Human and Bacterial Toll-Interleukin Receptor Domains Exhibit Distinct Dynamic Features and Functions
Abstract
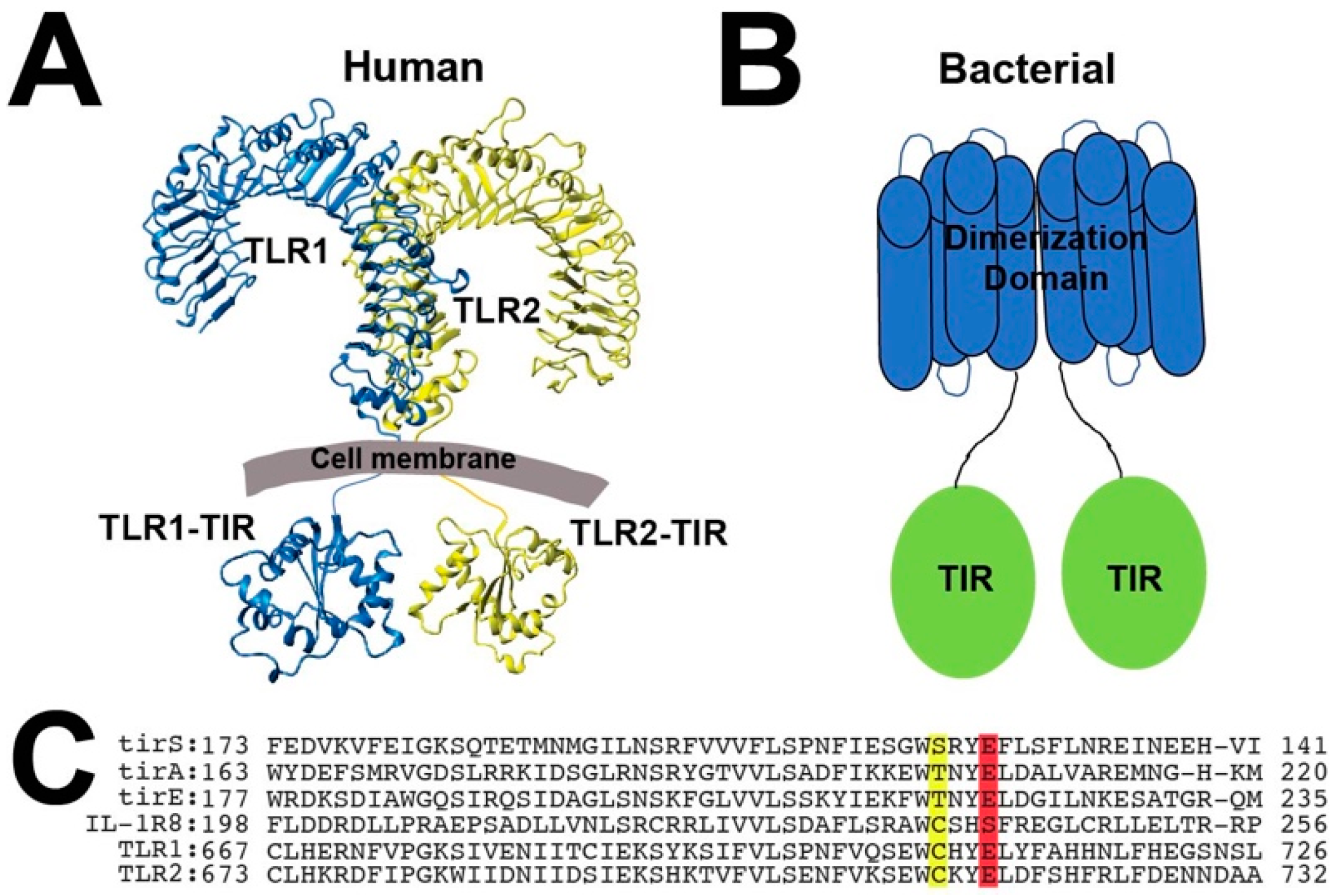
:1. Introduction
2. Results
2.1. Bacterial Proteins of tirS, tirA, and tirE All Cleave NAD+ and Related Coenzymes
2.2. Bacterial TIR Domains of tirS, tirA, and tirE Undergo Chemical Exchange with Dimerization Modulated by Their Coil-Coil Domains
2.3. The CC Domain Is Not Completely Helical
2.4. TLR1 and TLR2 TIR Domains form Disulfide-Linked Homodimers
2.5. TLR1 and TLR2 TIR Domains Form Disulfide-Linked Heterodimers
2.6. Mammalian Pro-inflammatory TIR Domains Exhibit a Range of Chemical Exchange

2.7. The IL-1R8-TIR Domain Undergoes a Slow Exchange
3. Discussion
4. Materials and Methods
4.1. Annotated Proteins and Expression Plasmids
4.2. Protein Expression and Purification
4.3. Nuclear Magnetic Resonance
4.4. Cross-Linking and GST Pull-Down Experiments
4.5. Mass Spectrometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Sato, Y.; Goto, Y.; Narita, N.; Hoon, D.S.B. Cancer Cells Expressing Toll-like Receptors and the Tumour Microenvironment. Cancer Microenviron. 2009, 2 (Suppl. S1), 204–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grivennikov, S.I.; Karin, M. Inflammation and oncogenesis: A vicious connection. Curr. Opin. Genet. Dev. 2010, 20, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayanan, K.B.; Park, H.H. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis 2015, 20, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Nyman, T.; Stenmark, P.; Flodin, S.; Johansson, I.; Hammarstrom, M.; Nordlund, P. The crystal structure of the human Toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J. Biol. Chem. 2008, 283, 11861–11865. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.W.; Tao, X.; Shen, B.H.; Horng, T.; Medzhitov, R.; Manley, J.L.; Tong, L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 2000, 408, 111–115. [Google Scholar] [CrossRef]
- Lin, Z.J.; Lu, J.; Zhou, W.H.; Shen, Y.Q. Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling. PLoS ONE 2012, 7, e34202. [Google Scholar] [CrossRef] [Green Version]
- Vyncke, L.; Bovijn, C.; Pauwels, E.; Van Acker, T.; Ruyssinck, E.; Burg, E.; Tavernier, J.; Peelman, F. Reconstructing the TIR Side of the Myddosome: A Paradigm for TIR-TIR Interactions. Structure 2016, 24, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Nada, M.; Ohnishi, H.; Tochio, H.; Kato, Z.; Kimura, T.; Kubota, K.; Yamamoto, T.; Kamatari, Y.O.; Tsutsumi, N.; Shirakawa, M.; et al. Molecular analysis of the binding mode of Toll/interleukin-1 receptor (TIR) domain proteins during TLR2 signaling. Mol. Immunol. 2012, 52, 108–116. [Google Scholar] [CrossRef]
- Carlsson, E.; Ding, J.L.; Byrne, B. SARM modulates MyD88-mediated TLR activation through BB-loop dependent TIR-TIR interactions. Biochim. Biophys. Acta-Mol. Cell Res. 2016, 1863, 244–253. [Google Scholar] [CrossRef]
