Untargeted LC-MS/MS-Based Multi-Informative Molecular Networking for Targeting the Antiproliferative Ingredients in Tetradium ruticarpum Fruit
Abstract
:1. Introduction
2. Results and Discussion
2.1. Antiproliferative Activity of Different Extracts of TR Fruit
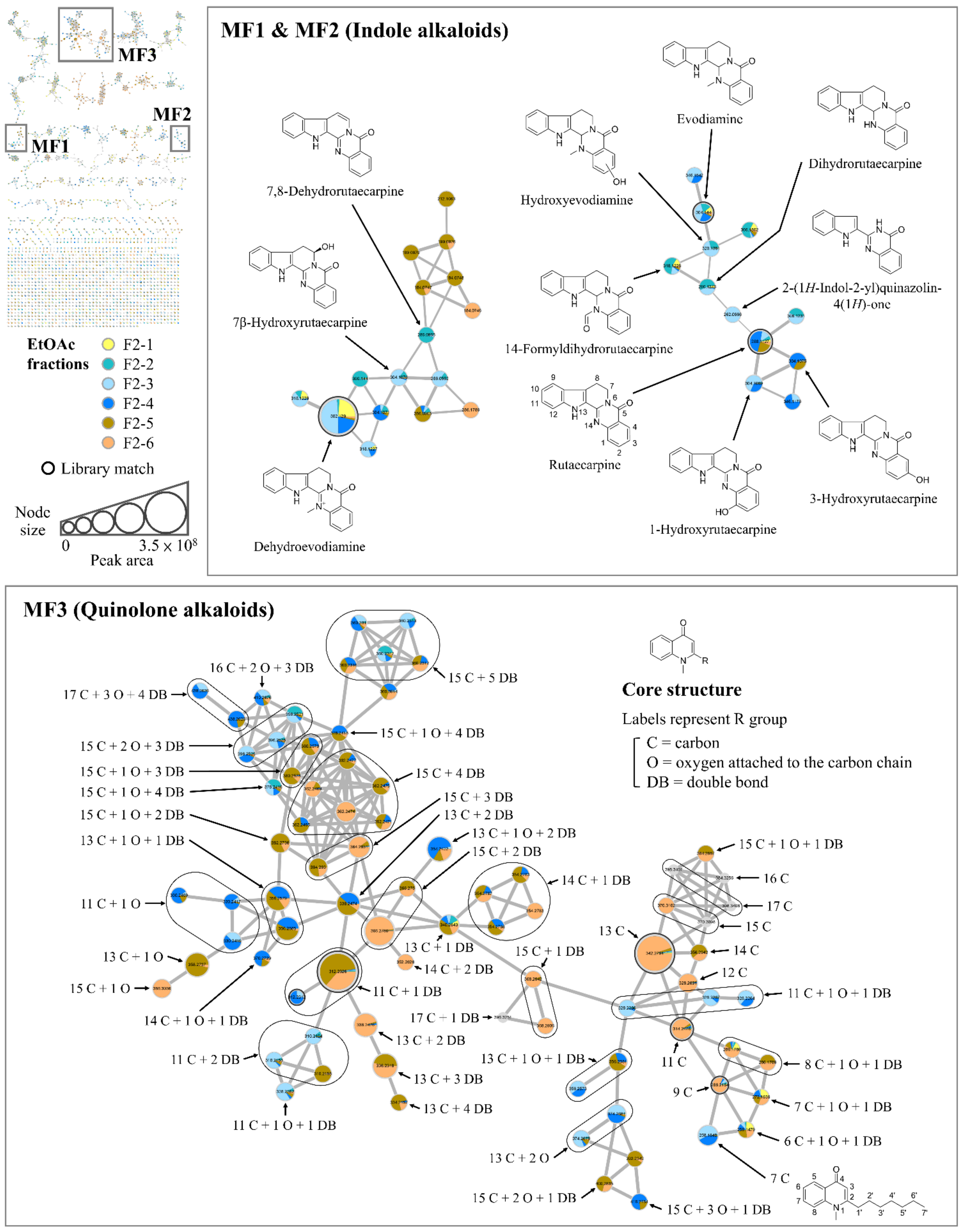
2.2. Overview of the Mass Spectral Molecular Network and Targeted Molecular Families of the EtOAc Fractions of TR Fruit
2.3. Targeting of the Characteristic Compounds of the Bioactive EtOAc Fraction F2-3 by Multi-Informative Molecular Networking
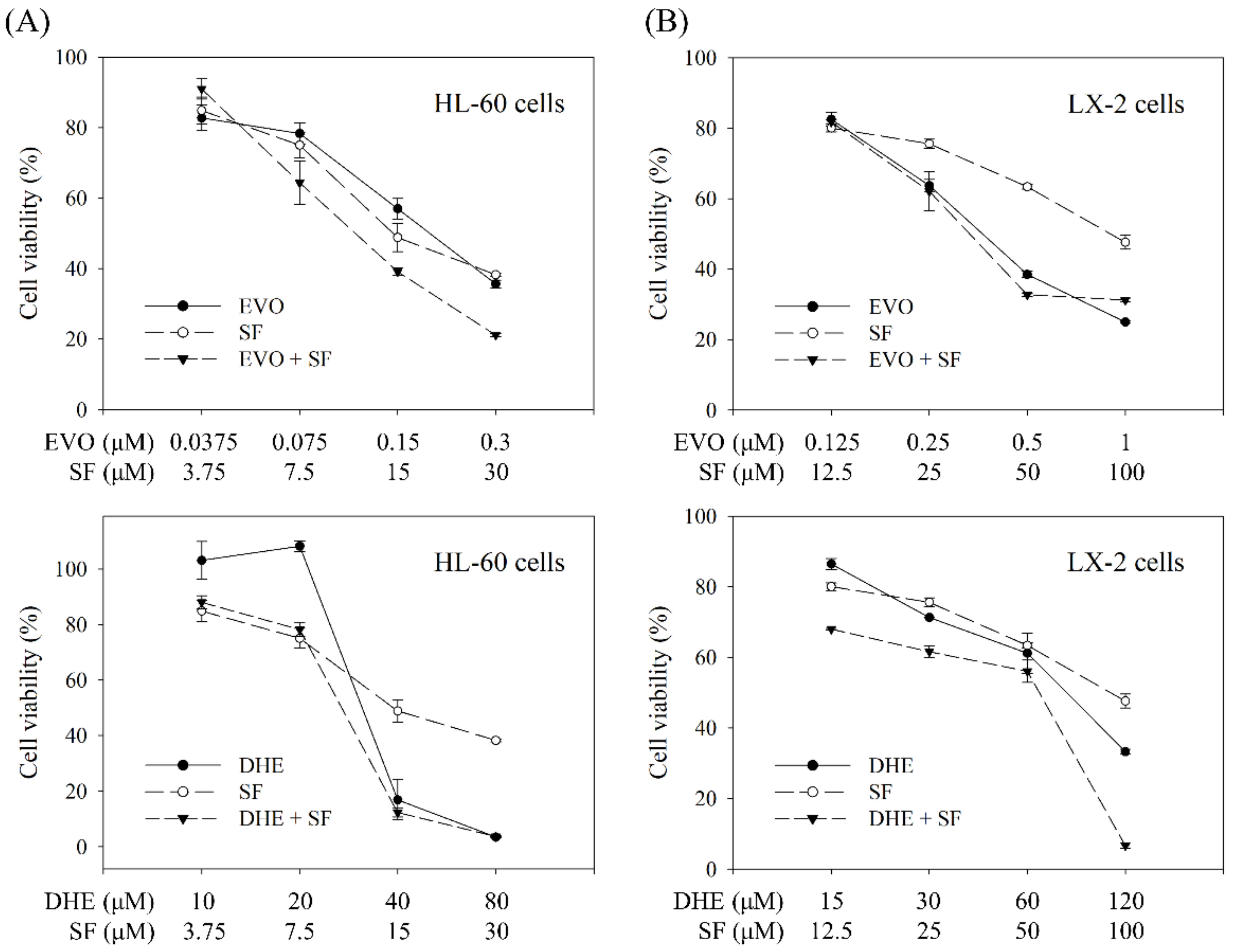
2.4. Individual and Combined Effects of the Targeted Compounds on Cell Viability
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Extraction and Isolation
3.3. LC-MS/MS Analysis
3.4. Compound Annotation
3.5. Molecular Networking
3.5.1. LC-MS/MS Data Preprocessing
