.41 | 1.0 | 0.051 | 0.12 | 0.93 | 2.1 | 0.41 | 1.7 | 0.30 | 0.79 |
| Dehydrovomifoliol | 1.1 | 2.2 | 0.72 | 1.1 | 1.8 | 2.2 | 1.7 | 1.9 | 1.04 | 1.2 |
| Isokaempferide | 0.040 | 0.061 | ND | - | 0.023 | 0.021 | 0.023 | 0.030 | ND | - |
| Isorhamnetin | 0.071 | 0.12 | 0.10 | 0.13 | 0.11 | 0.084 | 0.074 | 0.14 | 0.033 | 0.043 |
| Lumichrome | 0.33 | 1.1 | ND | - | 0.11 | 0.15 | 0.29 | 0.55 | 0.15 | 0.36 |
| Methyl Syringate | 0.78 | 1.5 | 0.33 | 0.53 | 0.65 | 1.1 | 0.32 | 0.49 | 1.4 | 1.9 |
| Phenyllactic acid | 1.8 | 2.7 | 0.96 | 1.0 | 4.7 | 4.1 | 2.8 | 3.0 | 1.4 | 1.2 |
| Tectochrysin | 0.049 | 0.091 | ND | - | 0.030 | 0.030 | 0.024 | 0.043 | ND | - |
| PLS-DA Model | Variable (m/z, Monoisotopic Mass) | VIP Value | Rt (min) | Molecular Formula | Δm/z [mDa] | mSigma | Name |
|---|---|---|---|---|---|---|---|
| All varieties | 284.0685 | 4.52 | 10.14 | C16H12O5 | 0.159 | 5.6 | Acacetin |
| 284.1048 | 4.29 | 9.75 | C17H16O4 | −0.222 | 6.9 | Phenethyl caffeate | |
| 242.1517 | 4.21 | 4.72 | C13H22O4 | −0.437 | 21.0 | Unknown 1 | |
| 166.0996 | 4.15 | 4.72 | C10H14O2 | 0.021 | 3.6 | Unknown 2 | |
| 508.1157 | 4.13 | 9.68 | C30H20O8 | −0.567 | 35.8 | [2M − H]− of Chrysin | |
| Blossom vs. Thyme | 508.1157 | 5.50 | 9.68 | C30H20O8 | −0.567 | 35.8 | [2M − H]− of Chrysin |
| 544.1364 | 5.35 | 7.25 | C30H24O10 | 1.912 | 38.8 | [2M − H]− of Pinobanksin | |
| 284.0685 | 5.06 | 10.14 | C16H12O5 | 0.159 | 5.6 | Acacetin | |
| 286.0841 | 4.65 | 6.97 | C16H14O5 | −0.125 | 11.2 | uknown 1 | |
| 314.0790 | 4.57 | 9.01 | C17H14O6 | 1.010 | 4.6 | Pinobanksin 3-O-acetate | |
| Blossom | 237.0999 | 3.34 | 4.07 | C9H17O7 | 0.495 | 32.3 | Unknown 1 |
| 284.1048 | 3.20 | 9.75 | C17H16O4 | −0.222 | 6.9 | Phenethyl caffeate | |
| 248.1047 | 2.57 | 9.45 | C14H16O4 | −0.257 | 15.2 | Prenyl caffeate | |
| 364.1522 | 2.56 | 4.89 | C19H24O7 | 0.612 | 17.9 | Unknown 2 | |
| 378.1673 | 2.49 | 5.05 | C20H26O7 | −0.032 | 36.6 | Unknown 3 | |
| Thyme | 212.0686 | 3.50 | 3.55 | C10H12O5 | 0.215 | 7.8 | Eudesmic acid |
| 282.1094 | 3.43 | 5.59 | C14H18O6 | 0.845 | 10.5 | Unknown 1 | |
| 242.0579 | 3.21 | 5.72 | C14H10O4 | −0.217 | 6.1 | Unknown 2 | |
| 284.0684 | 3.05 | 9.92 | C13H22O4 | 0.074 | 7.7 | 3-methylgalangin | |
| 242.1517 | 3.04 | 4.72 | C16H12O5 | −0.437 | 12.9 | Unknown 3 | |
| Fir | 130.0631 | 4.03 | 1.34 | C6H10O3 | −1.607 | 34.2 | unknown 1 |
| 124.0164 | 3.28 | 1.68 | C6H4O3 | 0.961 | 11.2 | unknown 2 | |
| 160.1090 | 2.92 | 5.61 | C8H16O3 | −0.755 | 14.9 | unknown 3 | |
| 62.9962 | 2.88 | 4.09 | - | - | - | unknown 4 | |
| 120.0416 | 2.87 | 2.55 | C4H8O4 | −1.835 | 32.2 | unknown 5 | |