- Rana, R.R.; Simpson, P.; Zhang, M.H.; Jennions, M.; Ukegbu, C.; Spear, A.M.; Alguel, Y.; Matthews, S.J.; Atkins, H.S.; Byrne, B. Yersinia pestis TIR-domain protein forms dimers that interact with the human adaptor protein MyD88. Microb. Pathog. 2011, 51, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Snyder, G.A.; Cirl, C.; Jiang, J.S.; Chen, K.; Waldhuber, A.; Smith, P.; Rommler, F.; Snyder, N.; Fresquez, T.; Durr, S.; et al. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 6985–6990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldhuber, A.; Snyder, G.A.; Rommler, F.; Cirl, C.; Muller, T.; Xiao, T.S.; Svanborg, C.; Miethke, T. A Comparative Analysis of the Mechanism of Toll-Like Receptor-Disruption by TIR-Containing Protein C from Uropathogenic Escherichia coli. Pathogens 2016, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Nimma, S.; Ve, T.; Williams, S.J.; Kobe, B. Towards the structure of the TIR-domain signalosome. Curr. Opin. Struct. Biol. 2017, 43, 122–130. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.S.; Kim, S.E.; Heo, J.Y.; Lee, M.E.; Kim, H.M.; Paik, S.G.; Lee, H.Y.; Lee, J.O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [Google Scholar] [CrossRef] [Green Version]
- Nanson, J.D.; Kobe, B.; Ve, T. Death, TIR, and RHIM: Self-assembling domains involved in innate immunity and cell-death signaling. J. Leukoc. Biol. 2019, 105, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Lo, Y.C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef] [Green Version]
- Sporny, M.; Guez-Haddad, J.; Lebendiker, M.; Ulisse, V.; Volf, A.; Mim, C.; Isupov, M.N.; Opatowsky, Y. Structural Evidence for an Octameric Ring Arrangement of SARM1. J. Mol. Biol. 2019, 431, 3591–3605. [Google Scholar] [CrossRef]
- Bratkowski, M.; Xie, T.; Thayer, D.A.; Lad, S.; Mathur, P.; Yang, Y.S.; Danko, G.; Burdett, T.C.; Danao, J.; Cantor, A.; et al. Structural and Mechanistic Regulation of the Pro-degenerative NAD Hydrolase SARM1. Cell Rep. 2020, 32, 107999. [Google Scholar] [CrossRef]
- Sporny, M.; Guez-Haddad, J.; Khazma, T.; Yaron, A.; Dessau, M.; Shkolnisky, Y.; Mim, C.; Isupov, M.N.; Zalk, R.; Hons, M.; et al. Structural basis for SARM1 inhibition and activation under energetic stress. Elife 2020, 9, e62021. [Google Scholar] [CrossRef]
- Figley, M.D.; Gu, W.X.; Nanson, J.D.; Shi, Y.; Sasaki, Y.; Cunnea, K.; Malde, A.K.; Jia, X.Y.; Luo, Z.Y.; Saikot, F.K.; et al. SARM1 is a metabolic sensor activated by an increased NMN/NAD(+) ratio to trigger axon degeneration. Neuron 2021, 109, 1118–1136. [Google Scholar] [CrossRef] [PubMed]
- Ve, T.; Vajjhala, P.R.; Hedger, A.; Croll, T.; DiMaio, F.; Horsefield, S.; Yu, X.; Lavrencic, P.; Hassan, Z.; Morgan, G.P.; et al. Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat. Struct. Mol. Biol. 2017, 24, 743–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirey, K.A.; Lai, W.; Brown, L.J.; Blanco, J.C.G.; Beadenkopf, R.; Wang, Y.J.; Vogel, S.N.; Snyder, G.A. Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease. Innate Immun. 2020, 26, 26–34. [Google Scholar] [CrossRef]
