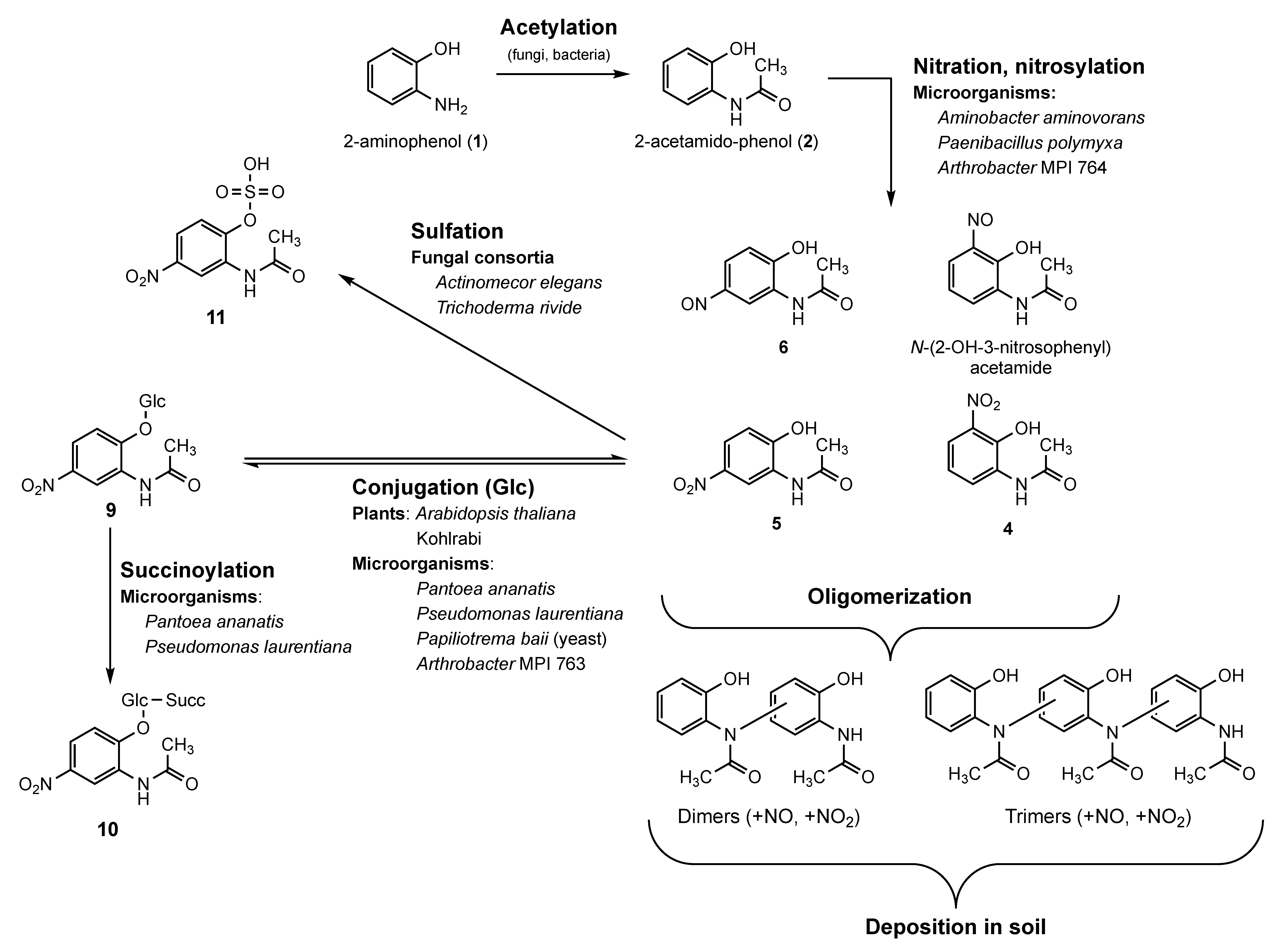
Bioactive Nitrosylated and Nitrated N-(2-hydroxyphenyl)acetamides and Derived Oligomers: An Alternative Pathway to 2-Amidophenol-Derived Phytotoxic Metabolites
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis, Purification, and Identification of Compounds
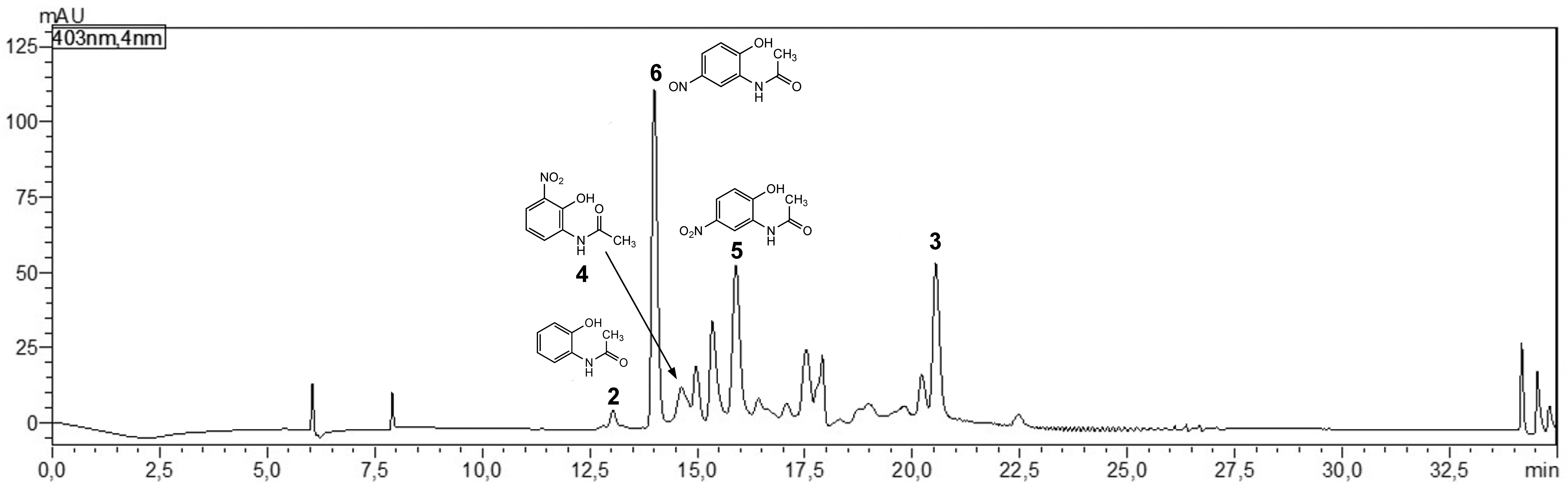
2.1.1. Identification of the Nitrated Derivatives of 2-Acetamido-phenol
2.1.2. Identification of N-(2-hydroxy-3-nitrophenyl)acetamide in the Fraction A4 of P. polymyxa Bacterial Culture Extract
2.1.3. Identification of N-(2-hydroxy-5-nitrosophenyl)acetamide in the Fraction A2 of P. polymyxa Bacterial Culture Extract
2.1.4. Presence of Dimers and Trimers Derived from 2-acetamido-phenol
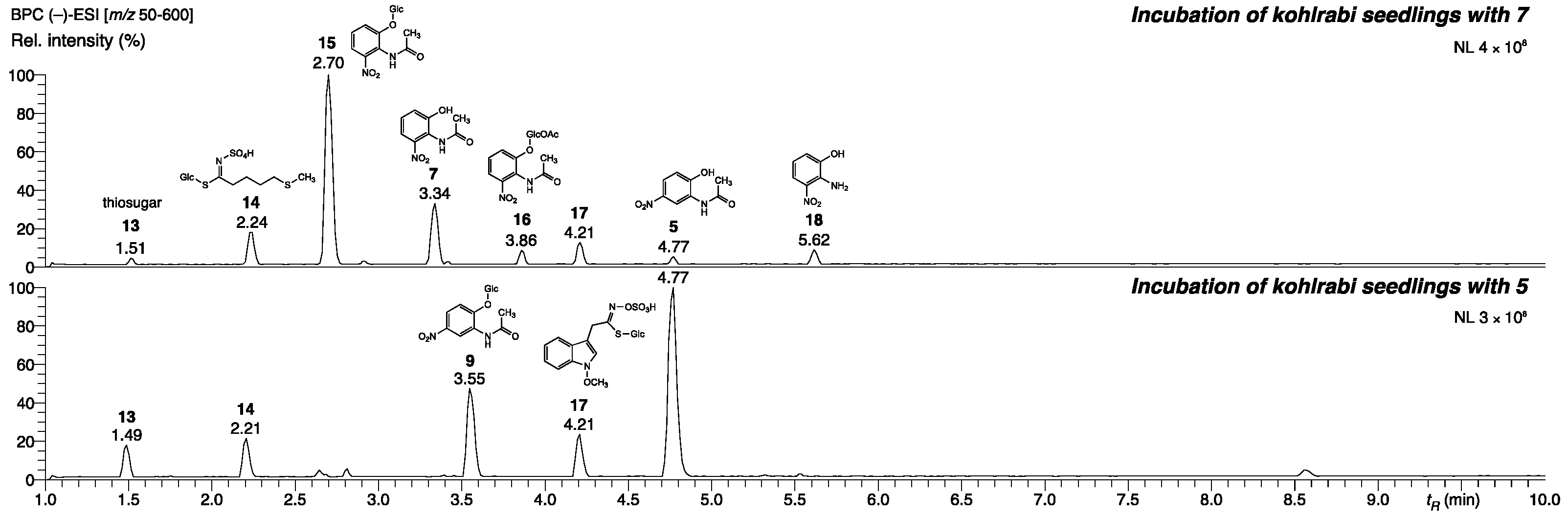
2.2. Bacterial and Fungal Detoxification Products Derived from N-(2-hydroxy-5-nitrophenyl)acetamide
2.3. Incubation of Plants with Nitrated Derivatives of 2-Acetamido-phenol
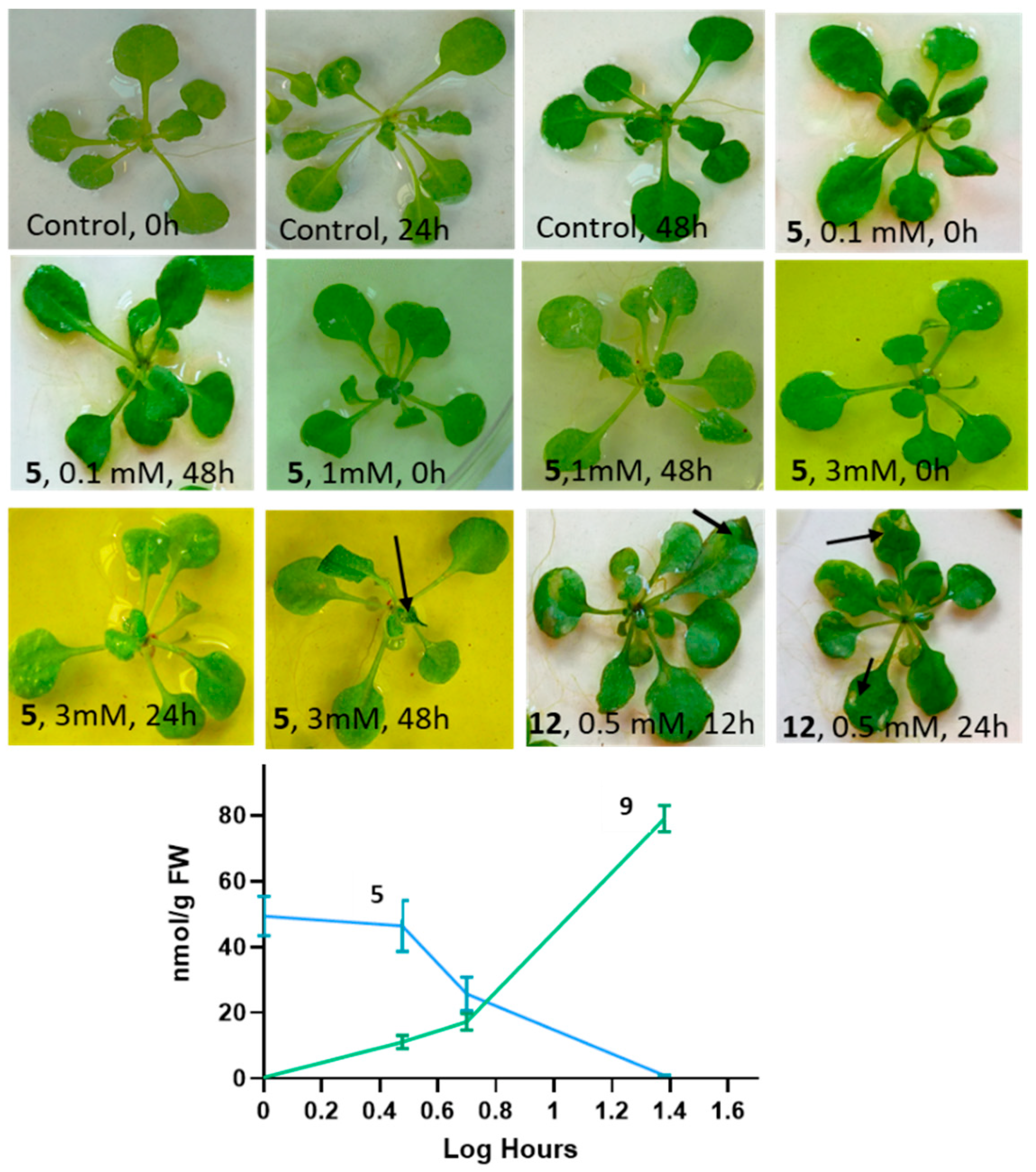
2.3.1. Effects of N-(2-hydroxy-5-nitrophenyl)acetamide on Arabidopsis Thaliana Phenotype and Accumulation of N-(2-hydroxy-5-nitrophenyl)acetamide Glucoside
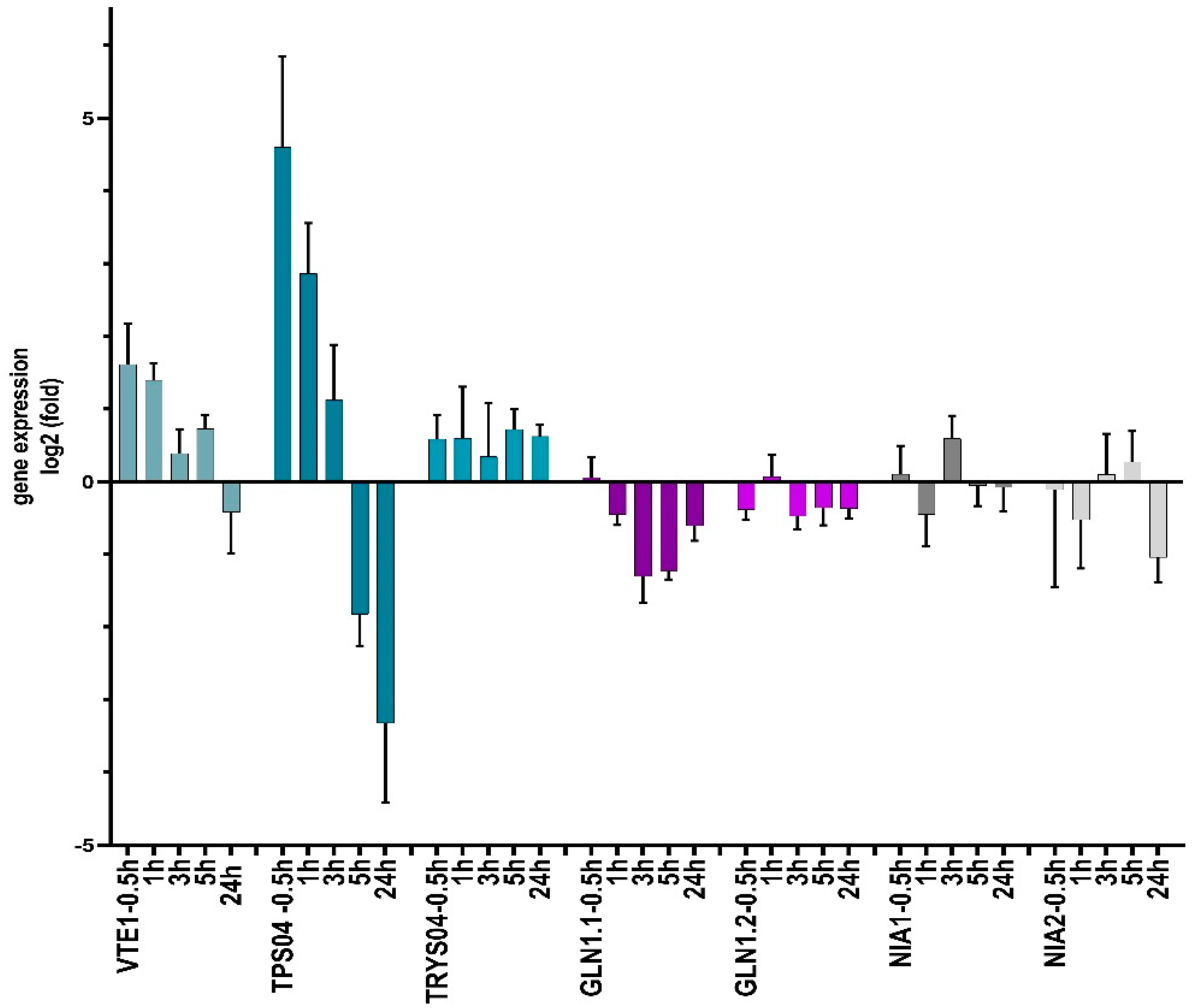
2.3.2. N-(2-hydroxy-5-nitrophenyl)acetamide Influences Gene Expression
2.4. Implication of Nitration of 2-Acetamido-phenol
3. Materials and Methods
3.1. Reference Substances and Compounds Used for Incubations
3.2. Purification of Aqueous Phase F49 (A. aminovorans) for NMR Measurements
3.3. Synthesis of N-(2-hydroxy-3-nitrophenyl)acetamide (4)
3.4. Synthesis of N-(2-hydroxy-6-nitrophenyl)acetamide (7)
3.5. Microorganisms and Plants
3.5.1. Microorganisms
3.5.2. Incubations of Microorganisms
3.5.3. Extraction of Culture Media and Microorganisms
3.5.4. Plants
3.5.5. Plant Incubation, Extraction of Roots, and Leaves of Plants
3.6. Analyses of Compounds in Microbial Culture Media and Plant Extracts
3.6.1. Sample Preparation
3.6.2. LC/HRMS Methods
3.6.3. isCID/MS2 Experiments in ESI Mode
3.6.4. NMR Measurements
