Transcriptomics and Metabolomics Analyses Reveal Defensive Responses and Flavonoid Biosynthesis of Dracaena cochinchinensis (Lour.) S. C. Chen under Wound Stress in Natural Conditions
Abstract
:1. Introduction
2. Results
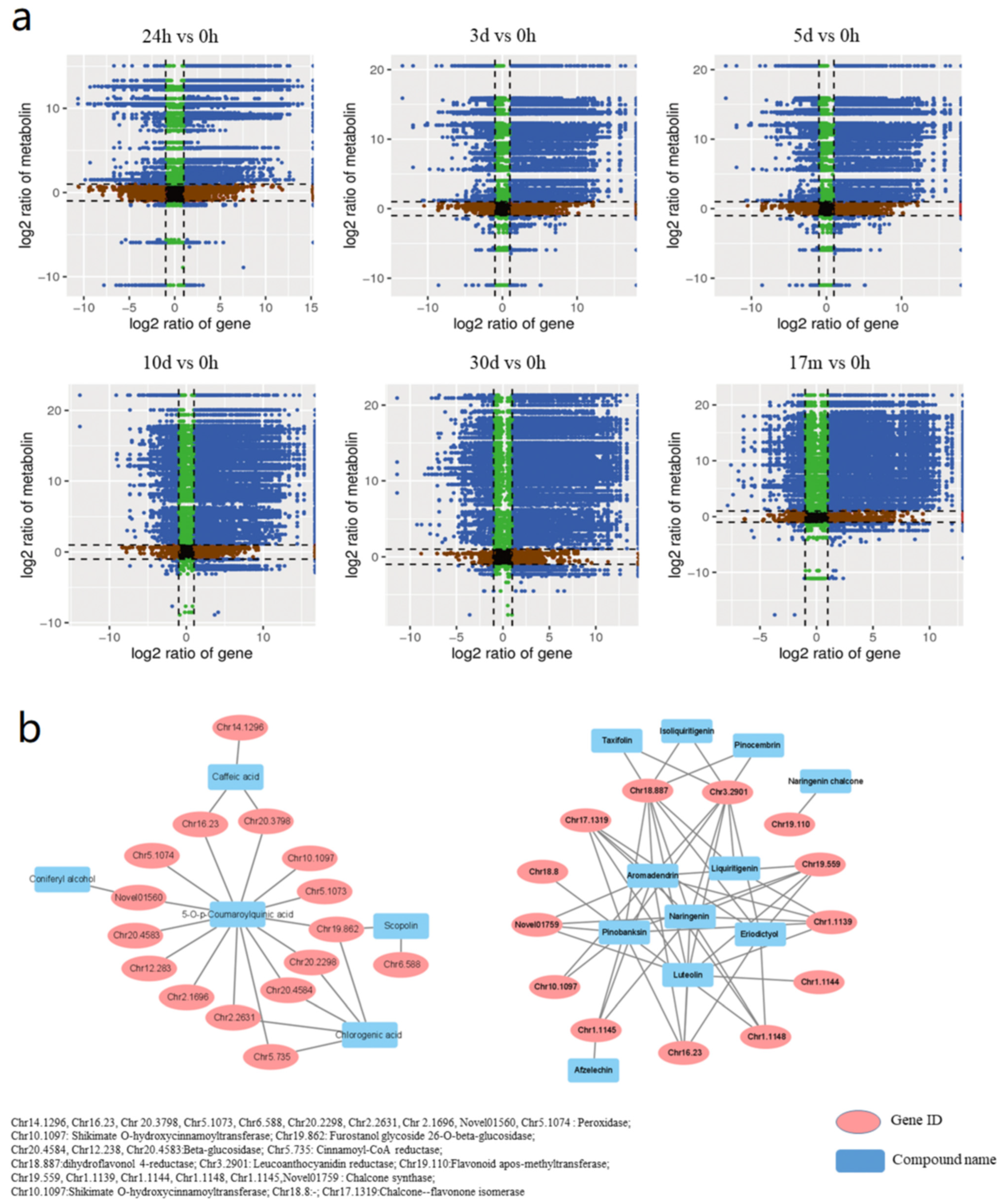
2.1. Identification of Secondary Metabolites and Analysis of Differential Metabolites in D. cochinchinensis Stems at Different Timepoints after Wounding
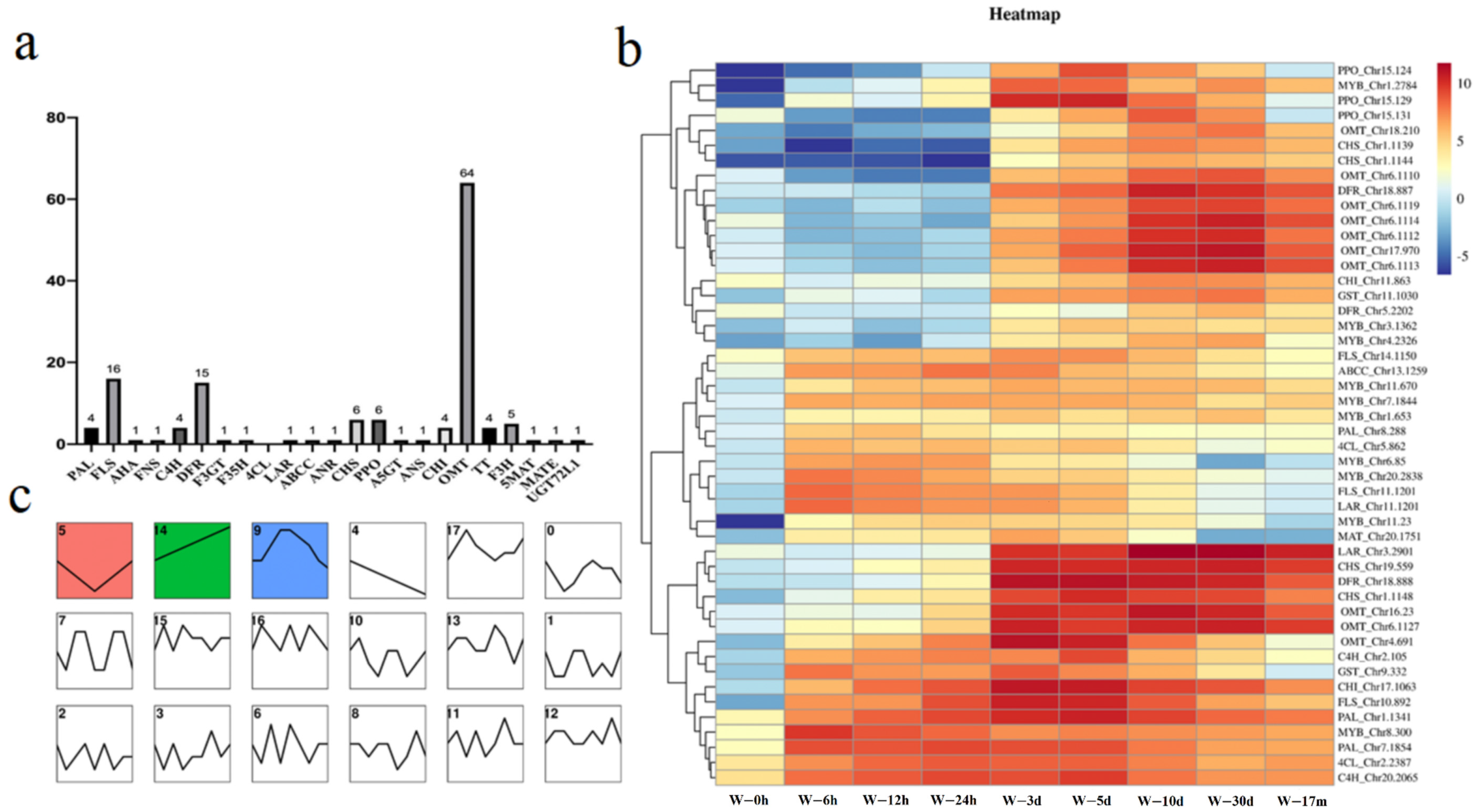
2.2. General Transcriptomics Analysis and Functional Enrichment of DEGs in D. cochinchinensis Stems at Different Times after Wounding
2.3. Wound-Activated Molecular Events in Different KEGG Pathways and Their Association
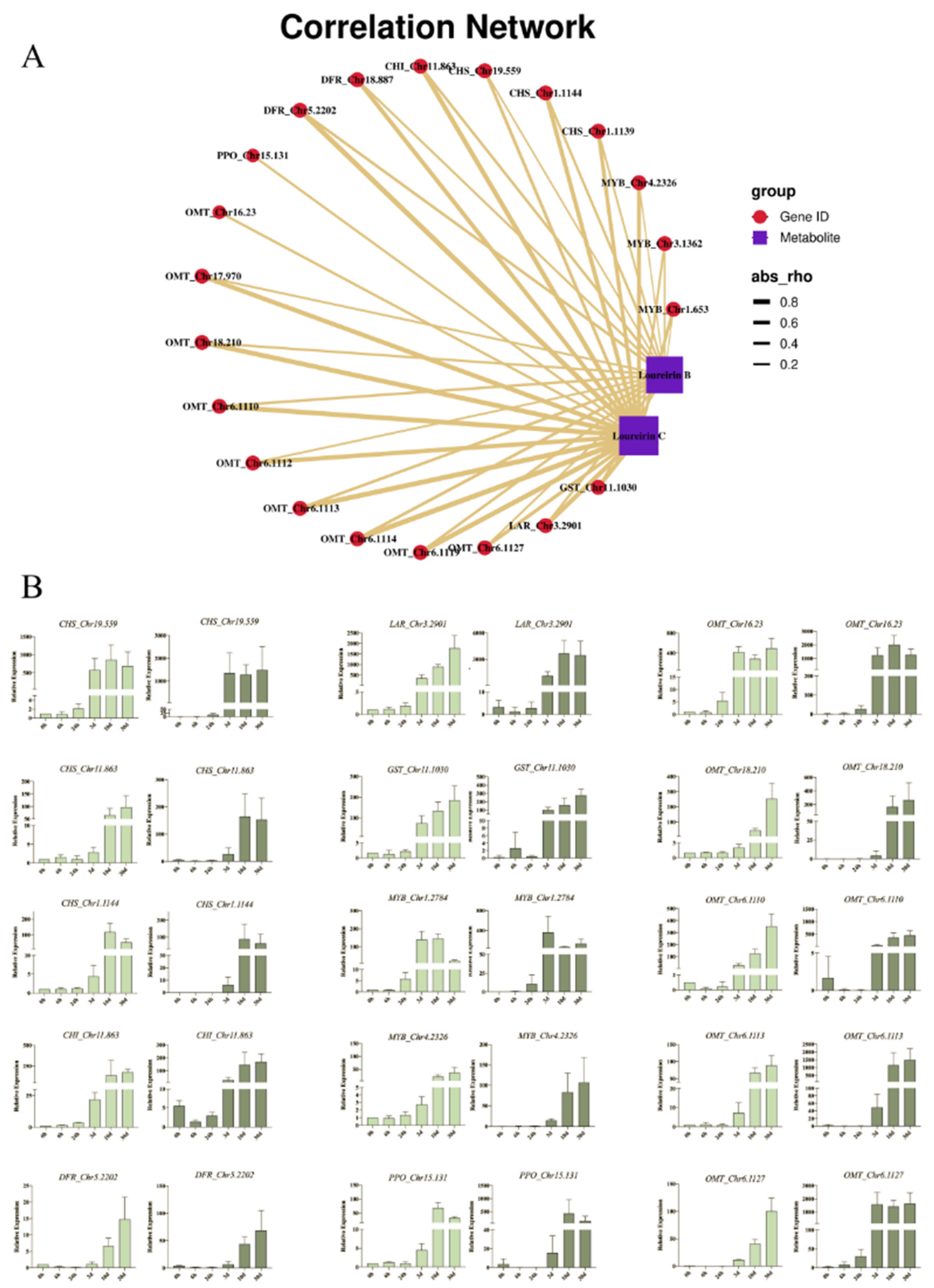
2.4. The Combined Analysis and Correlation Network of DEGs and DEMs in D. cochinchinensis Stems at Different Timepoints after Wounding
2.5. Flavonoids Biosynthesis Pathway in D. cochinchinensis Stems under Wound Stress
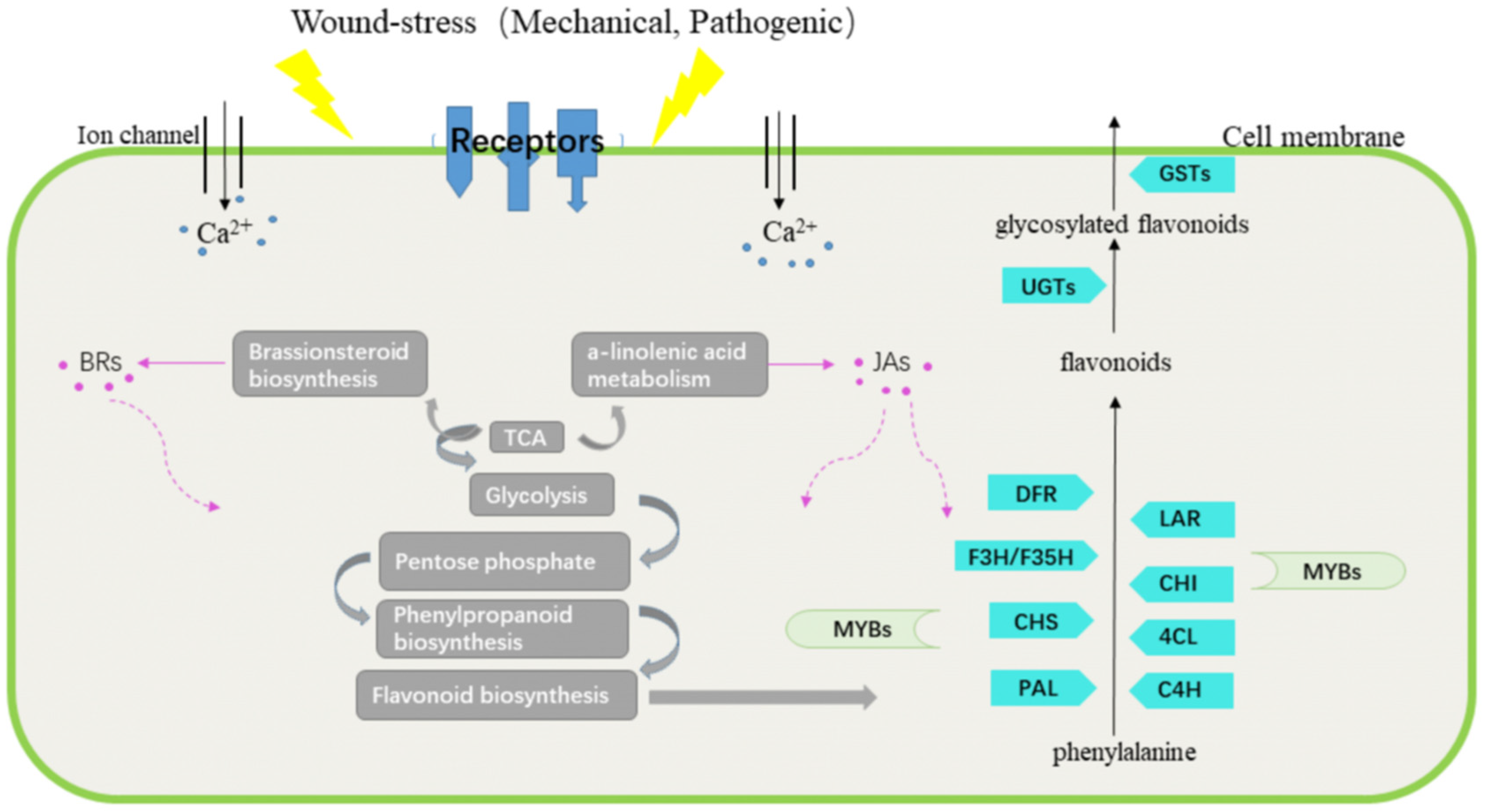
3. Discussion
3.1. Wound-Induced Responses of D. cochinchinensis Reveal Plants’ General Defense Mechanism against Wound Stress in Natural Conditions
3.2. Signals Triggering the Defensive Responses in D. cochinchinensis Stems against Mechanical Damage
3.3. Interplay between Wound Stress and Flavonoid Biosynthesis in D. cochinchinensis
3.4. Decoding the Flavonoid Biosynthesis Pathway in D. cochinchinensis Is the Key to Effectively Producing High-Quality Dragon’s Blood
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Secondary Metabolites Extraction
4.2.2. UPLC Conditions
4.2.3. ESI-Q TRAP-MS/MS
4.2.4. Secondary Metabolome Data Analysis
4.2.5. RNA Extraction and Sequencing
4.2.6. Transcriptome Data Analysis
4.2.7. Combined Analysis of Metabolome and Transcriptome
4.2.8. qRT-PCR Validation for Putative Genes Related to Flavonoid Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Breitsameter, L.; Küchenmeister, K.; Küchenmeister, F.; Isselstein, J. Tolerance to mechanical damage in ten herbaceous grassland plant species. Plant Soil Environ. 2012, 58, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Gao, Z.; Wang, G.; Wang, Y.; Zhang, W.; Lin, X. Classification and bioactivity of phytolectin. Med. J. Chin. People’s Health 2016, 23, 164–168. [Google Scholar]