3.5.2. Molecular Network Generation Using GNPS
3.6. Quantification of Targeted Compounds
3.7. Cell Culture and Culture Conditions
3.8. Cell Viability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, X.; Li, C.-G.; Chang, D.; Bensoussan, A. Current status and major challenges to the safety and efficacy presented by Chinese herbal medicine. Medicines 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Ma, Q.; Ye, L.; Piao, G. The traditional medicine and modern medicine from natural products. Molecules 2016, 21, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.-S.; Weng, J.-K. Demystifying traditional herbal medicine with modern approach. Nat. Plants 2017, 3, 17109. [Google Scholar] [CrossRef] [PubMed]
- Beniddir, M.A.; Kang, K.B.; Genta-Jouve, G.; Huber, F.; Rogers, S.; van der Hooft, J.J.J. Advances in decomposing complex metabolite mixtures using substructure- and network-based computational metabolomics approaches. Nat. Prod. Rep. 2021, 38, 1967–1993. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Wu, C.-H.; Moree, W.J.; Lamsa, A.; Medema, M.H.; Zhao, X.; Gavilan, R.G.; Aparicio, M.; Atencio, L.; Jackson, C.; et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 2013, 110, E2611–E2620. [Google Scholar] [CrossRef] [Green Version]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef] [PubMed]
- Olivon, F.; Allard, P.-M.; Koval, A.; Righi, D.; Genta-Jouve, G.; Neyts, J.; Apel, C.; Pannecouque, C.; Nothias, L.-F.; Cachet, X.; et al. Bioactive natural products prioritization using massive multi-informational molecular networks. ACS Chem. Biol. 2017, 12, 2644–2651. [Google Scholar] [CrossRef]
- Ministry of Health and Welfare. Taiwan Herbal Pharmacopeia, 4th ed.; Ministry of Health and Welfare: Taipei, Taiwan, 2021.
