Design, Synthesis and Biological Evaluation of [1,2,4]Triazolo[1,5-a]pyrimidine Indole Derivatives against Gastric Cancer Cells MGC-803 via the Suppression of ERK Signaling Pathway
Abstract
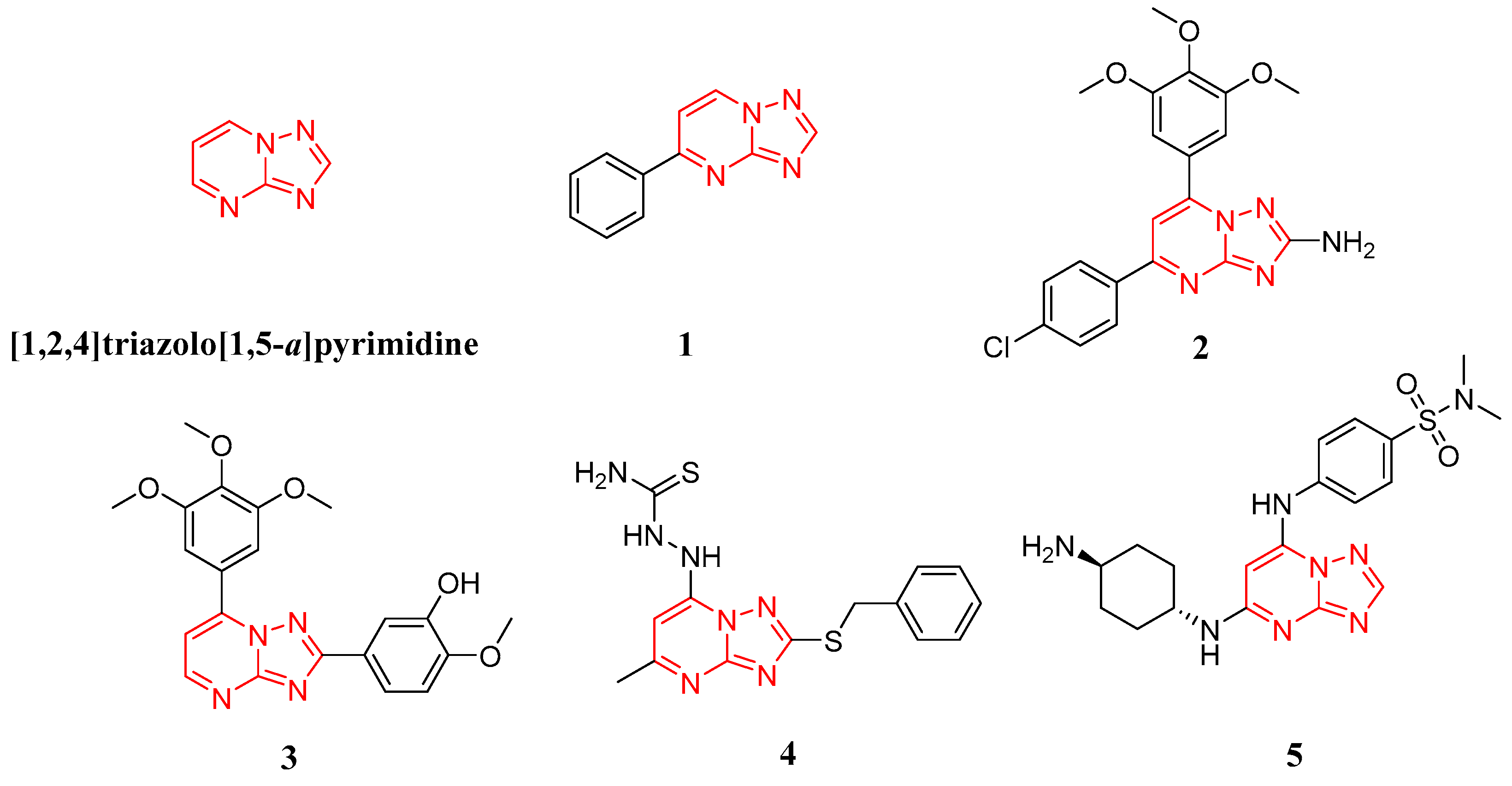
:1. Introduction
2. Results and Discussion
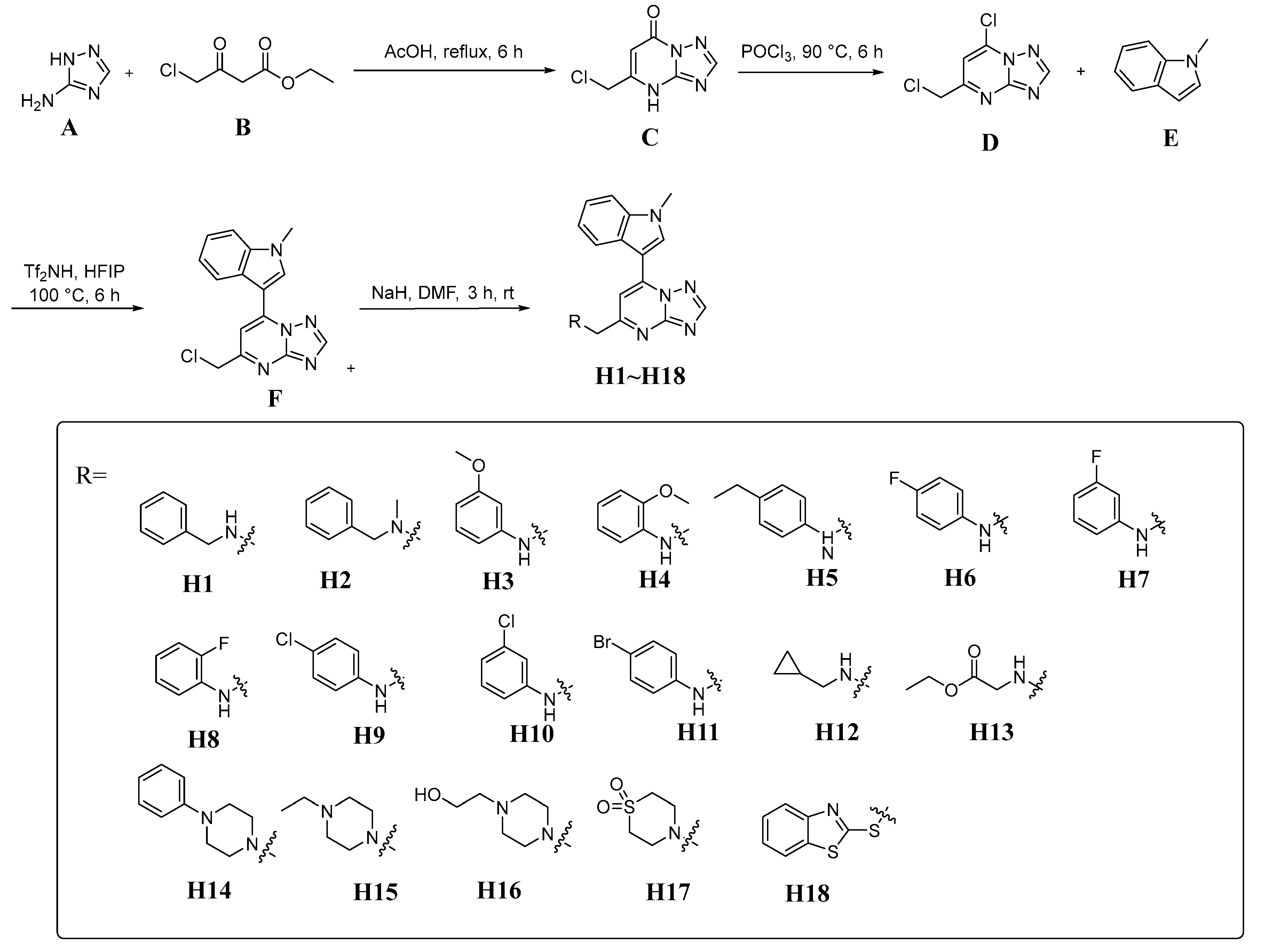
2.1. Chemistry
2.2. Biological Evaluation
Antiproliferative Activities of Compounds H1–H18
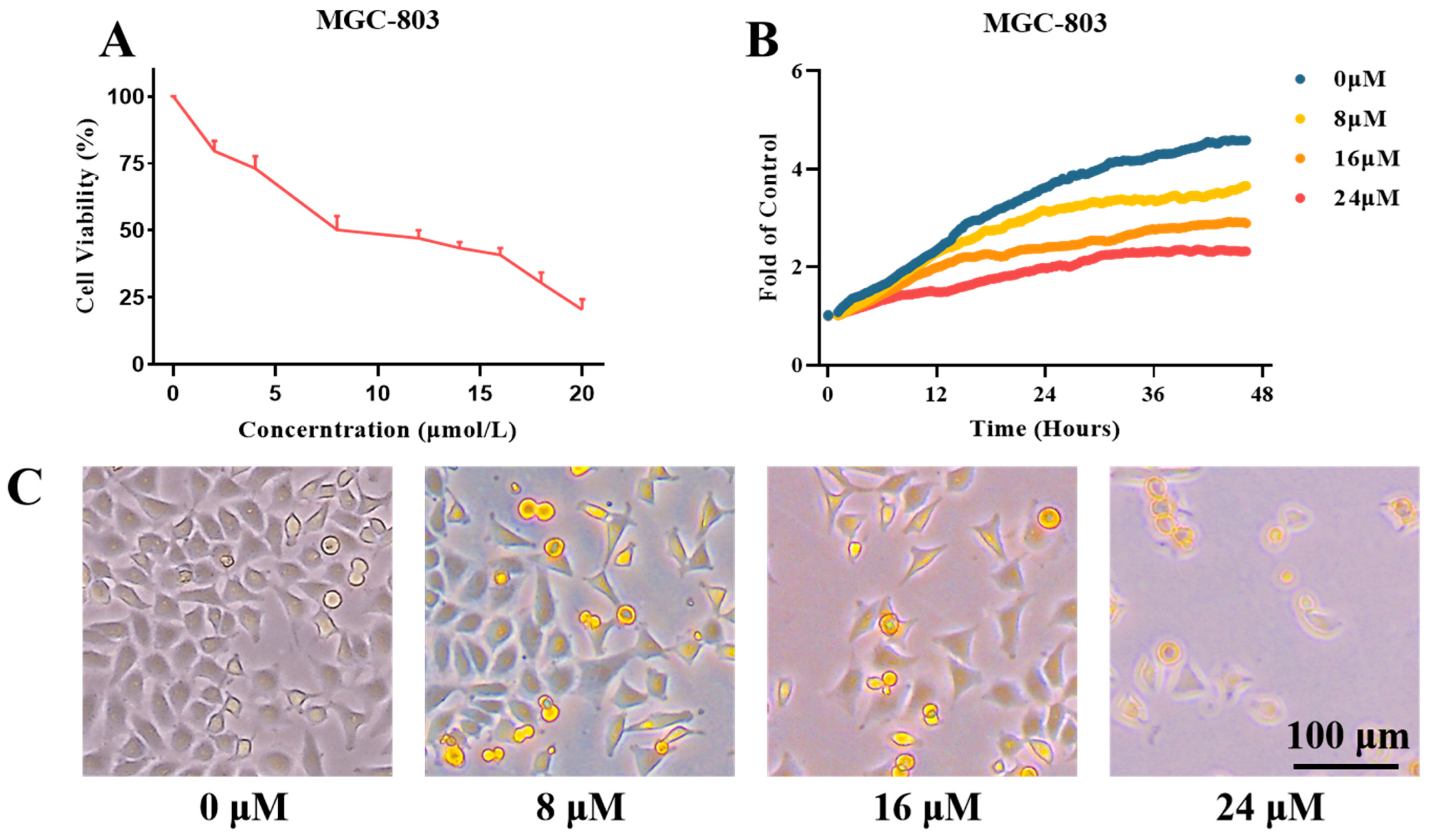
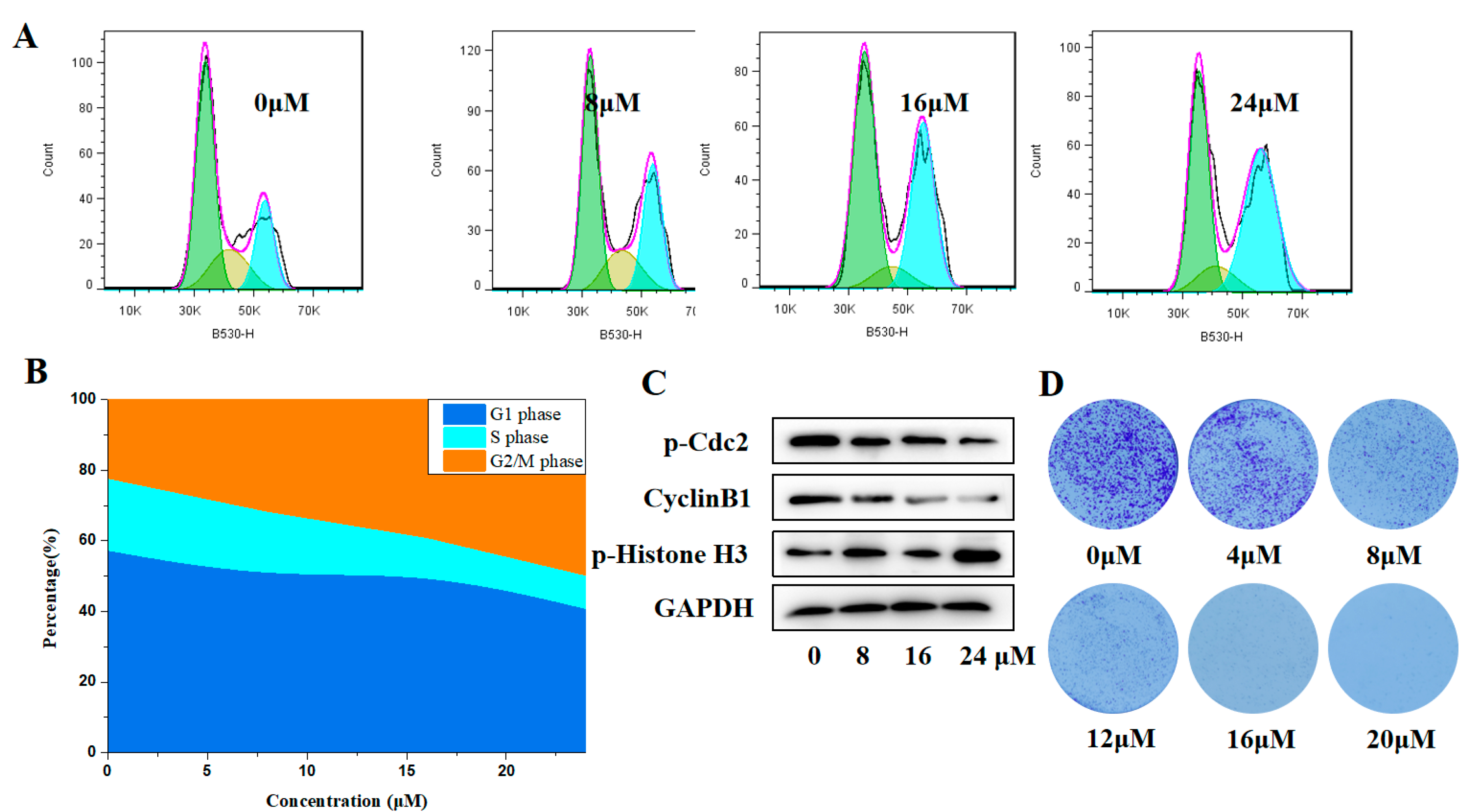
2.3. Inhibitory Effects of Compound H12 on MGC-803 Cells
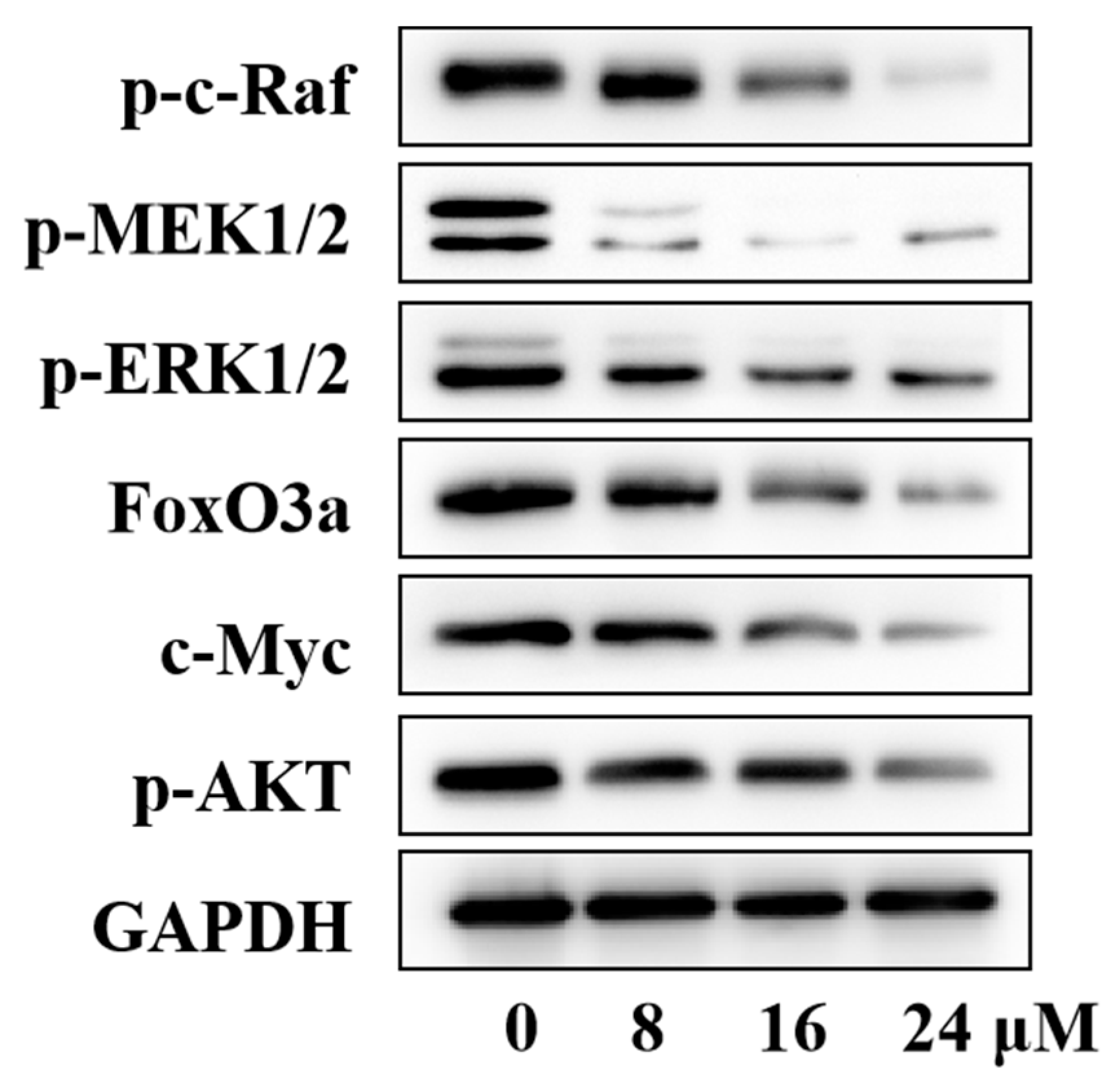
2.4. Inhibition of the ERK Signaling Pathway by Compound H12
2.5. Effects of Compound H11 on the Proliferation of MGC-803 Cells
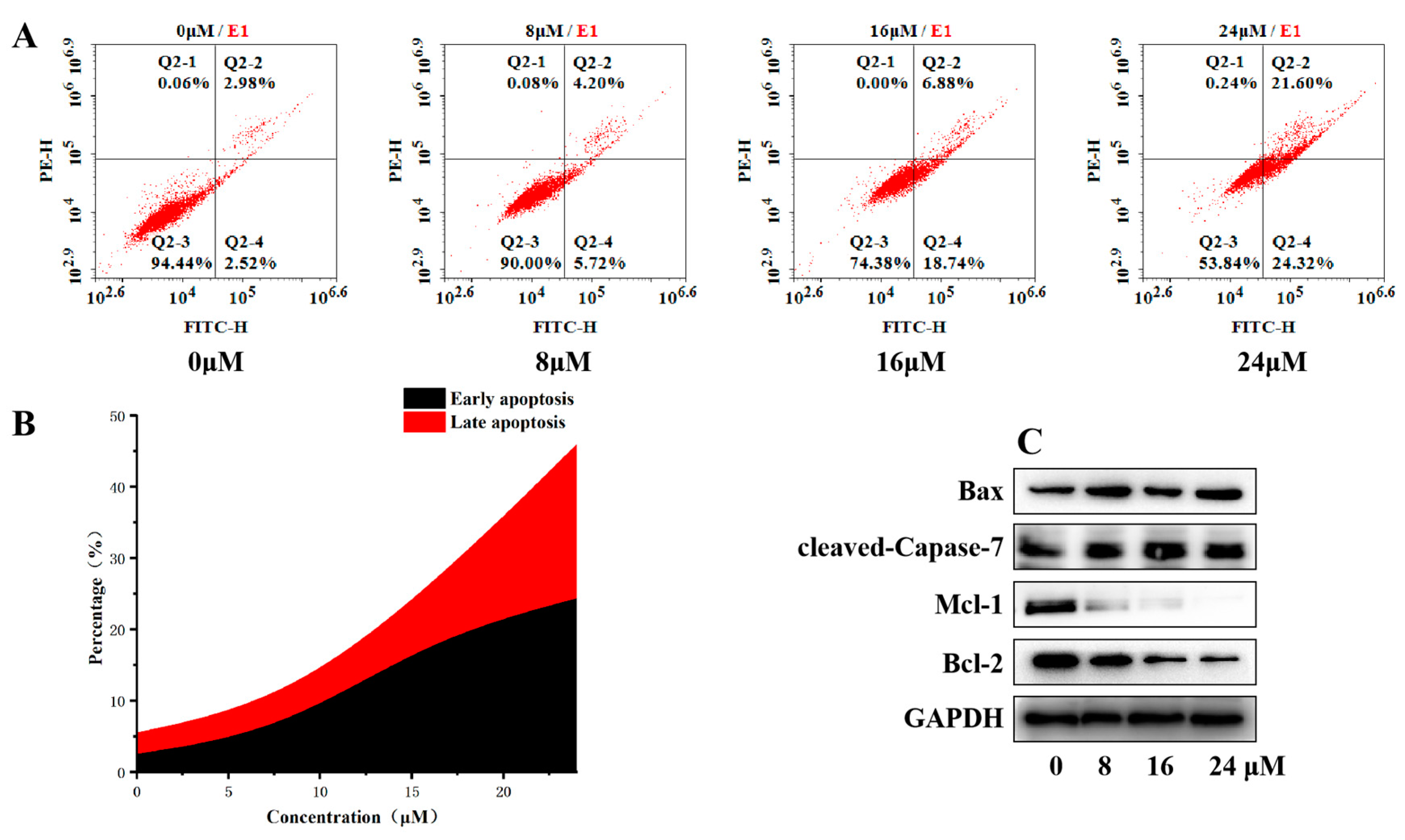
2.6. Effects of Compound H12 on the Apoptosis of MGC-803 Cells
3. Conclusions
4. Materials and Methods
4.1. Synthesis of Compound C
4.2. Synthesis of Compound D
4.3. Synthesis of Compound F
4.4. Synthesis of Compounds H1–H18
4.5. Cell Culture
4.6. MTT Assay
4.7. Colony Formation Assay
4.8. Western Blotting Assay
4.9. Annexin V-FITC/PI Double-Staining Assay for Apoptosis Detection
4.10. Cell Cycle Distribution Assay
4.11. Real-Time Cell Analyzer (RTCA) Cell Proliferation Detection Assay
