Establishing a Klebsiella pneumoniae-Based Cell-Free Protein Synthesis System
Abstract
:1. Introduction
2. Results and Discussion
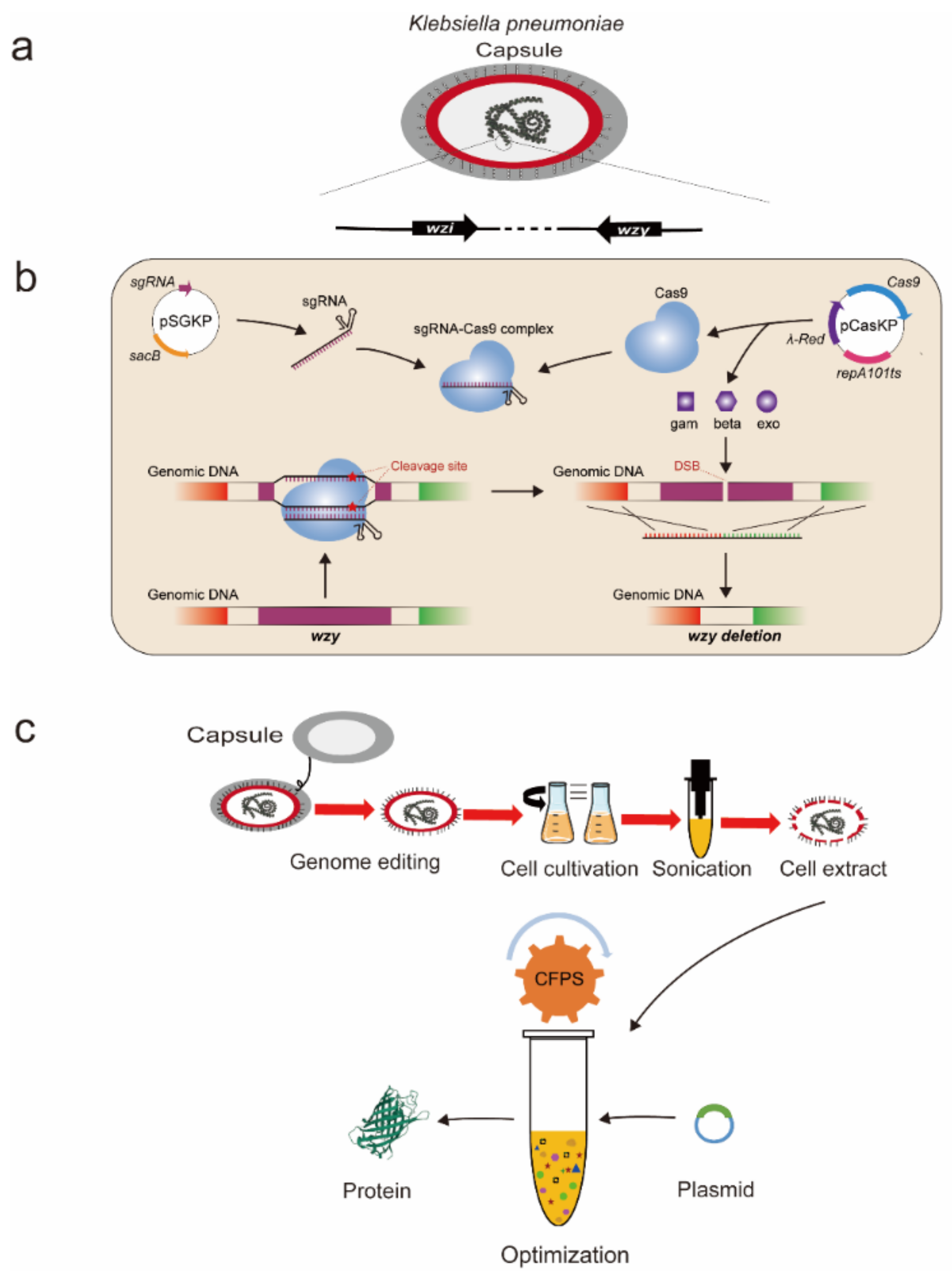
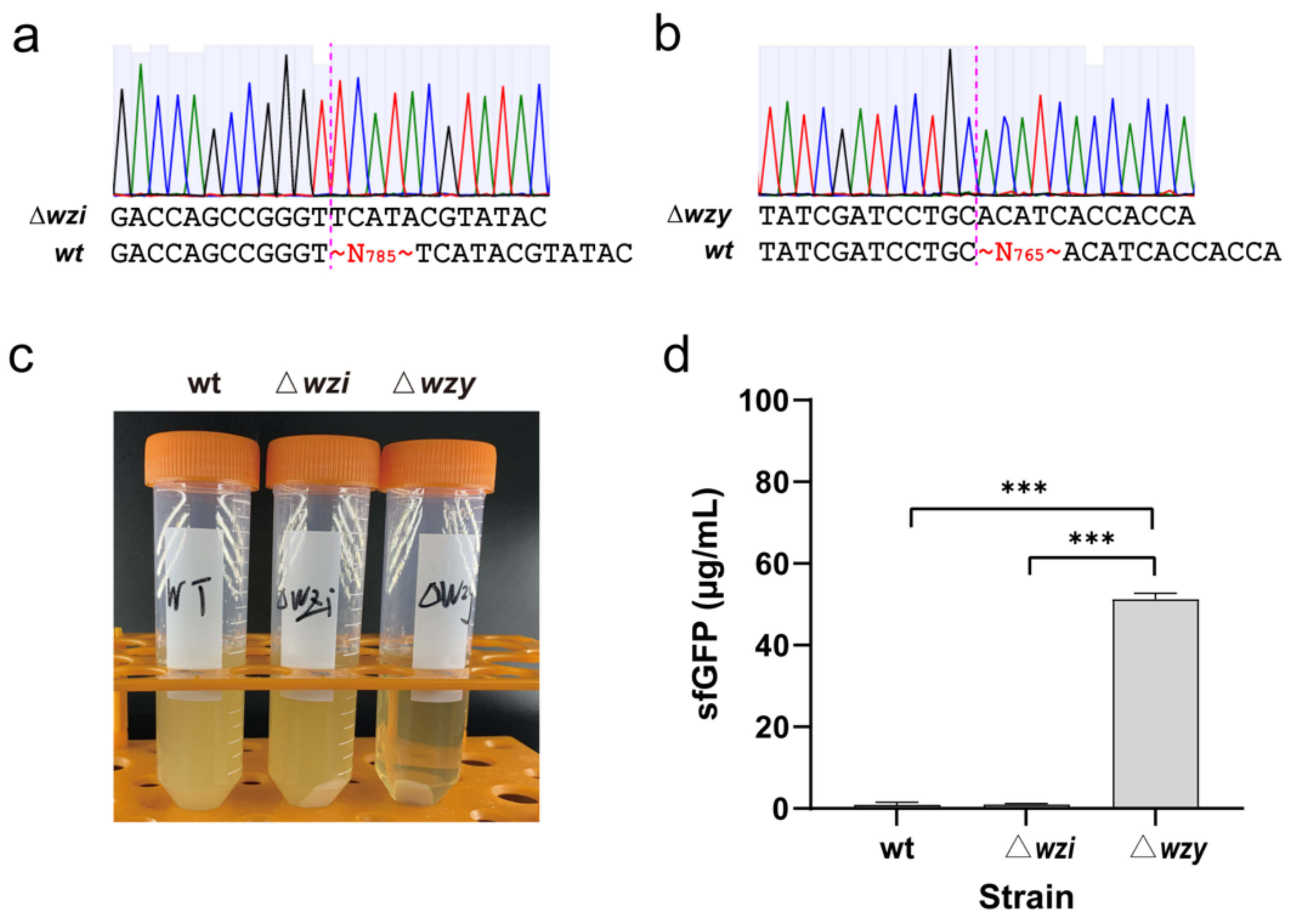
2.1. Genome Engineering of K. pneumoniae by Deleting Two Capsule-Associated Genes
2.2. Optimization of Cell Extract Preparation
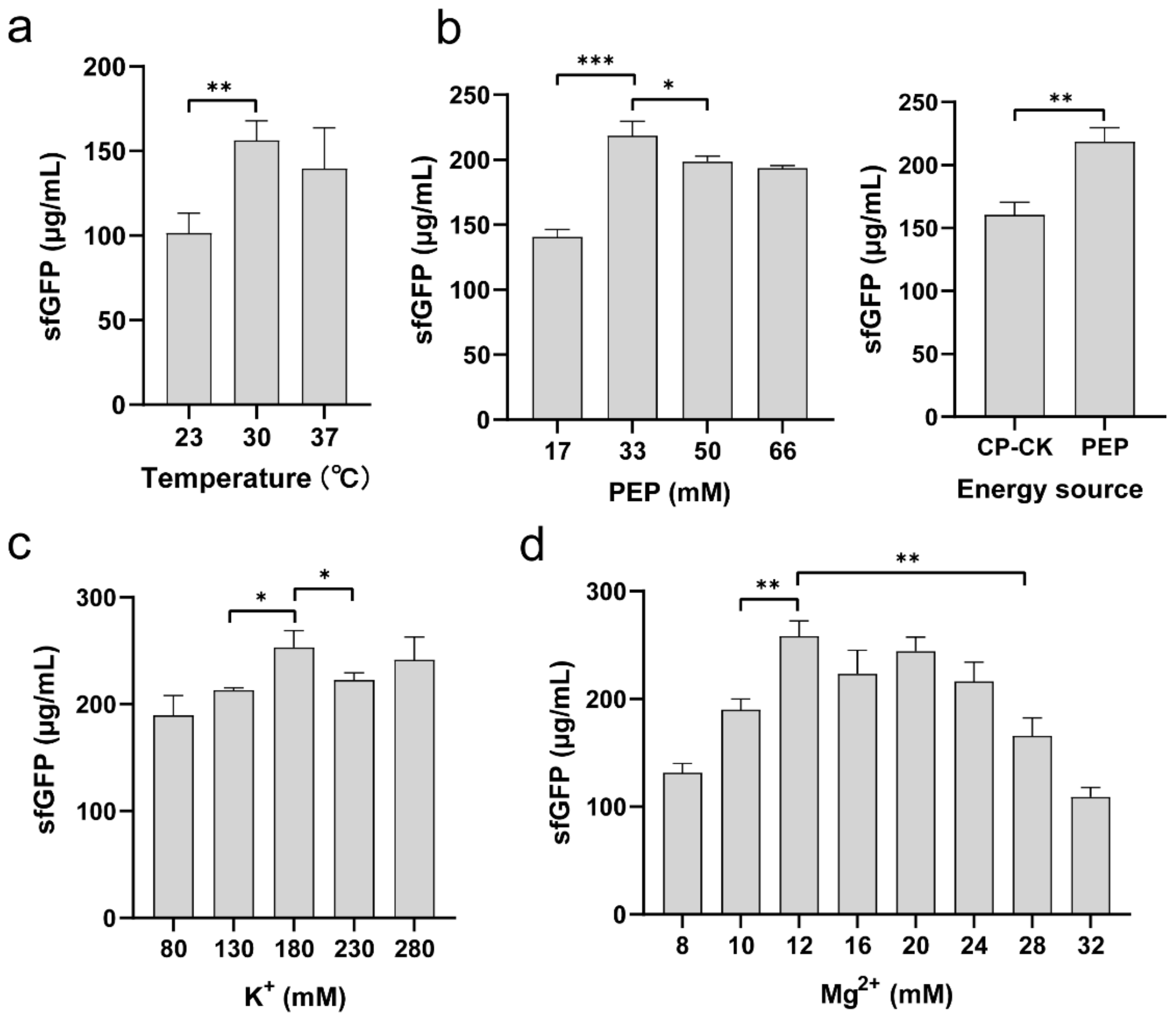
2.3. Optimization of Reaction Conditions in K. pneumoniae CFPS
3. Materials and Methods
3.1. Bacterial Strains, Culture Media, and Plasmids
3.2. Genome Engineering of the Wild-Type K. pneumoniae KP_1.6366 Strain
3.3. Preparation of Cell Extracts
3.4. Cell-Free Protein Synthesis (CFPS) Reactions
3.5. Protein Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Silverman, A.D.; Karim, A.S.; Jewett, M.C. Cell-free gene expression: An expanded repertoire of applications. Nat. Rev. Genet. 2020, 21, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Q.; Zhang, L.; Chen, M.; Li, J. Cell-free protein synthesis: Recent advances in bacterial extract sources and expanded applications. Biochem. Eng. J. 2019, 141, 182–189. [Google Scholar] [CrossRef]
- Brookwell, A.; Oza, J.P.; Caschera, F. Biotechnology applications of cell-free expression systems. Life 2021, 11, 1367. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Q.; Wu, C.; Jewett, M.C.; Li, J. Cell-free protein synthesis enables one-pot cascade biotransformation in an aqueous-organic biphasic system. Biotechnol. Bioeng. 2020, 117, 4001–4008. [Google Scholar] [CrossRef]
- Zhuang, L.; Huang, S.; Liu, W.Q.; Karim, A.S.; Jewett, M.C.; Li, J. Total in vitro biosynthesis of the nonribosomal macrolactone peptide valinomycin. Metab. Eng. 2020, 60, 37–44. [Google Scholar] [CrossRef]