- Li, M.; Wang, C. Traditional uses, phytochemistry, pharmacology, pharmacokinetics and toxicology of the fruit of Tetradium ruticarpum: A review. J. Ethnopharmacol. 2020, 263, 113231. [Google Scholar] [CrossRef]
- Shan, Q.-Y.; Sang, X.-N.; Hui, H.; Shou, Q.-Y.; Fu, H.-Y.; Hao, M.; Liu, K.-H.; Zhang, Q.-Y.; Cao, G.; Qin, L.-P. Processing and polyherbal formulation of Tetradium ruticarpum (A. Juss.) Hartley: Phytochemistry, pharmacokinetics, and toxicity. Front. Pharmacol. 2020, 11, 133. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Li, D.; Chu, C.; Li, X.; Wang, X.; Jia, Y.; Hua, H.; Xu, F. Antiproliferative effects of alkaloid evodiamine and its derivatives. Int. J. Mol. Sci. 2018, 19, 3403. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Shi, L.; Liang, T.; Ji, L.; Zhang, G.; Shen, Y.; Zhu, F.; Xu, L. Anti-tumor effect of evodiamine by inducing Akt-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 485, 54–61. [Google Scholar] [CrossRef]
- Liao, J.-F.; Chiou, W.-F.; Shen, Y.-C.; Wang, G.-J.; Chen, C.-F. Anti-inflammatory and anti-infectious effects of Evodia rutaecarpa (Wuzhuyu) and its major bioactive components. Chin. Med. 2011, 6, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Wu, D.-Z.; Yuan, J.-Y.; Zhang, R.-R.; Hu, Z.-B. Gastroprotective effect of Fructus Evodiae water extract on ethanol-induced gastric lesions in rats. Am. J. Chin. Med. 2006, 34, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yu, Y.; Choi, S.; Lee, H.; Yu, J.; Lee, J.-H.; Kim, W.-Y. Evodiamine eliminates colon cancer stem cells via suppressing Notch and Wnt signaling. Molecules 2019, 24, 4520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Qu, S.; Shi, Q.; He, D.; Jin, X. Evodiamine induces apoptosis and enhances TRAIL-induced apoptosis in human bladder cancer cells through mTOR/S6K1-mediated downregulation of Mcl-1. Int. J. Mol. Sci. 2014, 15, 3154–3171. [Google Scholar] [CrossRef] [Green Version]
- Yun, U.-J.; Bae, S.-J.; Song, Y.-R.; Kim, Y.-W. A critical YAP in malignancy of HCC is regulated by evodiamine. Int. J. Mol. Sci. 2022, 23, 1855. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, G.; Luan, S.; Luan, C.; Shao, H.; Dong, F.; Liu, X. Evodiamine inhibits the proliferation of leukemia cell line K562 by regulating peroxisome proliferators-activated receptor gamma (PPARγ) pathway. J. Recept. Signal Transduct. Res. 2016, 36, 422–428. [Google Scholar] [CrossRef]
- Huang, W.-C.; Hou, S.-M.; Wu, M.-P.; Hsia, C.-W.; Jayakumar, T.; Hsia, C.-H.; Bhavan, P.S.; Chung, C.-L.; Sheu, J.-R. Decreased human platelet activation and mouse pulmonary thrombosis by rutaecarpine and comparison of the relative effectiveness with BAY11-7082: Crucial signals of p38-NF-κB. Molecules 2022, 27, 476. [Google Scholar] [CrossRef]