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Luo, Y.; Zhang, S.; Liu, Z.-J.; Chen, W.; Fu, J.; Zeng, Q.-F.; Zhu, H.-L. Synthesis and antimicrobical evaluation of a novel class of 1,3,4-thiadiazole: Derivatives bearing 1,2,4-triazolo[1,5-a]pyrimidine moiety. Eur. J. Med. Chem. 2013, 64, 54–61. [Google Scholar] [CrossRef]
- Wang, H.; Lee, M.; Peng, Z.; Blázquez, B.; Lastochkin, E.; Kumarasiri, M.; Bouley, R.; Chang, M.; Mobashery, S. Synthesis and evaluation of 1,2,4-triazolo[1,5-a]pyrimidines as antibacterial agents against Enterococcus faecium. J. Med. Chem. 2015, 58, 4194–4203. [Google Scholar] [CrossRef] [Green Version]
- Oukoloff, K.; Lucero, B.; Francisco, K.R.; Brunden, K.R.; Ballatore, C. 1,2,4-Triazolo[1,5-a]pyrimidines in drug design. Eur. J. Med. Chem. 2019, 165, 332–346. [Google Scholar] [CrossRef]
- Desantis, J.; Massari, S.; Corona, A.; Astolfi, A.; Sabatini, S.; Manfroni, G.; Palazzotti, D.; Cecchetti, V.; Pannecouque, C.; Tramontano, E.; et al. 1,2,4-Triazolo[1,5-a]pyrimidines as a Novel Class of Inhibitors of the HIV-1 Reverse Transcriptase-Associated Ribonuclease H Activity. Molecules 2020, 25, 1183. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Li, C.; Chen, W.; Liu, T.; Yu, M.; Fu, L.; Sun, Y.; Liu, H.; De Clercq, E.; Pannecouque, C.; et al. Fused heterocycles bearing bridgehead nitrogen as potent HIV-1 NNRTIs. Part 3: Optimization of [1,2,4]triazolo[1,5-a]pyrimidine core via structure-based and physicochemical property-driven approaches. Eur. J. Med. Chem. 2015, 92, 754–765. [Google Scholar] [CrossRef]