3.6.5. Semi-Preparative Purification of the Major Compounds
3.7. Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sicker, D.; Frey, M.; Schulz, M.; Gierl, A. Role of Natural Benzoxazinones in the Survival Strategy of Plants. In International Review of Cytology, a Survey of Cell Biology; Kwang, W.J., Ed.; Academic Press: San Diego, CA, USA, 2000; Volume 198, pp. 319–346. [Google Scholar]
- Sicker, D.; Schulz, M. Benzoxazinones in plants: Occurrence, synthetic access and biological activity. In Natural Products Chemistry 27, Studies Bioactive Natural Products Part H; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 2002; pp. 185–232. [Google Scholar]
- Macías, F.A.; Marín, D.; Oliveros-Bastidas, A.; Castellano, D.; Simonet, A.M.; Molinillo, J.M. Structure−activity relationships (SAR) studies of benzoxazinones, their degradation products and analogues. Phytotoxicity on standard target species (STS). J. Agric. Food Chem. 2005, 53, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Macias, F.A.; Marin, D.; Oliveros-Bastidas, A.; Molinillo, J.M.G. Rediscovering the bioactivity and ecological role of 1,4-benzoxazinones. Nat. Prod. Rep. 2009, 26, 478–489. [Google Scholar] [CrossRef] [PubMed]
- Wouters, F.C.; Gershenzon, J.; Vassao, D.G. Plant defense and herbivore counter-defense: Benzoxazinoids and insect herbivores. J. Braz. Chem. Soc. 2016, 27, 1379–1397. [Google Scholar] [CrossRef] [PubMed]
- Voloshchuk, N.; Schütz, V.; Laschke, L.; Gryganskyi, A.P.; Schulz, M. The Trichoderma viride F-00612 consortium tolerates 2-amino-3H-phenoxazin-3-one and degrades nitrated benzo[d]oxazol-2(3H)-one. Chemoecology 2020, 30, 79–88. Available online: https://link.springer.com/article/10.1007/s00049-020-00300-w (accessed on 24 January 2020). [CrossRef]
- Frébortová, J.; Novák, O.; Frébort, I.; Jorda, R. Degradation of cytokinins by maize cytokinin dehydrogenase is mediated by free radicals generated by enzymatic oxidation of natural benzoxazinones. Plant J. 2010, 61, 467–481. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef]