- Nie, X.-Q.; Chen, H.-H.; Zhang, J.-Y.; Zhang, Y.-J.; Yang, J.-W.; Pan, H.-J.; Song, W.-X.; Murad, F.; He, Y.-Q.; Bian, K. Rutaecarpine ameliorates hyperlipidemia and hyperglycemia in fat-fed, streptozotocin-treated rats via regulating the IRS-1/PI3K/Akt and AMPK/ACC2 signaling pathways. Acta. Pharmacol. Sin. 2016, 37, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ge, J.; Zheng, Q.; Zhang, J.; Sun, R.; Liu, R. Evodiamine and rutaecarpine from Tetradium ruticarpum in the treatment of liver diseases. Phytomedicine 2020, 68, 153180. [Google Scholar] [CrossRef]
- Chiou, W.-F.; Liao, J.-F.; Chen, C.-F. Comparative study on the vasodilatory effects of three quinazoline alkaloids isolated from Evodia rutaecarpa. J. Nat. Prod. 1996, 59, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.-C.; Wang, Y.-H.; Liou, K.-T.; Chen, C.-M.; Chen, C.-H.; Wang, W.-Y.; Chang, S.; Hou, Y.-C.; Chen, K.-T.; Chen, C.-F.; et al. Anti-inflammatory effects and mechanisms of the ethanol extract of Evodia rutaecarpa and its bioactive components on neutrophils and microglial cells. Eur. J. Pharmacol. 2007, 555, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Bligh, S.W.A.; Smith, E. Quinolone alkaloids from Fructus euodiae show activity against methicillin-resistant Staphylococcus aureus. Phytother. Res. 2014, 28, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Zhang, M.; Zhang, W.; Ma, S.; Chen, G.; Shi, B. Antifungal activity of schinifoline against Candida albicans in Caenorhabditis elegans. Phyton 2019, 88, 317–324. [Google Scholar] [CrossRef]
- To, D.-C.; Bui, T.Q.; Nhung, N.T.A.; Tran, Q.-T.; Do, T.-T.; Tran, M.-H.; Hien, P.-P.; Ngu, T.-N.; Quy, P.-T.; Nguyen, T.-H.; et al. On the inhibitability of natural products isolated from Tetradium ruticarpum towards tyrosine phosphatase 1B (PTP1B) and α-glucosidase (3W37): An in vitro and in silico study. Molecules 2021, 26, 3691. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, C.; Luo, T.; Wang, J.; Tang, Y.; Chen, Z.; Yu, L. Limonin: A review of its pharmacology, toxicity, and pharmacokinetics. Molecules 2019, 24, 3679. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Itagaki, S.; Kurokawa, T.; Ogura, J.; Kobayashi, M.; Hirano, T.; Sugawara, M.; Iseki, K. In vitro and in vivo antioxidant properties of chlorogenic acid and caffeic acid. Int. J. Pharm. 2011, 403, 136–138. [Google Scholar] [CrossRef]
- da Cunha, F.M.; Duma, D.; Assreuy, J.; Buzzi, F.C.; Niero, R.; Campos, M.M.; Calixto, J.B. Caffeic acid derivatives: In vitro and in vivo anti-inflammatory properties. Free Radic. Res. 2004, 38, 1241–1253. [Google Scholar] [CrossRef]
- Hwang, S.J.; Kim, Y.-W.; Park, Y.; Lee, H.-J.; Kim, K.-W. Anti-inflammatory effects of chlorogenic acid in lipopolysaccharide-stimulated RAW 264.7 cells. Inflamm. Res. 2014, 63, 81–90. [Google Scholar] [CrossRef]
- Rasul, A.; Yu, B.; Zhong, L.; Khan, M.; Yang, H.; Ma, T. Cytotoxic effect of evodiamine in SGC-7901 human gastric adenocarcinoma cells via simultaneous induction of apoptosis and autophagy. Oncol. Rep. 2012, 27, 1481–1487. [Google Scholar]