- Wan, Y.H.; Wu, W.Y.; Guo, S.X.; He, S.J.; Tang, X.D.; Wu, X.Y.; Nandakumar, K.S.; Zou, M.; Li, L.; Chen, X.G.; et al. [1,2,4]Triazolo[1,5-a]pyrimidine derivative (Mol-5) is a new NS5-RdRp inhibitor of DENV2 proliferation and DENV2-induced inflammation. Acta Pharmacol. Sin. 2020, 41, 706–718. [Google Scholar] [CrossRef]
- Gilandoust, M.; Harsha, K.B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Pandey, V.; Lobie, P.E.; Basappa; Rangappa, K.S. Synthesis, characterization and cytotoxicity studies of 1,2,3-triazoles and 1,2,4-triazolo [1,5-a] pyrimidines in human breast cancer cells. Bioorganic. Med. Chem. Lett. 2018, 28, 2314–2319. [Google Scholar] [CrossRef]
- Kohandel, O.; Sheikhi-Mohammareh, S.; Oroojalian, F.; Memariani, T.; Mague, J.; Shiri, A. A Dimroth rearrangement approach for the synthesis of selenopheno[2,3-e][1,2,4]triazolo[1,5-c]pyrimidines with cytotoxic activity on breast cancer cells. Mol. Divers. 2021, 26, 1621–1633. [Google Scholar] [CrossRef]
- Mohamed, H.S.; Amin, N.H.; El-Saadi, M.T.; Abdel-Rahman, H.M. Design, synthesis, biological assessment, and in-Silico studies of 1,2,4-triazolo[1,5-a]pyrimidine derivatives as tubulin polymerization inhibitors. Bioorganic. Chem. 2022, 121, 105687. [Google Scholar] [CrossRef]