- Jang, T.H.; Park, H.H. Crystal Structure of TIR Domain of TLR6 Reveals Novel Dimeric Interface of TIR-TIR Interaction for Toll-Like Receptor Signaling Pathway. J. Mol. Biol. 2014, 426, 3305–3313. [Google Scholar] [CrossRef] [PubMed]
- Li, F.J.; Starrs, L.; Burgio, G. Tug of war between Acinetobacter baumannii and host immune responses. Pathog. Dis. 2018, 76, ftz004. [Google Scholar] [CrossRef] [Green Version]
- Newman, R.M.; Salunkhe, P.; Godzik, A.; Reed, J.C. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 2006, 74, 594–601. [Google Scholar] [CrossRef] [Green Version]
- Low, L.Y.; Mukasa, T.; Reed, J.C.; Pascual, J. Characterization of a TIR-like protein from Paracoccus denitrificans. Biochem. Biophys. Res. Commun. 2007, 356, 481–486. [Google Scholar] [CrossRef] [Green Version]
- Askarian, F.; van Sorge, N.M.; Sangvik, M.; Beasley, F.C.; Henriksen, J.R.; Sollid, J.U.E.; van Strijp, J.A.G.; Nizet, V.; Johannessen, M. A Staphylococcus aureus TO Domain Protein Virulence Factor Blocks TLR2-Mediated NF-KB Signaling. J. Innate Immun. 2014, 6, 485–498. [Google Scholar] [CrossRef]
- Patot, S.; Imbert, P.R.C.; Baude, J.; Simoes, P.M.; Campergue, J.B.; Louche, A.; Nijland, R.; Bes, M.; Tristan, A.; Laurent, F.; et al. The TIR Homologue Lies near Resistance Genes in Staphylococcus aureus, Coupling Modulation of Virulence and Antimicrobial Susceptibility. PLoS Pathog. 2017, 13, e1006092. [Google Scholar]
- Horsefield, S.; Burdett, H.; Zhang, X.X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.C.; Gilley, J.; Lai, J.S.; Rank, M.X.; et al. NAD(+) cleavage activity by animal an plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef] [Green Version]
- Essuman, K.; Summers, D.W.; Sasaki, Y.; Mao, X.R.; Yim, A.K.Y.; DiAntonio, A.; Milbrandt, J. TIR Domain Proteins Are an Ancient Family of NAD(+)-Consuming Enzymes. Curr. Biol. 2018, 28, 421–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, L.A.J. SIGIRR puts the brakes on Toll-like receptors. Nat. Immunol. 2003, 4, 823–824. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.; Low, L.Y.; Hsu, S.; Li, S.; Liu, T.; Santelli, E.; Le Negrate, G.; Reed, J.C.; Woods, V.L.; Pascual, J. Molecular Mimicry in Innate Immunity CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN. J. Biol. Chem. 2009, 284, 21386–21392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alaidarous, M.; Ve, T.; Casey, L.W.; Valkov, E.; Ericsson, D.J.; Ullah, M.O.; Schembri, M.A.; Mansell, A.; Sweet, M.J.; Kobe, B. Mechanism of Bacterial Interference with TLR4 Signaling by Brucella Toll/Interleukin-1 Receptor Domain-containing Protein TcpB. J. Biol. Chem. 2014, 289, 654–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, G.A.; Deredge, D.; Waldhuber, A.; Fresquez, T.; Wilkins, D.Z.; Smith, P.T.; Durr, S.; Cirl, C.; Jiang, J.S.; Jennings, W.; et al. Crystal Structures of the Toll/Interleukin-1 Receptor (TIR) Domains from the Brucella Protein TcpB and Host Adaptor TIRAP Reveal Mechanisms of Molecular Mimicry. J. Biol. Chem. 2014, 289, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Hafsa, N.E.; Wishart, D.S. CSI 2.0: A significantly improved version of the Chemical Shift Index. J. Biomol. NMR 2014, 60, 131–146. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Lange, O.F.; Rossi, P.; Sgourakis, N.G.; Song, Y.F.; Lee, H.W.; Aramini, J.M.; Ertekin, A.; Xiao, R.; Acton, T.B.; Montelione, G.T.; et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl. Acad. Sci. USA 2012, 109, 10873–10878. [Google Scholar] [CrossRef] [Green Version]
- Kimple, M.E.; Siderovski, D.P.; Sondek, J. Functional relevance of the disulfide-linked complex of the N-terminal PDZ domain of InaD with NorpA. Embo J. 2001, 20, 4414–4422. [Google Scholar] [CrossRef] [Green Version]
- Holcomb, J.; Jiang, Y.Y.; Guan, X.Q.; Trescott, L.; Lu, G.R.; Hou, Y.N.; Wang, S.; Brunzelle, J.; Sirinupong, N.; Li, C.Y.; et al. Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization. Biochem. Biophys. Res. Commun. 2014, 448, 169–174. [Google Scholar] [CrossRef]
- Spellmon, N.; Holcomb, J.; Niu, A.; Choudhary, V.; Sun, X.; Zhang, Y.; Wan, J.; Doughan, M.; Haden, S.; Hachem, F.; et al. Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF. Biochem. Biophys. Res. Commun. 2017, 485, 529–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, D.; Song, L.; Geng, S.Z.; Jiao, Y.; Zhou, X.H.; Song, H.Q.; Kang, X.L.; Zhou, Y.; Xu, X.L.; Sun, J.; et al. Salmonella Coiled-Coil- and TIR-Containing TcpS Evades the Innate Immune System and Subdues Inflammation. Cell Rep. 2019, 28, 804–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushpa, V.A.; Goncharuk, M.V.; Lin, C.; Zalevsky, A.O.; Talyzina, I.A.; Luginina, A.P.; Vakhrameev, D.D.; Shevtsov, M.B.; Goncharuk, S.A.; Arseniev, A.S.; et al. Modulation of Toll-like receptor 1 intracellular domain structure and activity by Zn2+ ions. Commun. Biol. 2021, 4, 1003. [Google Scholar] [CrossRef]
- Hughes, M.M.; Lavrencic, P.; Coll, R.C.; Ve, T.; Ryan, D.G.; Williams, N.C.; Menon, D.; Mansell, A.; Board, P.G.; Mobli, M.; et al. Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E6480–E6489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kendrick, A.A.; Holliday, M.; Isern, N.G.; Zhang, F.; Camilloni, C.; Huynh, C.; Vendruscolo, M.; Armstrong, G.S.; Eisenmesser, E.Z. The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide. Protein Sci. 2014, 23, 464–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, G.; Martinez, G.; Schleucher, J.; Wijmenga, S.S. Detection of nan-second internal motion and determination of the overall tumbling times independent of the time scale of internal motion in proteins from NMR relaxation data. J. Biomol. NMR 2003, 27, 291–312. [Google Scholar] [CrossRef] [PubMed]