- Mulder, C.; Wardle, D.A.; Durrett, M.S.; Bellingham, P.J. Leaf damage by herbivores and pathogens on New Zealand islands that differ in seabird densities. N. Z. J. Ecol. 2015, 39, 221–230. [Google Scholar]
- Yoshioka, M.; Adachi, A.; Sato, Y.; Doke, N.; Kondo, T.; Yoshioka, H. RNAi of the sesquiterpene cyclase gene for phytoalexin production impairs pre- and post-invasive resistance to potato blight pathogens. Mol. Plant Pathol. 2019, 20, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Kitano, T.; Yoshimoto, R.; Takata, R.; Ube, N.; Ueno, K.; Ueno, M.; Yabuta, Y.; Teraishi, M.; Holland, C.K.; et al. Natural variation in the expression and catalytic activity of a naringenin 7-O-methyltransferase influences antifungal defenses in diverse rice cultivars. Plant J. 2020, 101, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; De Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chen, H.; Yang, Y.; Zhang, Z.; Wei, J.; Meng, H.; Chen, W.; Feng, J.; Gan, B.; Chen, X.; et al. Whole-tree Agarwood-Inducing Technique: An Efficient Novel Technique for Producing High-Quality Agarwood in Cultivated Aquilaria sinensis Trees. Molecules 2013, 18, 3086–3106. [Google Scholar] [CrossRef]
- Fan, J.-Y.; Yi, T.; Sze-To, C.-M.; Zhu, L.; Peng, W.-L.; Zhang, Y.-Z.; Zhao, Z.-Z.; Chen, H.-B. A Systematic Review of the Botanical, Phytochemical and Pharmacological Profile of Dracaena cochinchinensis, a Plant Source of the Ethnomedicine “Dragon’s Blood”. Molecules 2014, 19, 10650–10669. [Google Scholar] [CrossRef] [Green Version]
- Gupta, D.; Bleakley, B.; Gupta, R.K. Dragon’s blood: Botany, chemistry and therapeutic uses. J. Ethnopharmacol. 2008, 115, 361–380. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, X.; Yao, R.; Li, C.; Zhang, Z.; Xu, Y.; Wei, J.-H. Dragon’s Blood from Dracaena Worldwide: Species, Traditional Uses, Phytochemistry and Pharmacology. Am. J. Chin. Med. 2021, 49, 1315–1367. [Google Scholar] [CrossRef] [PubMed]
- Al-Awthan, Y.S.; Bahattab, O.S. Phytochemistry and Pharmacological Activities of Dracaena cinnabari Resin. BioMed Res. Int. 2021, 2021, 8561696. [Google Scholar] [CrossRef] [PubMed]
- He, T.-C.; Wang, D.-W.; Zheng, S.-M.; Yan, Y.-M.; Jiao, Y.-B.; Cheng, Y.-X.; Wang, F. Antifungal and wound healing promotive compounds from the resins of Dracaena cochinchinensis. Fitoterapia 2021, 151, 104904. [Google Scholar] [CrossRef] [PubMed]