- Schulz, M.; Marocco, A.; Tabaglio, V.; Macias, F.A.; Molinillo, J.M.G. Benzoxazinoids in rye allelopathy—From discovery to application in sustainable weed control and organic farming. J. Chem. Ecol. 2013, 39, 154–174. [Google Scholar] [CrossRef]
- Schulz, M.; Sicker, D.; Schackow, O.; Hennig, L.; Hofmann, D.; Disko, U.; Ventura, M.; Basyuk, K. 6-Hydroxy-5-nitrobenzo[d]oxazol- 2(3H)-one—A degradable derivative of natural 6-hydroxybenzoxazolin-2(3H)-one produced by Pantoea ananatis. Commun. Integr. Biol. 2017, 10, e1302633. [Google Scholar] [CrossRef][Green Version]
- Friebe, A.; Vilich, V.; Hennig, L.; Kluge, M.; Sicker, D. Detoxification of benzoxazolinone allelochemicals from wheat by Gaeumannomyces graminis var. tritici, G. graminis var. graminis, G. graminis var. avenae, and Fusarium culmorum. Appl. Environ. Microbiol. 1998, 64, 2386–2391. [Google Scholar]
- Fomsgaard, I.S.; Mortensen, A.G.; Carlsen, S.C.K. Microbial transformation products of benzoxazolinone and benzoxazinone allelochemicals—A review. Chemosphere 2004, 54, 1025–1038. [Google Scholar] [CrossRef]
- Glenn, A.E.; Davis, C.B.; Gao, M.; Gold, S.E.; Mitchell, T.R.; Proctor, R.H.; Stewart, J.E.; Snook, M.E. Two horizontally transferred xenobiotic resistance gene clusters associated with detoxification of benzoxazolinones by Fusarium Species. PLoS ONE 2016, 11, e0147486. [Google Scholar] [CrossRef]
- Friebe, A.; Wieland, I.; Schulz, M. Tolerance of Avena sativa to the allelochemical benzoxazolinone. Degradation of BOA by root-colonizing bacteria. Angew. Bot. 1996, 70, 150–154. [Google Scholar]
- Gagliardo, R.W.; Chilton, W.S. Soil transformation of 2(3H)-benzoxazolinone of rye into phytotoxic 2-amino-3H-phenoxazin- 3-one. J. Chem. Ecol. 1992, 18, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Zikmundová, M.; Drandarov, K.; Bigler, L.; Hesse, M.; Werner, C. Biotransformation of 2-benzoxazolinone and 2-hydroxy-1,4-benzoxazin-3-one by endophytic fungi isolated from Aphelandra tetragona. Appl. Environ. Microbiol. 2002, 68, 4863–4870. [Google Scholar] [CrossRef] [PubMed]