- Yang, F.; Yu, L.Z.; Diao, P.C.; Jian, X.E.; Zhou, M.F.; Jiang, C.S.; You, W.W.; Ma, W.F.; Zhao, P.L. Novel [1,2,4]triazolo[1,5-a]pyrimidine derivatives as potent antitubulin agents: Design, multicomponent synthesis and antiproliferative activities. Bioorganic. Chem. 2019, 92, 103260. [Google Scholar] [CrossRef]
- Huo, X.S.; Jian, X.E.; Ou-Yang, J.; Chen, L.; Yang, F.; Lv, D.X.; You, W.W.; Rao, J.J.; Zhao, P.L. Discovery of highly potent tubulin polymerization inhibitors: Design, synthesis, and structure-activity relationships of novel 2,7-diaryl-[1,2,4]triazolo[1,5-a]pyrimidines. Eur. J. Med. Chem. 2021, 220, 113449. [Google Scholar] [CrossRef]
- Safari, F.; Bayat, M.; Nasri, S.; Karami, S. Synthesis and evaluation of anti-tumor activity of novel triazolo[1,5-a] pyrimidine on cancer cells by induction of cellular apoptosis and inhibition of epithelial-to-mesenchymal transition process. Bioorg. Med. Chem. Lett. 2020, 30, 127111. [Google Scholar] [CrossRef]
- Oukoloff, K.; Kovalevich, J.; Cornec, A.S.; Yao, Y.; Owyang, Z.A.; James, M.; Trojanowski, J.Q.; Lee, V.M.; Smith, A.B., III; Brunden, K.R.; et al. Design, synthesis and evaluation of photoactivatable derivatives of microtubule (MT)-active [1,2,4]triazolo[1,5-a]pyrimidines. Bioorg. Med. Chem. Lett. 2018, 28, 2180–2183. [Google Scholar] [CrossRef]
- Zhang, N.; Ayral-Kaloustian, S.; Nguyen, T.; Afragola, J.; Hernandez, R.; Lucas, J.; Gibbons, J.; Beyer, C. Synthesis and SAR of [1,2,4]triazolo[1,5-a]pyrimidines, a class of anticancer agents with a unique mechanism of tubulin inhibition. J. Med. Chem. 2007, 50, 319–327. [Google Scholar] [CrossRef]