- Copeland, C.E.; Langlois, A.; Kim, J.; Kwon, Y.C. The cell-free system: A new apparatus for affordable, sensitive, and portable healthcare. Biochem. Eng. J. 2021, 175, 108124. [Google Scholar] [CrossRef]
- Ji, X.; Liu, W.Q.; Li, J. Recent advances in applying cell-free systems for high-value and complex natural product biosynthesis. Curr. Opin. Microbiol. 2022, 67, 102142. [Google Scholar] [CrossRef]
- Tian, X.; Liu, W.Q.; Xu, H.; Ji, X.; Liu, Y.; Li, J. Cell-free expression of NO synthase and P450 enzyme for the biosynthesis of an unnatural amino acid L-4-nitrotryptophan. Synth. Syst. Biotechnol. 2022, 7, 775–783. [Google Scholar] [CrossRef]
- Schwarz, D.; Junge, F.; Durst, F.; Frölich, N.; Schneider, B.; Reckel, S.; Sobhanifar, S.; Dötsch, V.; Bernhard, F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007, 2, 2945–2957. [Google Scholar] [CrossRef]
- Min, S.E.; Lee, K.H.; Park, S.W.; Yoo, T.H.; Oh, C.H.; Park, J.H.; Yang, S.Y.; Kim, Y.S.; Kim, D.M. Cell-free production and streamlined assay of cytosol-penetrating antibodies. Biotechnol. Bioeng. 2016, 113, 2107–2112. [Google Scholar] [CrossRef]
- Li, J.; Lawton, T.J.; Kostecki, J.S.; Nisthal, A.; Fang, J.; Mayo, S.L.; Rosenzweig, A.C.; Jewett, M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 2016, 11, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Albayrak, C.; Swartz, J.R. Direct polymerization of proteins. ACS Synth. Biol. 2014, 3, 353–362. [Google Scholar] [CrossRef]
- Zemella, A.; Thoring, L.; Hoffmeister, C.; Kubick, S. Cell-free protein synthesis: Pros and cons of prokaryotic and eukaryotic systems. ChemBioChem 2015, 16, 2420–2431. [Google Scholar] [CrossRef] [Green Version]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A user’s guide to cell-free protein synthesis. Methods Protoc. 