- Xin, N.; Li, Y.-J.; Li, Y.; Dai, R.-J.; Meng, W.-W.; Chen, Y.; Schlappi, M.; Deng, Y.-L. Dragon’s Blood extract has antithrombotic properties, affecting platelet aggregation functions and anticoagulation activities. J. Ethnopharmacol. 2011, 135, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-F.; Song, M.-F.; Zhang, Y.; Zhang, Z.-L. Transcriptome profiling reveals candidate flavonoid-related genes during formation of dragon’s blood from Dracaena cochinchinensis (Lour.) S.C.Chen under conditions of wounding stress. J. Ethnopharmacol. 2021, 273, 113987. [Google Scholar] [CrossRef] [PubMed]
- Al-Okaishi, A. Local Management System of Dragon’s Blood Tree (Dracaena cinnabari Balf. f.) Resin in Firmihin Forest, Socotra Island, Yemen. Forests 2020, 11, 389. [Google Scholar] [CrossRef] [Green Version]
- Hubálková, I.; Maděra, P.; Volařík, D. Growth dynamics of Dracaena cinnabari under controlled conditions as the most effective way to protect endangered species. Saudi J. Biol. Sci. 2017, 24, 1445–1452. [Google Scholar] [CrossRef] [Green Version]
- Nadezhdina, N.; Plichta, R.; Nadezhdin, V.; Gebauer, R.; Jupa, R.; Habrova, H.; Madera, P. A comparative structural and functional study of leaf traits and sap flow in Dracaena cinnabari and Dracaena draco seedlings. Funct. Plant Biol. 2015, 42, 1092. [Google Scholar] [CrossRef]
- Sun, J.; Liu, J.-N.; Fan, B.; Chen, X.; Pang, D.; Zheng, J.; Zhang, Q.; Zhao, Y.; Xiao, W.; Tu, P.; et al. Phenolic constituents, pharmacological activities, quality control, and metabolism of Dracaena species: A review. J. Ethnopharmacol. 2019, 224, 112138. [Google Scholar] [CrossRef]
- Xia, D.; Zhu, J.; Li, H.; Dong, G.; Ying, W.; Fu, W.; Peng, S. Cloning and Expression Analysis of 4-coumaric Acid Coenzyme A Ligase from Dracaena cambodiana. Mol. Plant Breed. 2019, 23, 7656–7662. [Google Scholar]
- Zhu, J.-H.; Cao, T.-J.; Dai, H.-F.; Li, H.-L.; Guo, D.; Mei, W.-L.; Peng, S.-Q. De Novo transcriptome characterization of Dracaena cambodiana and analysis of genes involved in flavonoid accumulation during formation of dragon’s blood. Sci. Rep. 2016, 6, 38315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-H.; Li, H.-L.; Guo, D.; Wang, Y.; Dai, H.-F.; Mei, W.-L.; Peng, S.