- Wang, S.; Li, Z.-R.; Suo, F.-Z.; Yuan, X.-H.; Yu, B.; Liu, H.-M. Synthesis, structure-activity relationship studies and biological characterization of new [1,2,4]triazolo[1,5-a]pyrimidine-based LSD1/KDM1A inhibitors. Eur. J. Med. Chem. 2019, 167, 388–401. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, L.J.; Zheng, Y.C.; Shen, D.D.; Miao, E.F.; Qiao, X.P.; Zhao, L.J.; Liu, Y.; Huang, R.; Yu, B.; et al. Design, synthesis and biological evaluation of [1,2,4]triazolo[1,5-a]pyrimidines as potent lysine specific demethylase 1 (LSD1/KDM1A) inhibitors. Eur. J. Med. Chem. 2017, 125, 940–951. [Google Scholar] [CrossRef]
- Richardson, C.M.; Williamson, D.S.; Parratt, M.J.; Borgognoni, J.; Cansfield, A.D.; Dokurno, P.; Francis, G.L.; Howes, R.; Moore, J.D.; Murray, J.B.; et al. Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: Protein structure-guided design and SAR. Bioorg. Med. Chem. Lett. 2006, 16, 1353–1357. [Google Scholar] [CrossRef]
- Sravanthi, T.V.; Manju, S.L. Indoles–A promising scaffold for drug development. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2016, 91, 1–10. [Google Scholar] [CrossRef]
- Chadha, N.; Silakari, O. Indoles as therapeutics of interest in medicinal chemistry: Bird’s eye view. Eur. J. Med. Chem. 2017, 134, 159–184. [Google Scholar] [CrossRef]