- Nold, C.; Nold, M.; Rudloff, I.; Lo, C.; Mangan, N.; Rotter, B.; Zepp, J.; Azam, T.; Li, S.Z.; Mansell, A.; et al. Interleukin 37 employs the IL-1 family orphan receptor SIGIRR and the alpha chain of the IL-18 receptor to inhibit innate immunity. J. Immunol. 2013, 190, 354–365. [Google Scholar]
- Eisenmesser, E.Z.; Gottschlich, A.; Redzic, J.S.; Paukovich, N.; Nix, J.C.; Azam, T.; Zhang, L.; Zhao, R.; Kieft, J.S.; The, E.; et al. Interleukin-37 monomer is the active form for reducing innate immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 5514–5522. [Google Scholar] [CrossRef] [Green Version]
- Lech, M.; Garlanda, C.; Mantovani, A.; Kirschning, C.J.; Schlondorff, D.; Anders, H.J. Different roles of TIR8/SIGIRR on toll-like receptor signaling in intrarenal antigen-presenting cells and tubular epithelial cells. Kidney Int. 2007, 72, 182–192. [Google Scholar] [CrossRef] [Green Version]
- Moreau, C.; Kirchberger, T.; Zhang, B.; Thomas, M.P.; Weber, K.; Guse, A.H.; Potter, B.V.L. Aberrant Cyclization Affords a C-6 Modified Cyclic Adenosine 5 ‘-Diphosphoribose Analogue with Biological Activity in Jurkat T Cells. J. Med. Chem. 2012, 55, 1478–1489. [Google Scholar] [CrossRef]
- Bentham, A.R.; Zdrzalek, R.; De la Concepcion, J.C.; Banfield, M.J. Uncoiling CNLs: Structure/Function Approaches to Understanding CC Domain Function in Plant NLRs. Plant Cell Physiol. 2018, 59, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Vijay, K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018, 59, 391–412. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, L.; Chin, C.C.; Rajarathnam, K. Role of intramolecular disulfides in stability and structure of a noncovalent homodimer. Biophys. J. 2007, 93, 2129–2134. [Google Scholar] [CrossRef] [Green Version]
- Plugis, N.M.; Weng, N.; Zhao, Q.; Palanski, B.A.; Maecker, H.T.; Habtezion, A.; Khosla, C. Interleukin 4 is inactivated via selective disulfide-bond reduction by extracellular. Proc. Natl. Acad. Sci. USA 2018, 115, 8781–8786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karki, R.; Zhang, Y.; Igwe, O.J. Activation of c-Src: A hub for exogenous pro-oxidant-mediated activation of Toll-like receptor 4 signaling. Free. Radic. Biol. Med. 2014, 71, 256–269. [Google Scholar] [CrossRef] [Green Version]
- Goldring, M.B.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 471–478. [Google Scholar] [CrossRef]
- Tamaki, Y.; Takakubo, Y.; Hirayama, T.; Konttinen, Y.T.; Goodman, S.B.; Yamakawa, M.; Takagi, M. Expression of Toll-like Receptors and Their Signaling Pathways in Rheumatoid Synovitis. J. Rheumatol. 2011, 38, 810–820. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.J.; Wang, Y.S.; Huang, Y.F.; Yang, H.Z.; Wang, J.P.; Hu, Z.W. Toll-like receptor 2 mediates invasion via activating NF-kappa B in MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 2009, 379, 1027–1032. [Google Scholar] [CrossRef]