-Q. Transcriptome-wide identification and expression analysis of glutathione S-transferase genes involved in flavonoids accumulation in Dracaena cambodiana. Plant Physiol. Biochem. 2016, 104, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-H.; Xia, D.-N.; Xu, J.; Guo, D.; Li, H.-L.; Wang, Y.; Mei, W.-L.; Peng, S.-Q. Identification of the bHLH gene family in Dracaena cambodiana reveals candidate genes involved in flavonoid biosynthesis. Ind. Crop. Prod. 2020, 150, 112407. [Google Scholar] [CrossRef]
- Alseekh, S.; de Souza, L.P.; Benina, M.; Fernie, A.R. The style and substance of plant flavonoid decoration; towards defining both structure and function. Phytochemistry 2020, 174, 112347. [Google Scholar] [CrossRef] [PubMed]
- Pucker, B.; Reiher, F.; Schilbert, H.M. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants 2020, 9, 1103. [Google Scholar] [CrossRef]
- Pucker, B. Automatic identification and annotation of MYB gene family members in plants. BMC Genom. 2022, 23, 220. [Google Scholar] [CrossRef]
- Zhang, Y.; Swart, C.; Alseekh, S.; Scossa, F.; Jiang, L.; Obata, T.; Graf, A.; Fernie, A.R. The Extra-Pathway Interactome of the TCA Cycle: Expected and Unexpected Metabolic Interactions. Plant Physiol. 2018, 177, 966–979. [Google Scholar] [CrossRef] [Green Version]
- Gaupels, F.; Sarioglu, H.; Beckmann, M.; Hause, B.; Spannagl, M.; Draper, J.; Lindermayr, C.; Durner, J. Deciphering Systemic Wound Responses of the Pumpkin Extrafascicular Phloem by Metabolomics and Stable Isotope-Coded Protein Labeling. Plant Physiol. 2012, 160, 2285–2299. [Google Scholar] [CrossRef] [Green Version]
- Geigenberger, P. Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 2003, 6, 247–256. [Google Scholar] [CrossRef]
- Mandal, M.K.; Chanda, B.; Xia, Y.; Yu, K.; Sekine, K.; Gao, Q.-M.; Selote, D.; Kachroo, A.; Kachroo, P. Glycerol-3-phosphate and systemic immunity. Plant Signal. Behav. 2011, 6, 1871–1874. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhang, X.; Yang, Y.; Sui, C.; Xu, Y.; Wei, J. Interxylary phloem and xylem rays are the structural foundation of agarwood resin formation in the stems of Aquilaria sinensis. Trees 2018, 33, 533–542. [Google Scholar] [CrossRef]