- Kaushik, N.K.; Kaushik, N.; Attri, P.; Kumar, N.; Kim, C.H.; Verma, A.K.; Choi, E.H. Biomedical importance of indoles. Molecules 2013, 18, 6620–6662. [Google Scholar] [CrossRef]
- Liu, X.; Pang, X.J.; Liu, Y.; Liu, W.B.; Li, Y.R.; Yu, G.X.; Zhang, Y.B.; Song, J.; Zhang, S.Y. Discovery of Novel Diarylamide N-Containing Heterocyclic Derivatives as New Tubulin Polymerization Inhibitors with Anti-Cancer Activity. Molecules 2021, 26, 4047. [Google Scholar] [CrossRef]
- Song, J.; Guan, Y.F.; Liu, W.B.; Song, C.H.; Tian, X.Y.; Zhu, T.; Fu, X.J.; Qi, Y.Q.; Zhang, S.Y. Discovery of novel coumarin-indole derivatives as tubulin polymerization inhibitors with potent anti-gastric cancer activities. Eur. J. Med. Chem. 2022, 238, 114467. [Google Scholar] [CrossRef]
- Ahmadian-Fard-Fini, S.; Ghanbari, D.; Amiri, O.; Salavati-Niasari, M. Electro-spinning of cellulose acetate nanofibers/Fe/carbon dot as photoluminescence sensor for mercury (II) and lead (II) ions. Carbohydr. Polym. 2020, 229, 115428. [Google Scholar] [CrossRef]
- Monsef, R.; Salavati-Niasari, M. Hydrothermal architecture of Cu(5)V(2)O(10) nanostructures as new electro-sensing catalysts for voltammetric quantification of mefenamic acid in pharmaceuticals and biological samples. Biosens. Bioelectron. 2021, 178, 113017. [Google Scholar] [CrossRef]