- Schütz, V.; Bigler, L.; Girel, S.; Laschke, L.; Sicker, D.; Schulz, M. Conversions of benzoxazinoids and downstream metabolites by soil microorganisms. Front. Ecol. Evol. 2019, 7, 238. [Google Scholar] [CrossRef]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The role of reporting standards for metabolite annotation and identification in metabolomic studies. GigaScience 2013, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Chaleckis, R.; Meister, I.; Zhang, P.; Wheelock, C.E. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. [Google Scholar] [CrossRef]
- Niessen, W.M.A. Tandem mass spectrometry of organic nitro and halogen compounds: Competition between losses of molecules and of radicals. IJMS 2021, 460, 116496. [Google Scholar] [CrossRef]
- Potter, D.W.; Miller, D.W.; Hinson, J.A. Identification of acetaminophen polymerization products catalyzed by horseradish peroxidase. J. Biol. Chem. 1985, 260, 12174–12180. [Google Scholar] [CrossRef]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Rice, A.M.; Long, Y.; King, S.B. Nitroaromatic antibiotics as nitrogen oxide sources. Biomolecules 2021, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Ju, K.S.; Parales, R.E. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol. Mol. Biol. Rev. 2010, 74, 250–272. [Google Scholar] [CrossRef]
- Arnaiz, A.; Rosa-Diaz, I.; Romero-Puertas, M.C.; Sandalio, L.M.; Diaz, I. Nitric oxide, an essential intermediate in the plant–herbivore interaction. Front. Plant Sci. 2021, 11, 2029. [Google Scholar] [CrossRef]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 11. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Padilla, M.N.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitric oxide release from nitro-fatty acids in Arabidopsis roots. Plant Signal. Behav. 2016, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Mun, B.-G.; Imran, Q.M.; Lee, S.-U.; Adamu, T.A.; Shahid, M.; Kim, K.-M.; Yun, B.-W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef]