- Noordermeer, M.A.; Veldink, G.A.; Vliegenthart, J.F. Fatty acid hydroperoxide lyase: A plant cytochrome p450 enzyme involved in wound healing and pest resistance. Chembiochem 2001, 2, 494–504. [Google Scholar] [CrossRef]
- Xue, Y.; Song, M.F.; Zhang, Y.; Zhang, Z.L.; Li-Juan, Z. Study on Signaling Molecules Concentration Change during Process of Resina Draconis by Injury Induced. Mod. Chin. Med. 2019, 5, 18–27. [Google Scholar]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 2009, 47, 1–8. [Google Scholar] [CrossRef]
- Kachroo, A.; Lapchyk, L.; Fukushige, H.; Hildebrand, D.; Klessig, D.; Kachroo, P. Plastidial. Fatty Acid Signaling Modulates Salicylic Acid– and Jasmonic Acid–Mediated Defense Pathways in the Arabidopsis ssi2 Mutant. Plant Cell 2003, 15, 2952–2965. [Google Scholar] [CrossRef] [Green Version]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef]
- Tamari, G.; Borochov, A.; AUom, R.; Weiss, D. Methyl jasmonate induces pigmentation and flavonoid gene expression in petunia corollas: A possible role in wound response. Physiol. Plant 1995, 94, 45–50. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, Q.; Yuan, L.; Jiang, Y.; Huang, Y.; Sun, M.; Tang, S.; Luo, K. Isolation and promoter analysis of a chalcone synthase gene PtrCHS4 from Populus trichocarpa. Plant Cell Rep. 2011, 30, 1661–1671. [Google Scholar] [CrossRef]
- Naoumkina, M.; Farag, M.A.; Sumner, L.W.; Tang, Y.; Liu, C.-J.; Dixon, R.A. Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2007, 104, 17909–17915. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Yang, F.; Zhang, S.; Sun, S.; Miao, X.; Shi, Z. The OsmiR396–Os GRF 8–OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef] [Green Version]
- Scollo, S.; Folch, A.; Costa, A. A parametric and comparative study of different tephra fallout models. J. Volcanol. Geotherm. Res. 2003, 176, 199–211. [Google Scholar] [CrossRef]
- Wang, X. Structure, function, and engineering of enzymes in isoflavonoid biosynthesis. Funct. Integr. Genom. 2011, 11, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Jura-Morawiec, J.; Oskolski, A.; Simpson, P. Revisiting the anatomy of the monocot cambium, a novel meristem. Planta 2021, 254, 6. [Google Scholar] [CrossRef] [PubMed]