2019, 2, 24. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, H.; Kwon, Y.C.; Jewett, M.C. Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng. 2017, 114, 1343–1353. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Needham, H.; Polizzi, K.M.; Freemont, P.S. Streptomyces venezuelae TX-TL—A next generation cell-free synthetic biology tool. Biotechnol. J. 2017, 12, 1600678. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Liu, W.Q.; Li, J. Translation related factors improve the productivity of a Streptomyces-based cell-free protein synthesis system. ACS Synth. Biol. 2020, 9, 1221–1224. [Google Scholar] [CrossRef]
- Moore, S.J.; Lai, H.E.; Chee, S.M.; Toh, M.; Coode, S.; Chengan, K.; Capel, P.; Corre, C.; de los Santos, E.L.; Freemont, P.S. A Streptomyces venezuelae cell-free toolkit for synthetic biology. ACS Synth. Biol. 2021, 10, 402–411. [Google Scholar] [CrossRef]
- Xu, H.; Yang, C.; Tian, X.; Chen, Y.; Liu, W.Q.; Li, J. Regulatory part engineering for high-yield protein synthesis in an all-Streptomyces-based cell-free expression system. ACS Synth. Biol. 2022, 11, 570–578. [Google Scholar] [CrossRef]
- Kelwick, R.; Webb, A.J.; MacDonald, J.T.; Freemont, P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 2016, 38, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, J.; Jewett, M.C. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol. 2018, 3, ysy003. [Google Scholar] [CrossRef]
- Des Soye, B.J.; Davidson, S.R.; Weinstock, M.T.; Gibson, D.G.; Jewett, M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 2018, 7, 2245–2255. [Google Scholar] [CrossRef]