- Hu, B.; Flemetakis, E.; Rennenberg, H. Pedospheric microbial nitric oxide production challenges root symbioses. Trends Plant Sci. 2021, 26, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-specific NO synthesis by Arabidopsis thaliana nitrate reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef]
- Sánchez-Vicente, I.; Fernández-Espinosa, M.G.; Lorenzo, O. Nitric oxide molecular targets: Reprogramming plant development upon stress. J. Exp. Bot. 2019, 70, 4441–4460. [Google Scholar] [CrossRef]
- Kanwischer, M.; Porfirova, S.; Bergmuller, E.; Dörmann, P. Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol. 2005, 137, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, K.; Inoue, E.; Watanabe-Takahashi, A.; Obara, M.; Yamaya, T.; Takahashi, H. Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J. Biol. Chem. 2004, 279, 16598–16605. [Google Scholar] [CrossRef]
- Yao, J.; Luo, J.-S.; Xiao, Y.; Zhang, Z. The plant defensin gene AtPDF2.1 mediates ammonium metabolism by regulating glutamine synthetase activity in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 557. [Google Scholar] [CrossRef]
- Herde, M.; Gärtner, K.; Köllner, T.G.; Fode, B.; Boland, W.; Gershenzon, J.; Gatz, C.; Tholl, D. Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile c16-homoterpene TMTT. Plant Cell 2008, 20, 1152–1168. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Halitschke, R.; Li, D.; Paetz, C.; Su, H.; Heiling, S.; Xu, S.; Baldwin, I. Controlled hydroxylations of diterpenoids allow for plant chemical defense without autotoxicity. Science 2021, 371, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Desel, C.; Hubbermann, E.M.; Schwarz, K.; Krupinska, K. Nitration of γ-tocopherol in plant tissues. Planta 2007, 226, 1311–1322. [Google Scholar] [CrossRef] [PubMed]