- Garnock-Jones, K.P. Panobinostat: First global approval. Drugs 2015, 75, 695–704. [Google Scholar] [CrossRef]
- Greig, S.L. Osimertinib: First Global Approval. Drugs 2016, 76, 263–273. [Google Scholar] [CrossRef]
- Lucas, S. The Pharmacology of Indomethacin. Headache 2016, 56, 436–446. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, Y.; Li, G.; Zhang, Z.; Ma, L.; Cheng, B.; Chen, J. Discovery of Novel Benzimidazole and Indazole Analogues as Tubulin Polymerization Inhibitors with Potent Anticancer Activities. J. Med. Chem. 2021, 64, 4498–4515. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Yuan, X.; Yu, G.; Li, Y.; Zhang, Y.; Song, J.; Li, W.; Zhang, S. Design, Synthesis and Anticancer Activity Studies of Novel Quinoline-Indole Derivatives. Chin. J. Org. Chem. 2021, 41, 3617–3624. [Google Scholar] [CrossRef]
- Zhang, D.; Pang, X.; Liu, H.; Zhang, Q. Design, Synthesis and Anticancer Activity Studies of Novel Indole-Pyrimidine Biaryl Derivatives. Chin. J. Org. Chem. 2021, 41, 267–275. [Google Scholar] [CrossRef]
- Degirmenci, U.; Wang, M.; Hu, J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- Jian, S.; Gao, Q.L.; Wu, B.W.; Li, D.; Shi, L.; Zhu, T.; Lou, J.F.; Jin, C.Y.; Zhang, Y.B.; Zhang, S.Y.; et al. Novel tertiary sulfonamide derivatives containing benzimidazole moiety as potent anti-gastric cancer agents: Design, synthesis and SAR studies. Eur. J. Med. Chem. 2019, 183, 111731. [Google Scholar] [CrossRef]