| Oak | 222.0891 | 4.73 | 4.34 | C12H14O4 | 1.808 | 13.7 | unknown 1 |
| 192.0424 | 4.47 | 4.95 | C10H8O4 | 1.486 | 7.7 | scopoletin | |
| 278.1278 | 4.43 | 4.43 | C19H18O2 | 1.619 | 17.8 | unknown 2 | |
| 170.0215 | 4.26 | 1.27 | C7H6O5 | −0.002 | 12.7 | gallic acid | |
| 292.0211 | 4.25 | 3.47 | C20H4O3 | −1.151 | 31.0 | unknown 3 | |
| Pine | 378.1673 | 2.74 | 5.05 | C20H26O7 | −0.032 | 36.6 | unknown 1 |
| 516.1989 | 2.63 | 4.20 | C27H32O10 | 1.281 | 46.0 | unknown 2 | |
| 264.0786 | 2.53 | 7.08 | C17H12O3 | 0.066 | 4.8 | Unknown 3 | |
| 350.1364 | 2.43 | 4.25 | C18H22O7 | −0.275 | 4.8 | Unknown 4 | |
| 304.0582 | 2.39 | 4.88 | C15H12O7 | −0.037 | 23.6 | Taxifolin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koulis, G.A.; Tsagkaris, A.S.; Katsianou, P.A.; Gialouris, P.-L.P.; Martakos, I.; Stergiou, F.; Fiore, A.; Panagopoulou, E.I.; Karabournioti, S.; Baessmann, C.; et al. Thorough Investigation of the Phenolic Profile of Reputable Greek Honey Varieties: Varietal Discrimination and Floral Markers Identification Using Liquid Chromatography–High-Resolution Mass Spectrometry. Molecules 2022, 27, 4444. https://doi.org/10.3390/molecules27144444
Koulis GA, Tsagkaris AS, Katsianou PA, Gialouris P-LP, Martakos I, Stergiou F, Fiore A, Panagopoulou EI, Karabournioti S, Baessmann C, et al. Thorough Investigation of the Phenolic Profile of Reputable Greek Honey Varieties: Varietal Discrimination and Floral Markers Identification Using Liquid Chromatography–High-Resolution Mass Spectrometry. Molecules. 2022; 27(14):4444. https://doi.org/10.3390/molecules27144444
Chicago/Turabian StyleKoulis, Georgios A., Aristeidis S. Tsagkaris, Panagiota A. Katsianou, Panagiotis-Loukas P. Gialouris, Ioannis Martakos, Fotis Stergiou, Alberto Fiore, Eleni I. Panagopoulou, Sofia Karabournioti, Carsten Baessmann, and et al. 2022. "Thorough Investigation of the Phenolic Profile of Reputable Greek Honey Varieties: Varietal Discrimination and Floral Markers Identification Using Liquid Chromatography–High-Resolution Mass Spectrometry" Molecules 27, no. 14: 4444. https://doi.org/10.3390/molecules27144444
APA StyleKoulis, G. A., Tsagkaris, A. S., Katsianou, P. A., Gialouris, P.-L. P., Martakos, I., Stergiou, F., Fiore, A., Panagopoulou, E. I., Karabournioti, S., Baessmann, C., van der Borg, N., Dasenaki, M. E., Proestos, C., & Thomaidis, N. S. (2022). Thorough Investigation of the Phenolic Profile of Reputable Greek Honey Varieties: Varietal Discrimination and Floral Markers Identification Using Liquid Chromatography–High-Resolution Mass Spectrometry. Molecules, 27(14), 4444. https://doi.org/10.3390/molecules27144444