- Huang, X.; Zhang, Y.-B.; Yang, X.-W. Indoloquinazoline alkaloids from Euodia rutaecarpa and their cytotoxic activities. J. Asian Nat. Prod. Res. 2011, 13, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, W.; Yang, X.-W. New cytotoxic quinolone alkaloids from fruits of Evodia rutaecarpa. Fitoterapia 2012, 83, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Li, Z.-L.; Li, D.-H.; Sun, Y.-T.; Shan, D.-T.; Bai, J.; Pei, Y.-H.; Jing, Y.-K.; Hua, H.-M. Quinolone and indole alkaloids from the fruits of Euodia rutaecarpa and their cytotoxicity against two human cancer cell lines. Phytochemistry 2015, 109, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-X.; Zan, K.; Shi, S.-P.; Zeng, K.-W.; Jiang, Y.; Guan, Y.; Xiao, C.-L.; Gao, H.-Y.; Wu, L.-J.; Tu, P.-F. Quinolone alkaloids with antibacterial and cytotoxic activities from the fruits of Evodia rutaecarpa. Fitoterapia 2013, 89, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, X.; Liu, B.; Zhang, L.; Fan, Z.; Ji, Y. Screening and identification of hepatotoxic component in Evodia rutaecarpa based on spectrum–effect relationship and UPLC-Q-TOFMS. Biomed. Chromatogr. 2016, 30, 1975–1983. [Google Scholar] [CrossRef]
- Yang, S.; Tian, M.; Yuan, L.; Deng, H.; Wang, L.; Li, A.; Hou, Z.; Li, Y.; Zhang, Y. Analysis of E. rutaecarpa alkaloids constituents in vitro and in vivo by UPLC-Q-TOF-MS combined with diagnostic fragment. J. Anal. Methods Chem. 2016, 2016, 4218967. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.K.; Lee, D.W.; Jeon, T.W.; Jin, C.H.; Kim, G.H.; Jun, I.H.; Lee, D.J.; Kim, S.I.; Kim, D.H.; Jahng, Y.; et al. Characterization of the Phase II metabolites of rutaecarpine in rat by liquid chromatography-electrospray ionization-tandem mass spectrometry. Xenobiotica 2005, 35, 1135–1145. [Google Scholar] [CrossRef]
- Cabral, R.S.A.; Allard, P.-M.; Marcourt, L.; Young, M.C.M.; Queiroz, E.F.; Wolfender, J.-L. Targeted isolation of indolopyridoquinazoline alkaloids from Conchocarpus fontanesianus based on molecular networks. J. Nat. Prod. 2016, 79, 2270–2278. [Google Scholar] [CrossRef]
- Ling, Y.; Hu, P.; Zhang, L.; Jin, H.; Chen, J.; Tao, Z.; Huang, L.; Ren, R. Identification and structural characterization of acylgluconic acids, flavonol glycosides, limonoids and alkaloids from the fruits of Evodia rutaecarpa by high performance liquid chromatography coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry. J. Chromatogr. Sci. 2016, 54, 1593–1604. [Google Scholar]
- Sun, Q.; Xie, L.; Song, J.; Li, X. Evodiamine: A review of its pharmacology, toxicity, pharmacokinetics and preparation researches. J. Ethnopharmacol. 2020, 262, 113164. [Google Scholar] [CrossRef]
- Dong, G.; Wang, S.; Miao, Z.; Yao, J.; Zhang, Y.; Guo, Z.; Zhang, W.; Sheng, C. New tricks for an old natural product: Discovery of highly potent evodiamine derivatives as novel antitumor agents by systemic structure–activity relationship analysis and biological evaluations. J. Med. Chem. 2012, 55, 7593–7613. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Bi, K.; Wu, S.; Li, Y.; Huang, Y.; Sheng, C.; Dong, G. Water-soluble derivatives of evodiamine: Discovery of evodiamine-10-phosphate as an orally active antitumor lead compound. Eur. J. Med. Chem. 2021, 220, 113544. [Google Scholar] [CrossRef] [PubMed]