- Hubálková, I. Prediction of Dragon’s Blood Tree (Dracaena cinnabari Balf.) Stand Sample Density on Soqotra Island. J. Landsc. Ecol. 2011, 4, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Ghose, K.; Selvaraj, K.; McCallum, J.; Kirby, C.W.; Sweeney-Nixon, M.; Cloutier, S.J.; Deyholos, M.; Datla, R.; Fofana, B. Identification and functional characterization of a flax UDP-glycosyltransferase glucosylating secoisolariciresinol (SECO) into secoisolariciresinol monoglucoside (SMG) and diglucoside (SDG). BMC Plant Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [Green Version]
- Campos, L.; López-Gresa, M.P.; Fuertes, D.; Bellés, J.M.; Rodrigo, I.; Lisón, P. Tomato glycosyltransferase Twi1 plays a role in flavonoid glycosylation and defence against virus. BMC Plant Biol. 2019, 19, 450. [Google Scholar] [CrossRef] [Green Version]
- Jura-Morawiec, J.; Tulik, M. Dragon’s blood secretion and its ecological significance. Chemoecology 2016, 26, 101–105. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, X.; Gao, Y.; Zong, G.; Wang, D.; Liu, M.; Fei, S.; Wei, Y.; Yin, Z.; Chen, J.; et al. Identification of a UDP-Glucosyltransferase favouring substrate- and regio-specific biosynthesis of flavonoid glucosides in Cyclocarya paliurus. Phytochemistry 2019, 163, 75–88. [Google Scholar] [CrossRef]
- Hollman, P.; Katan, M. Dietary Flavonoids: Intake, Health Effects and Bioavailability. Food Chem. Toxicol. 1999, 37, 937–942. [Google Scholar] [CrossRef]
- Wang, T.; Wang, C.J.; Wang, H.; Liang, Y.Q.; Li, Z.Q. Research Survey on the Clinical Application of Dragon’s blood. Med. Inov. China 2015, 12, 154–156. [Google Scholar]
- Zhang, Z.; Yang, Y.; Wei, J.H.; Meng, H.; Sui, C.; Chen, H.Q. Advances in studies on mechanism of agarwood formation in Aquilaria sinensis and its hypothesis of agarwood formation induced by defense response. Chin. Tradit. Herb. Drugs 2010, 1, 157–159. [Google Scholar]
- Zhao, X.; Wang, C.; Meng, H.; Yu, Z.; Yang, M.; Wei, J. Dalbergia odorifera: A review of its traditional uses, phytochemistry, pharmacology, and quality control. J. Ethnopharmacol. 2020, 248, 112328. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Hu, Q.; Luo, J.; Li, Y.; Lu, C.; Chen, X.; Hu, H. Loureirin B, an essential component of Sanguis Draxonis, inhibits Kv1.3 channel and suppresses cytokine release from Jurkat T cells. Cell Biosci. 2014, 4, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.H.; Zhang, K.J.; Zhang, Z.L.; Liu, Y.; Lv, F.F.; Sun, P.W.; Gao, S.X.; Wang, Q.L.; Yu, C.C.; Jiang, J.M.; et al. The Chromosome-level Genome of Dracaena cochinchinesis Provides Insights into its Biological Features and the Mechanism of Dragon’s Blood formation. bioRxiv 2022. [Google Scholar] [CrossRef]