- Selmar, D.; Abouzeid, S.; Radwan, A.; Hijazin, T.; Yahyazadeh, M.; Laura Lewerenz, L.; Nowak, M.; Kleinwächter, M. Horizontal Natural Product Transfer: A Novel Attribution in Allelopathy. In Co-Evolution of Secondary Metabolites. Reference Series in Phytochemistry; Mérillon, J.M., Ramawat, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Lewerenz, L.; Hijazin, T.; Abouzeid, S.; Hänsch, R.; Selmar, D. Pilot study on the uptake and modification of harmaline in acceptor plants: An innovative approach to visualize the interspecific transfer of natural products. Phytochemistry 2020, 174, 112362. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Keiluweit, M.; Wanzek, T.; Kleber, M.; Nico, P.; Fendorf, S. Anaerobic microsites have an unaccounted role in soil carbon stabilization. Nat. Commun. 2017, 8, 1771. [Google Scholar] [CrossRef]
- Bardi, L.; Nari, L.; Morone, C.; Faga, M.G.; Malusà, E. Possible role of high temperature and soil biological fertility on kiwifruit early decline syndrome. Front. Agron. 2020, 2, 580659. [Google Scholar] [CrossRef]
- Inglett, P.W.; Reddy, R.; Corstanje, R. Encyclopedia of Soils in the Environment; Hillel, D., Ed.; Anaerobic Soils; Academic Press: Cambridge, MA, USA, 2005; pp. 71–78. [Google Scholar]
- Madhusoodanan, J. Microbiota munch on medications, causing big effects on drug activity. Proc. Natl. Acad. Sci. USA 2020, 117, 9135–9137. [Google Scholar] [CrossRef] [PubMed]
- Barbosa da Costa, N.; Hébert, M.-P.; Fugère, V.; Terrat, Y.; Fussmann, G.; Gonzalez, A.; Shapiro, B. A Glyphosate-Based Herbicide Cross-Selects for Antibiotic Resistance Genes in Bacterioplankton Communities. mSystems 2022, 7, e01482-21. [Google Scholar] [CrossRef] [PubMed]
- Rismondo, J.; Schulz, L.M. Not Just Transporters: Alternative Functions of ABC Transporters in Bacillus subtilis and Listeria monocytogenes. Microorganisms 2021, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Crooks, C.R.; Wright, J.; Callery, P.S.; Moreton, J.E. Synthesis and preliminary biological studies of 4- and 5-[2-hydroxy-3-(isopropylamino)propoxy]benzimidazoles: Selective β2 adrenergic blocking agents. J. Med. Chem. 1979, 22, 210–214. [Google Scholar] [CrossRef]