- Baruah, B.; Dasu, K.; Vaitilingam, B.; Mamnoor, P.; Venkata, P.P.; Rajagopal, S.; Yeleswarapu, K.R. Synthesis and cytotoxic activity of novel quinazolino-β-carboline-5-one derivatives. Bioorg. Med. Chem. 2004, 12, 1991–1994. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Pena-Pereira, F.; Wojnowski, W.; Tobiszewski, M. AGREE—Analytical GREEnness metric approach and software. Anal. Chem. 2020, 92, 10076–10082. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [Green Version]
- van der Hooft, J.J.J.; Wandy, J.; Barrett, M.P.; Burgess, K.E.V.; Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. USA 2016, 113, 13738–13743. [Google Scholar] [CrossRef] [Green Version]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Ernst, M.; Kang, K.B.; Caraballo-Rodriguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liu, X.; Shan, Y.; Xu, S.; Feng, X.; Wang, Q.-Z. A new quinolone alkaloid from the fruits of Tetradium ruticarpum. Nat. Prod. Res. 2021, 35, 222–227. [Google Scholar] [CrossRef]

| Sample | IC50 (μg/mL) | ||
|---|---|---|---|
| HL-60 | T24 | LX-2 | |
| 50% EtOH extract | 0.45 ± 0.14 | 3.31 ± 0.43 | 5.66 ± 0.24 |
| 50% MeOH extract | >20 | 18.68 ± 0.83 | >20 |
| MeOH extract | 0.39 ± 0.05 | 0.54 ± 0.15 | 3.49 ± 1.47 |
| EtOAc extract | 0.23 ± 0.14 | 0.32 ± 0.27 | 0.47 ± 0.03 |
| Hexane extract | 0.45 ± 0.09 | 1.49 ± 0.15 | 3.56 ± 0.74 |
| Fractions of EtOAc extract | |||
| F1 | >20 | >20 | >20 |
| F2 | 0.80 ± 0.03 | 5.99 ± 0.68 | 8.46 ± 2.10 |
| F2-1 | 5.32 ± 0.12 | 19.01 ± 0.26 | >20 |
| F2-2 | 2.77 ± 0.21 | 14.74 ± 0.55 | 14.19 ± 1.13 |
| F2-3 | 0.07 ± 0.01 | 0.15 ± 0.05 | 0.39 ± 0.07 |
| F2-4 | 1.98 ± 0.17 | 2.98 ± 0.73 | 2.58 ± 0.28 |
| F2-5 | 1.96 ± 0.03 | 10.97 ± 0.78 | 10.43 ± 1.32 |
| F2-6 | 0.51 ± 0.03 | 2.72 ± 0.09 | 2.17 ± 0.05 |
| F3 | 2.70 ± 0.90 | 10.64 ± 1.18 | >20 |
| F4 | 0.73 ± 0.02 | 2.82 ± 0.14 | 2.17 ± 0.19 |
| F5 | 3.25 ± 0.05 | 3.52 ± 0.42 | 7.77 ± 0.12 |
| Doxorubicin | 0.17 ± 0.06 | 0.13 ± 0.03 | 0.10 ± 0.03 |
| No. | RT (Min) | Name | Molecular Formula | Calculated m/z [M+H]+ | Observed m/z [M+H]+ | Error (ppm) | Cosine Score a | Fragment Ions (Relative Abundance in %) b | Peak Area (%) c | Ref. d |
|---|---|---|---|---|---|---|---|---|---|---|
| Indole alkaloids | ||||||||||
| 1 | 6.22 | Dehydroevodiamine | C19H15N3O | 302.1288 | 302.1290 | 0.66 | T: 0.996 | 302.1284 (4), 287.1056 (8), 286.0978 (100), 272.0810 (2), 258.1022 (3) | 19.04 | R; T; Yang et al. (2016) [37] |
| 2 | 8.87 | Evodiamine | C19H17N3O | 304.1444 | 304.1440 | −1.32 | T: 0.988 G: 0.857 | 171.0920 (38), 161.0710 (22), 154.0650 (8), 144.0809 (26), 134.0603 (100), 116.0499 (55), 106.0655 (59), 91.0546 (48), 79.0546 (25), 77.0389 (21) | 3.31 | R; T; G; Ling et al. (2016) [40] |