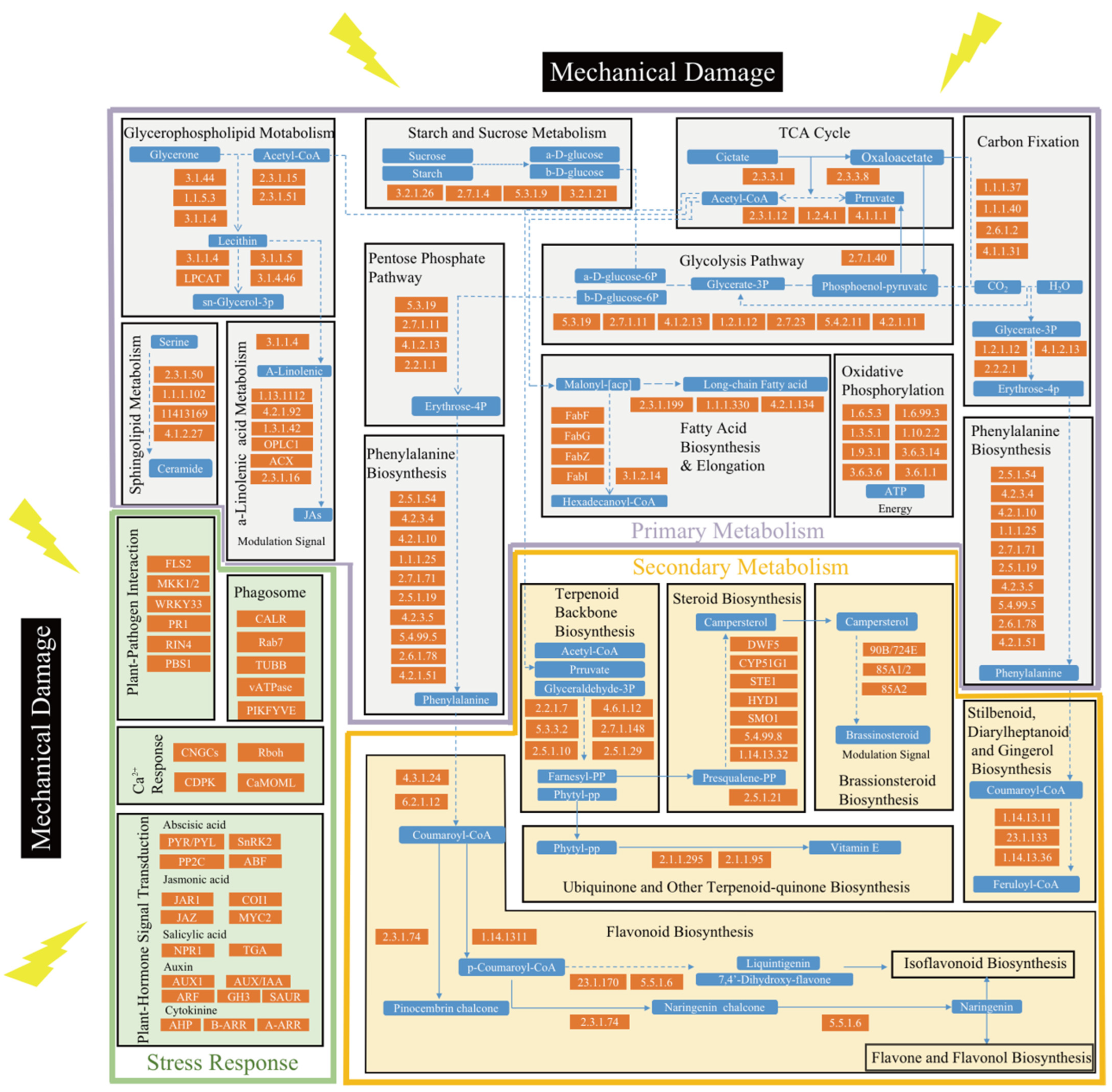

| Pathway | Enriched Number of DEGs/DEMs | Gene Symbol | Compound | |||||
|---|---|---|---|---|---|---|---|---|
| 1 Day | 3 Days | 5 Days | 10 Days | 30 Days | 17 Months | |||
| Phenylalanine metabolism | 15/1 | 8/1 | 10/1 | 15/1 | 8/1 | 4/1 | GOT1, hisC, PAL, AOC3, HPD | Cinnamic acid, salicylic acid |
| Ubiquinone and other terpenoid-quinone biosynthesis | 21/2 | 17/3 | 18/3 | 17/3 | 16/3 | 9/3 | CYP73A, 4CL, HPD, VTE3, VTE1, E2.1.1.95, HPT | Cinnamic acid, p-coumaric acid, 4-hydroxybenzoic acid, 3-[(1-carboxyvinyl)oxy]benzoic acid |
| Stilbenoid, diarylheptanoid, and gingerol biosynthesis | 7/2 | 7/3 | 8/4 | 7/4 | 6/4 | 5/4 | CYP73A, HCT, CYP98A, E2.1.1.104 | Resveratrol, 5-O-p-coumaroylquinic acid, chlorogenic acid, piceatannol |
| Aminoacyl-tRNA biosynthesis | 28/1 | 3/1 | 5/1 | 6/1 | 4/1 | - | EARS, VARS, gatA, LARS, AARS, DARS1, NARS, RARS, glyQ, PARS, TARS, HARS, FARSA, YARS, WARS | 10-Formyltetrahydrofuran |
| Isoquinoline alkaloid biosynthesis | 12/1 | 7/2 | 10/1 | 13/1 | 7/1 | 2/1 | GOT2, TYDC, E1.10.3.1, AOC3 | p-Coumaric acid, Protocatechualdehyde |
| One-carbon pool by folate | 10/1 | 6/1 | 7/1 | 13/1 | 8/1 | - | gcvT, glyA, MTHFS, MTHFD1L, metF | 10-Formyltetrahydrofuran |
| Flavonoid biosynthesis | 11/4 | 16/12 | 20/18 | 17/19 | 16/19 | 13/17 | CYP73A, CHS, E5.5.1.6, CYP75B1, ANR, LAR, HCT, C3’H, E2.1.1.104 | 7,4′-Dihydroxyflavone, pinocembrin, isoliquiritigenin, liquiritigenin, apigenin, galangin, naringenin chalcone, pinobanksin, naringenin, afzelechin, epiafzelechin, luteolin, eriodictyol, aromadendrin, quercetin, dihydroquercetin, 5-O-p-coumaroylquinic acid, chlorogenic acid, naringenin-7-O-glucoside, neohesperidin, butin |
| Tyrosine metabolism | 18/1 | 12/2 | 17/1 | 26/4 | 16/4 | 3/4 | FAH, maiA, HPD, ADH1_7, AOC3, GOT1, TDC-1, E1.10.3.1 | 2,5-Dihydroxybenzaldehyde, tyrosol,2,5-dihydroxybenzoic acid, gentisic acid, homogentisic acid, p-coumaric acid |