- Siebers, M.; Rohr, T.; Ventura, M.; Schütz, V.; Thies, S.; Kovacic, F.; Jaeger, K.-E.; Berg, M.; Dörmann, P.; Schulz, M. Disruption of microbial community composition and identification of plant growth promoting microorganisms after exposure of soil to rapeseed-derived glucosinolates. PLoS ONE 2018, 13, e0200160. [Google Scholar] [CrossRef]
- Kia, S.H.; Schulz, M.; Ayah, E.; Schouten, A.; Müllenborn, C.; Paetz, C.; Schneider, B.; Hofmann, D.; Disko, U.; Tabaglio, V.; et al. Abutilon theophrasti’s Defense against the allelochemical benzoxazolin-2(3h)-one: Support by Actinomucor elegans. J. Chem. Ecol. 2014, 40, 1286–1298. [Google Scholar] [CrossRef]
- Schulz, M.; Hofmann, D.; Sicker, D.; Hennig, L.; Schütz, V.; Preusche, M.; Thiele, B. Pantoea ananatis converts MBOA to 6-methoxy-4-nitro-benzoxazolin-2 (3H)-one (NMBOA) for cooperative degradation with its native root colonizing microbial consortium. Nat. Prod. Commun. 2018, 13, 1934578X1801301010. [Google Scholar] [CrossRef]
- Stahl, E.; Hartmann, M.; Scholten, N.; Zeier, J. A role for tocopherol biosynthesis in Arabidopsis basal immunity to bacterial infection. Plant Physiol. 2019, 181, 1008–1028. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; D’Auria, J.C.; Farooq, A.; Pichersky, E.; Gershenzon, J. Biosynthesis and Emission of terpenoid volatiles from Arabidopsis Flowers. Plant Cell. 2003, 15, 481–494. [Google Scholar] [CrossRef]
- Liu, C.; Sun, Q.; Zhao, L.; Li, Z.; Peng, Z.; Zhang, J.H. Heterologous expression of the transcription factor EsNAC1 in Arabidopsis enhances abiotic stress resistance and retards growth by regulating the expression of different target genes. Front. Plant Sci. 2018, 9, 1495. [Google Scholar] [CrossRef]
- Costa-Broseta, Á.; Castillo, M.; León, J. Post-translational modifications of nitrate reductases autoregulates nitric oxide biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 549. [Google Scholar] [CrossRef] [PubMed]

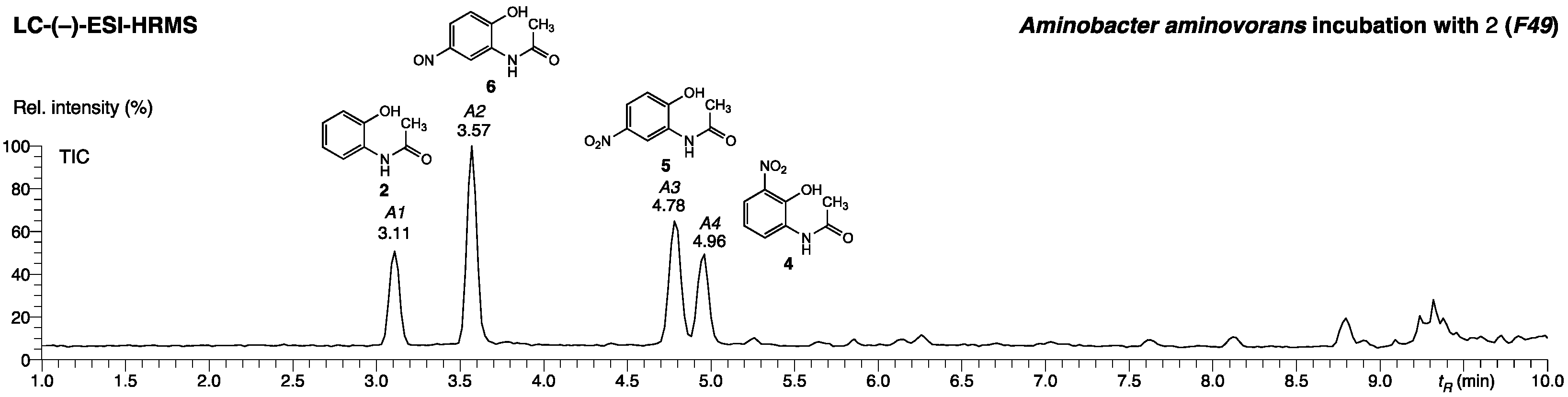
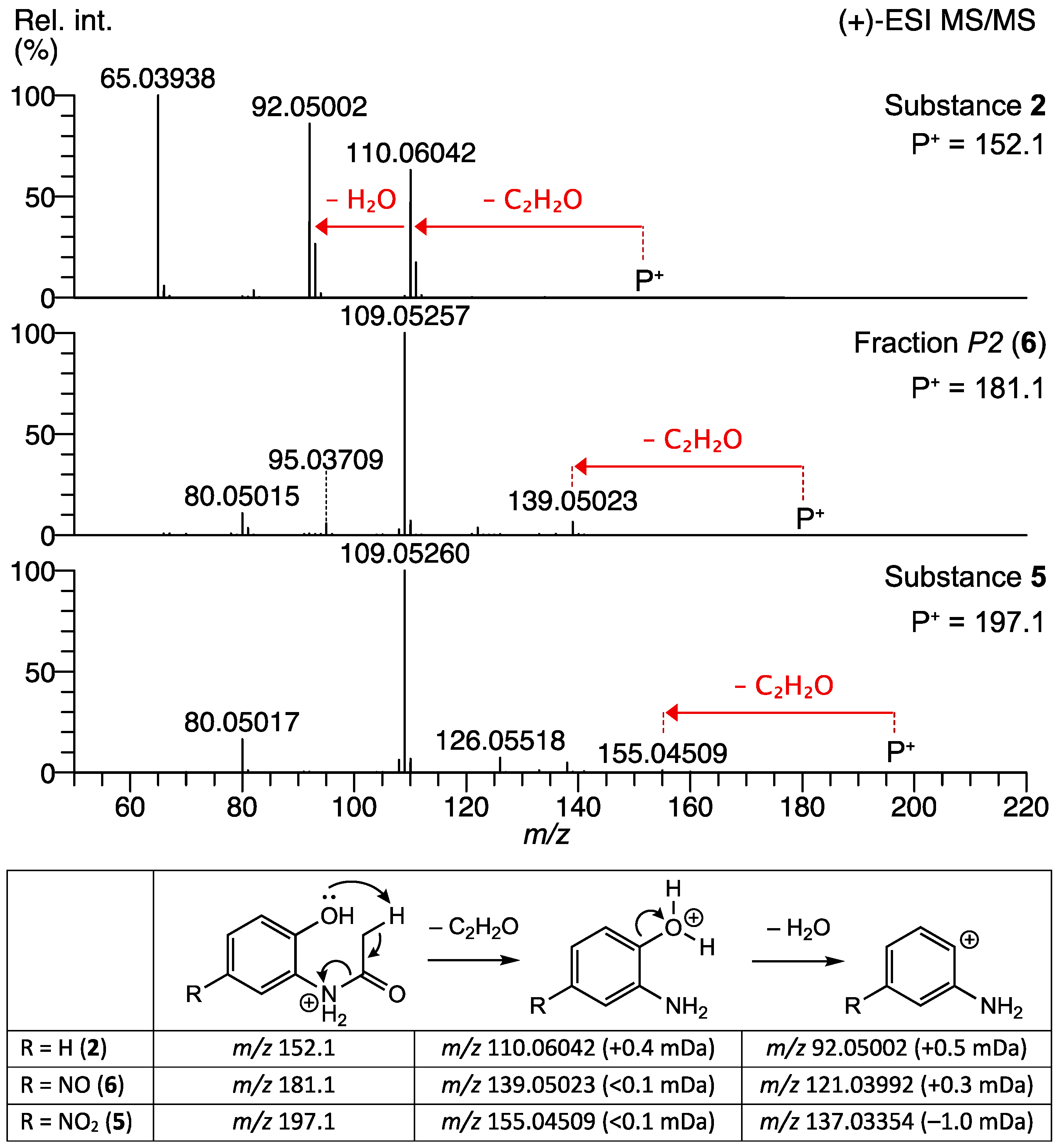