- Tye, H.; Kennedy, C.L.; Najdovska, M.; McLeod, L.; McCormack, W.; Hughes, N.; Dev, A.; Sievert, W.; Ooi, C.H.; Ishikawa, T.; et al. STAT3-Driven Upregulation of TLR2 Promotes Gastric Tumorigenesis Independent of Tumor Inflammation. Cancer Cell 2012, 22, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Lv, X.X.; Hua, F.; Lin, H.; Sun, W.; Cao, W.B.; Fu, X.M.; Xie, J.; Yu, J.J.; Li, Z.; et al. Targeting acute myeloid leukemia with a proapoptotic peptide conjugated to a toll-like receptor 2-mediated cell-penetrating peptide. Int. J. Cancer 2014, 134, 692–702. [Google Scholar] [CrossRef] [Green Version]
- Uno, K.; Kato, K.; Shimosegawa, T. Novel role of toll-like receptors in Helicobacter pylori—Induced gastric malignancy. World J. Gastroenterol. 2014, 20, 5244–5251. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Overview of the interleukin-1 family of ligands and receptors. Semin. Immunol. 2013, 25, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Paukovich, N.; Xue, M.J.; Elder, J.R.; Redzic, J.S.; Blue, A.; Pike, H.; Miller, B.G.; Pitts, T.M.; Pollock, D.D.; Hansen, K.; et al. Biliverdin Reductase B Dynamics Are Coupled to Coenzyme Binding. J. Mol. Biol. 2018, 430, 3234–3250. [Google Scholar] [CrossRef] [PubMed]
- Paukovich, N.; Redzic, J.S.; Chi, C.; Rahkola, J.T.; Issaian, A.; Blue, A.; Hansen, K.C.; Janoff, E.N.; Eisenmesser, E. Streptococcus pneumoniae G5 domains bind different ligands. Protein Sci. 2019, 28, 1797–1805. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Rahkola, J.; Redzic, J.S.; Chi, Y.C.; Tran, N.; Holyoak, T.; Zheng, H.J.; Janoff, E.; Eisenmesser, E. Mechanism and inhibition of Streptococcus pneumoniae IgA1 protease. Nat. Commun. 2020, 11, 6063. [Google Scholar] [CrossRef]
- Ranjbarian, F.; Sharma, S.; Falappa, G.; Taruschio, W.; Chabes, A.; Hofer, A. Isocratic HPLC analysis for the simultaneous determination of dNTPs, rNTPs and ADP in biological samples. Nucleic Acids Res. 2022, 50, e18. [Google Scholar] [CrossRef]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic Analysis and Visualization Engine for LC-MS Data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef] [Green Version]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.; Redzic, J.S.; Nemkov, T.; Saviola, A.J.; Dzieciatkowska, M.; Hansen, K.C.; D’Alessandro, A.; Dinarello, C.; Eisenmesser, E.Z. Human and Bacterial Toll-Interleukin Receptor Domains Exhibit Distinct Dynamic Features and Functions. Molecules 2022, 27, 4494. https://doi.org/10.3390/molecules27144494
Lee E, Redzic JS, Nemkov T, Saviola AJ, Dzieciatkowska M, Hansen KC, D’Alessandro A, Dinarello C, Eisenmesser EZ. Human and Bacterial Toll-Interleukin Receptor Domains Exhibit Distinct Dynamic Features and Functions. Molecules. 2022; 27(14):4494. https://doi.org/10.3390/molecules27144494
Chicago/Turabian StyleLee, Eunjeong, Jasmina S. Redzic, Travis Nemkov, Anthony J. Saviola, Monika Dzieciatkowska, Kirk C. Hansen, Angelo D’Alessandro, Charles Dinarello, and Elan Z. Eisenmesser. 2022. "Human and Bacterial Toll-Interleukin Receptor Domains Exhibit Distinct Dynamic Features and Functions" Molecules 27, no. 14: 4494. https://doi.org/10.3390/molecules27144494