| Flavone and flavonol biosynthesis | 2/1 | 3/4 | 5/7 | 5/9 | 4/8 | 3/8 | CYP75B1, E2.1.1.76 | Apigenin, acacetin, luteolin, quercetin, 3,7-di-O-methylquercetin, laricitrin, cosmosiin, cynaroside, astragalin |
| Phenylpropanoid biosynthesis | 46/5 | 36/9 | 57/7 | 61/9 | 40/8 | 24/6 | PAL, CYP73A, F5H, 4CL, CSE, HCT, C3’H, CYP73A, E2.1.1.104, REF1, CCR, CAD, E1.11.1.7 | Cinnamic acid, p-coumaryl alcohol, p-coumaric acid, caffeic acid, coniferyl alcohol, 5-O-p-coumaroylquinic acid, coniferin, chlorogenic acid, 1-O-Sinapoyl-d-glucose, scopolin, ferulic acid |
| Glycine, serine, and threonine metabolism | - | 17/1 | 21/1 | 33/1 | 15/1 | 3/1 | PGAM, gpmI, hprA, HPR2_3, serA, serB, SDS, trpA, glyA, betB, PIPOX, GGAT, AGXT2, gcvT, GCSH, DLD, GLDC, ltaE, AOC3, SDS | Betaine |
| Arginine and proline metabolism | - | 10/1 | 14/2 | 25/2 | 15/1 | - | proB, proA, PRODH, P4HA, E4.1.1.19, aguA, GOT1, speD, PAO4, ALDH, speE, MPAO, SMOX | p-Coumaroylputrescine, N-feruloylputrescine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Gao, S.; Zhang, Y.; Zhang, Z.; Wang, Q.; Xu, Y.; Wei, J. Transcriptomics and Metabolomics Analyses Reveal Defensive Responses and Flavonoid Biosynthesis of Dracaena cochinchinensis (Lour.) S. C. Chen under Wound Stress in Natural Conditions. Molecules 2022, 27, 4514. https://doi.org/10.3390/molecules27144514
Liu Y, Gao S, Zhang Y, Zhang Z, Wang Q, Xu Y, Wei J. Transcriptomics and Metabolomics Analyses Reveal Defensive Responses and Flavonoid Biosynthesis of Dracaena cochinchinensis (Lour.) S. C. Chen under Wound Stress in Natural Conditions. Molecules. 2022; 27(14):4514. https://doi.org/10.3390/molecules27144514
Chicago/Turabian StyleLiu, Yang, Shixi Gao, Yuxiu Zhang, Zhonglian Zhang, Qiuling Wang, Yanhong Xu, and Jianhe Wei. 2022. "Transcriptomics and Metabolomics Analyses Reveal Defensive Responses and Flavonoid Biosynthesis of Dracaena cochinchinensis (Lour.) S. C. Chen under Wound Stress in Natural Conditions" Molecules 27, no. 14: 4514. https://doi.org/10.3390/molecules27144514
APA StyleLiu, Y., Gao, S., Zhang, Y., Zhang, Z., Wang, Q., Xu, Y., & Wei, J. (2022). Transcriptomics and Metabolomics Analyses Reveal Defensive Responses and Flavonoid Biosynthesis of Dracaena cochinchinensis (Lour.) S. C. Chen under Wound Stress in Natural Conditions. Molecules, 27(14), 4514. https://doi.org/10.3390/molecules27144514