- Aw, R.; Polizzi, K.M. Biosensor-assisted engineering of a high-yield Pichia pastoris cell-free protein synthesis platform. Biotechnol. Bioeng. 2019, 116, 656–666. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Lin, X.; Wang, T.; Guo, W.; Lu, Y. Development and comparison of cell-free protein synthesis systems derived from typical bacterial chassis. Bioresour. Bioprocess. 2021, 8, 58. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, J.; Lu, X.; Zhang, Z.; Yang, Y.; Sun, S.; Kostas, E.T.; Shi, J.; Gao, M.; Baganz, F.; et al. 2,3-Dihydroxyisovalerate production by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 2020, 104, 6601–6613. [Google Scholar] [CrossRef]
- Walker, K.A.; Miller, V.L. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr. Opin. Microbiol. 2020, 54, 95–102. [Google Scholar] [CrossRef]
- Jung, M.Y.; Mazumdar, S.; Shin, S.H.; Yang, K.S.; Lee, J.; Oh, M.K. Improvement of 2,3-butanediol yield in Klebsiella pneumoniae by deletion of the pyruvate formate-lyase gene. Appl. Environ. Microbiol. 2014, 80, 6195–6203. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Wang, C.; Wei, D.; Shi, J.; Kim, C.H.; Jiang, B.; Han, Z.; Hao, J. Gluconic acid production by gad mutant of Klebsiella pneumoniae. World J. Microbiol. Biotechnol. 2016, 32, 132. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Tian, P. Engineering plasmid-free Klebsiella Pneumoniae for production of 3-hydroxypropionic acid. Curr. Microbiol. 2017, 74, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Lama, S.; Seol, E.; Park, S. Development of Klebsiella pneumoniae J2B as microbial cell factory for the production of 1,3-propanediol from glucose. Metab. Eng. 2020, 62, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Yuminaga, Y.; Shi, J.P.; Hao, J. Non-capsulated mutants of a chemical-producing Klebsiella pneumoniae strain. Biotechnol. Lett. 2018, 40, 679–687. [Google Scholar] [CrossRef]