| 3 | 7.93 | 1-Hydroxyrutaecarpine | C18H13N3O2 | 304.1081 | 304.1069 | −3.95 | NA | 304.1081 (100), 302.0919 (17), 289.0844 (31), 287.0812 (22), 261.1007 (11), 260.0814 (15), 169.0758 (20), 161.0343 (14), 136.0393 (9), 142.0650 (15) | 1.29 | M; S; Zhao et al. (2015) [34]; Cabral et al. (2016) [39] |
| 4 | 7.69 | 7β-Hydroxyrutaecarpine | C18H13N3O2 | 304.1081 | 304.1076 | −1.64 | NA | 286.0975 (100), 285.0898 (21), 258.1027 (28), 257.0949 (30), 167.0606 (43), 140.0493 (10), 130.0650 (6) | 1.10 | M; S; Li et al. (2016) [36] |
Quinolone alkaloids Compound name represents R group. C = Carbon; O = Oxygen attached to the carbon chain; DB = Double bond Compound name represents R group. C = Carbon; O = Oxygen attached to the carbon chain; DB = Double bond | ||||||||||
| 5 | 9.01 | 7 C (Schinifoline) | C17H23NO | 258.1852 | 258.1848 | −1.55 | NA | 258.1847 (7), 186.0911 (16), 173.0840 (100), 172.0752 (5), 158.0599 (14), 144.0802 (5), 132.0570 (14), 131.0492 (6) | 4.21 | R; Wang et al. (2013) [35] |
| 6 | 8.12 | 11 C + 1 O +1 DB | C21H29NO2 | 328.2271 | 328.2262 | −2.74 | NA | 310.2162 (20), 200.1065 (5), 187.0986 (8), 186.0915 (80), 173.0838 (100), 159.0674 (7) | 3.17 | M; S |
| 7 | 8.84 9.01 | 11 C + 1 O +1 DB | C21H29NO2 | 328.2271 | 328.2266 328.2267 | −1.52 −1.22 | NA | 328.2270 (69), 200.1065 (5), 186.0914 (69), 173.0838 (100), 172.0752 (5) | 1.31 1.30 | M; S |
| 8 | 7.80 8.02 | 13 C + 2 O | C23H35NO3 | 374.2690 | 374.2678 374.2681 | −3.21 −2.40 | NA | 356.2585 (100), 338.2481 (28), 286.1793 (6), 200.1066 (5), 186.0915 (42), 173.0838 (56) | 1.16 2.32 | M; S |
| 9 | 7.90 | 15 C + 2 O + 3 DB | C25H33NO3 | 396.2533 | 396.2528 | −1.26 | NA | 212.1064 (4), 200.1066 (7), 187.0985 (9), 186.0915 (100), 173.0839 (60) | 1.55 | M; S |
| Compound | IC50 (μM) | |
|---|---|---|
| HL-60 | LX-2 | |
| Evodiamine | 0.21 ± 0.02 | 0.39 ± 0.02 |
| Dehydroevodiamine | 33.58 ± 1.61 | 80.82 ± 6.51 |
| Schinifoline | 14.83 ± 2.02 | 92.28 ± 7.97 |
| Doxorubicin | 0.32 ± 0.10 | 0.18 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, C.-H.; Cheng, Y.-C.; Chang, Y.-C.; Kung, T.-H.; Chen, Y.-L.; Lai, K.-H.; Hsieh, H.-L.; Chen, C.-Y.; Hwang, T.-L.; Yang, Y.-L. Untargeted LC-MS/MS-Based Multi-Informative Molecular Networking for Targeting the Antiproliferative Ingredients in Tetradium ruticarpum Fruit. Molecules 2022, 27, 4462. https://doi.org/10.3390/molecules27144462
Su C-H, Cheng Y-C, Chang Y-C, Kung T-H, Chen Y-L, Lai K-H, Hsieh H-L, Chen C-Y, Hwang T-L, Yang Y-L. Untargeted LC-MS/MS-Based Multi-Informative Molecular Networking for Targeting the Antiproliferative Ingredients in Tetradium ruticarpum Fruit. Molecules. 2022; 27(14):4462. https://doi.org/10.3390/molecules27144462
Chicago/Turabian StyleSu, Chun-Han, Yu-Chieh Cheng, Yu-Chia Chang, Ting-Hsuan Kung, Yu-Li Chen, Kuei-Hung Lai, Hsi-Lung Hsieh, Chun-Yu Chen, Tsong-Long Hwang, and Yu-Liang Yang. 2022. "Untargeted LC-MS/MS-Based Multi-Informative Molecular Networking for Targeting the Antiproliferative Ingredients in Tetradium ruticarpum Fruit" Molecules 27, no. 14: 4462. https://doi.org/10.3390/molecules27144462