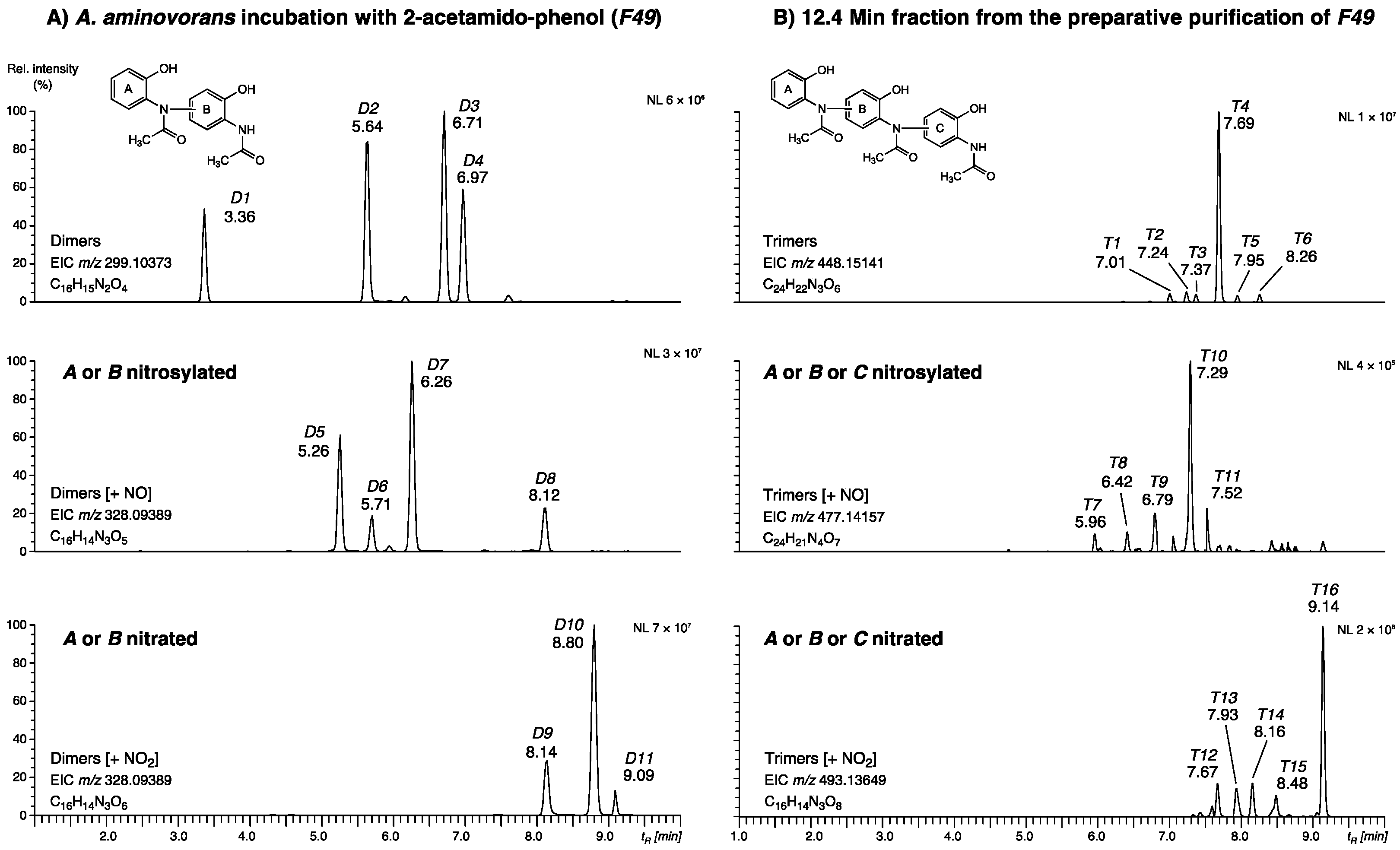
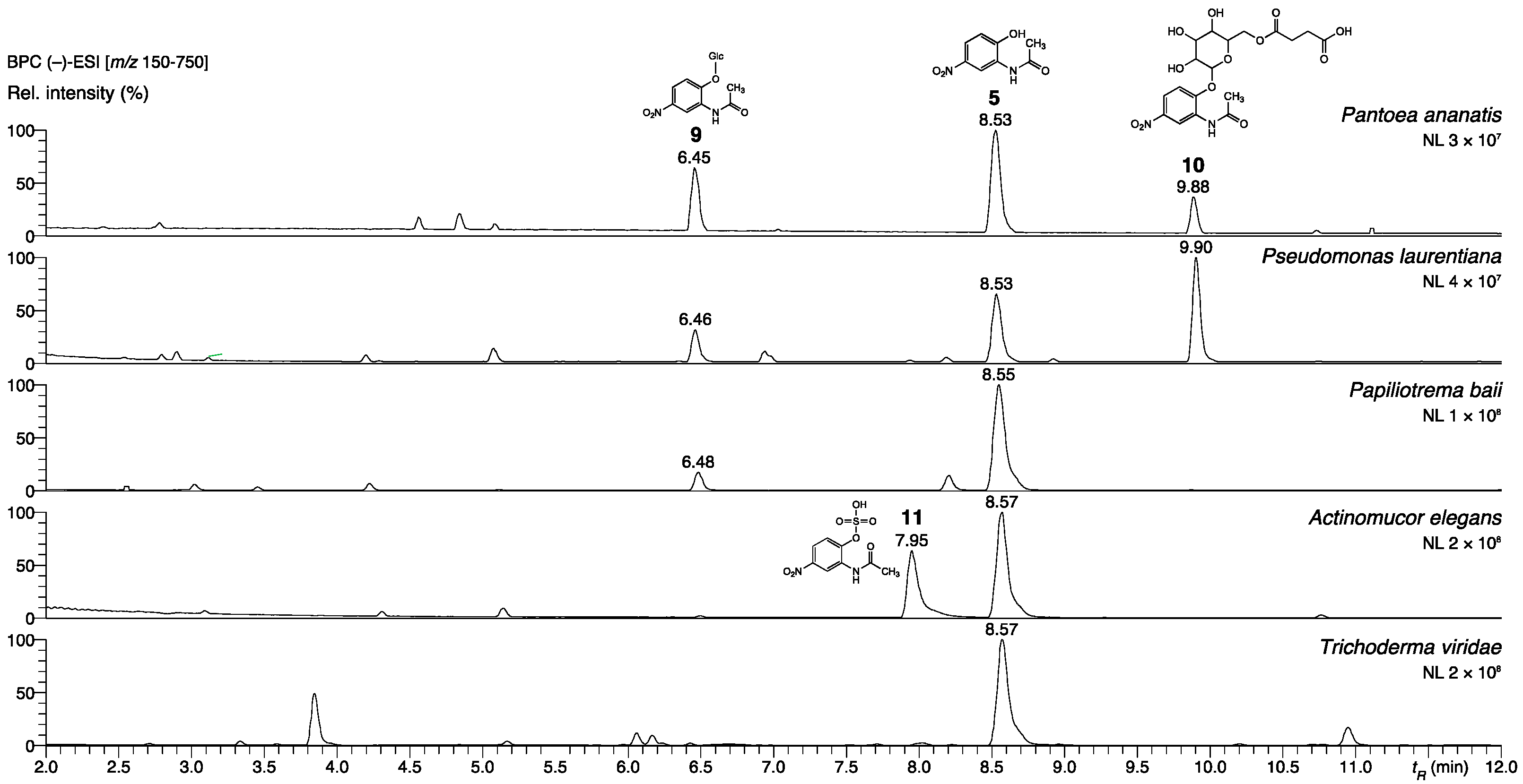
| Peak | tR [min] | Compound | Structure | (–)-ESI (m/z) Deviation [mDa] | ID Level | Reference |
|---|---|---|---|---|---|---|
| A1 | 3.11 | 2-Acetamido-phenol (2) | 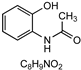 | 150.05621 0.2 | 1 | Commercially available |
| A2 | 3.57 | N-(2-Hydroxy-5-nitrosophenyl)acetamide (6) | 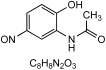 | 179.04637 0.2 | 2 | New structure |
| A3 | 4.78 | N-(2-Hydroxy-5-nitrophenyl)acetamide (5) |  | 195.04134 0.2 | 1 | Commercially available |
| A4 | 4.96 | N-(2-Hydroxy-3-nitrophenyl)acetamide (4) | 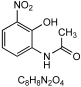 | 195.04136 0.2 | 1 | Synthesized compound |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girel, S.; Schütz, V.; Bigler, L.; Dörmann, P.; Schulz, M. Bioactive Nitrosylated and Nitrated N-(2-hydroxyphenyl)acetamides and Derived Oligomers: An Alternative Pathway to 2-Amidophenol-Derived Phytotoxic Metabolites. Molecules 2022, 27, 4786. https://doi.org/10.3390/molecules27154786
Girel S, Schütz V, Bigler L, Dörmann P, Schulz M. Bioactive Nitrosylated and Nitrated N-(2-hydroxyphenyl)acetamides and Derived Oligomers: An Alternative Pathway to 2-Amidophenol-Derived Phytotoxic Metabolites. Molecules. 2022; 27(15):4786. https://doi.org/10.3390/molecules27154786
Chicago/Turabian StyleGirel, Sergey, Vadim Schütz, Laurent Bigler, Peter Dörmann, and Margot Schulz. 2022. "Bioactive Nitrosylated and Nitrated N-(2-hydroxyphenyl)acetamides and Derived Oligomers: An Alternative Pathway to 2-Amidophenol-Derived Phytotoxic Metabolites" Molecules 27, no. 15: 4786. https://doi.org/10.3390/molecules27154786
APA StyleGirel, S., Schütz, V., Bigler, L., Dörmann, P., & Schulz, M. (2022). Bioactive Nitrosylated and Nitrated N-(2-hydroxyphenyl)acetamides and Derived Oligomers: An Alternative Pathway to 2-Amidophenol-Derived Phytotoxic Metabolites. Molecules, 27(15), 4786. https://doi.org/10.3390/molecules27154786