- Brisse, S.; Passet, V.; Haugaard, A.B.; Babosan, A.; Kassis-Chikhani, N.; Struve, C.; Decre, D. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 2013, 51, 4073–4078. [Google Scholar] [CrossRef] [Green Version]
- Chuang, Y.P.; Fang, C.T.; Lai, S.Y.; Chang, S.C.; Wang, J.T. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2006, 193, 645–654. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.T.; Lai, S.Y.; Yi, W.C.; Hsueh, P.R.; Liu, K.L. The Function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J. Infect. Dis. 2010, 201, 1268–1269. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, S.; Chen, W.; Song, L.; Zhang, Y.; Shen, Z.; Yu, F.; Li, M.; Ji, Q. CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 2018, 84, e01834-18. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.C.; Jewett, M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 2015, 5, 8663. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Liu, Y.; Liu, W.Q.; Wu, C.; Li, J. Designing modular cell-free systems for tunable biotransformation of L-phenylalanine to aromatic compounds. Front. Bioeng. Biotechnol. 2021, 9, 730663. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, W.Q.; Li, J. Establishing a eukaryotic Pichia pastoris cell-free protein synthesis system. Front. Bioeng. Biotechnol. 2020, 8, 536. [Google Scholar] [CrossRef]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef]
- Dopp, J.L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef]
- Xie, S.; Shen, B.; Zhang, C.; Huang, X.; Zhang, Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE 2014, 9, e100448. [Google Scholar] [CrossRef]

| Plasmid | Description | Source |
|---|---|---|
| pCasKP-apr | Expression of Cas9 and λ-Red proteins in K. pneumoniae | Addgene # 117231 |
| pSGKP-km | Expression of sgRNA in K. pneumonia | Addgene # 117233 |
| pSGKP-km_wzi | pSGKP-km derivative with wzi spacer | This study |
| pSGKP-km_wzy | pSGKP-km derivative with wzy spacer | This study |
| pBECKP-km | Expression of APOBEC1-nCas9 fusion protein and sgRNA in K. pneumonia | Addgene # 117235 |
| pJL1-sfGFP | Expression of the reporter protein sfGFP in CFPS | Addgene # 69496 |
| Key Parameters | Description |
|---|---|
| Cell extract preparation | |
| Strain | K. pneumonia Δwzy, a capsule-deficient strain |
| Cultivation | 500 mL of 2× YTPG, 34 °C, 250 rpm |
| Collection | OD600 = 4 |
| Lysis | Sonication, input energy: 800 J |
| CFPS reaction | |
| Total volume | 15 μL in 1.5 mL Eppendorf tube |
| Cell extract | 6 μL per reaction (40%, v/v) |
| Energy | PEP, 33 mM |
| Mg2+ ion | 12 mM |
| K+ ion | 180 mM |
| Temperature | 30 °C |
| Other components | See “Section 3.4.” |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Yang, M.; Zhao, W.; Ding, Y.; Wang, Y.; Li, J. Establishing a Klebsiella pneumoniae-Based Cell-Free Protein Synthesis System. Molecules 2022, 27, 4684. https://doi.org/10.3390/molecules27154684
Yang C, Yang M, Zhao W, Ding Y, Wang Y, Li J. Establishing a Klebsiella pneumoniae-Based Cell-Free Protein Synthesis System. Molecules. 2022; 27(15):4684. https://doi.org/10.3390/molecules27154684
Chicago/Turabian StyleYang, Chen, Miaomiao Yang, Wanhua Zhao, Yue Ding, Yu Wang, and Jian Li. 2022. "Establishing a Klebsiella pneumoniae-Based Cell-Free Protein Synthesis System" Molecules 27, no. 15: 4684. https://doi.org/10.3390/molecules27154684