- Zhao, Q.; Assimopoulou, A.N.; Klauck, S.M.; Damianakos, H.; Chinou, I.; Kretschmer, N.; Rios, J.L.; Papageorgiou, V.P.; Bauer, R.; Efferth, T. Inhibition of c-MYC with involvement of ERK/JNK/MAPK and AKT pathways as a novel mechanism for shikonin and its derivatives in killing leukemia cells. Oncotarget 2015, 6, 38934–38951. [Google Scholar] [CrossRef]
- Santarpia, L.; Lippman, S.M.; El-Naggar, A.K. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 103–119. [Google Scholar] [CrossRef] [Green Version]
- Cagnol, S.; Chambard, J.C. ERK and cell death: Mechanisms of ERK-induced cell death–Apoptosis, autophagy and senescence. FEBS J. 2010, 277, 2–21. [Google Scholar] [CrossRef]
- Song, J.; Wang, S.H.; Song, C.H.; Zhang, W.X.; Zhu, J.X.; Tian, X.Y.; Fu, X.J.; Xu, Y.; Jin, C.Y.; Zhang, S.Y. Discovery of N-benzylarylamide derivatives as novel tubulin polymerization inhibitors capable of activating the Hippo pathway. Eur. J. Med. Chem. 2022, 240, 114583. [Google Scholar] [CrossRef]
- Li, Y.-R.; Liu, F.-F.; Liu, W.-B.; Zhang, Y.-F.; Tian, X.-Y.; Fu, X.-J.; Xu, Y.; Song, J.; Zhang, S.-Y. A novel aromatic amide derivative SY-65 co-targeted tubulin and histone deacetylase 1 with potent anticancer activity in vitro and in vivo. Biochem. Pharmacol. 2022, 201, 115070. [Google Scholar] [CrossRef]
- Sun, Y.-X.; Song, J.; Kong, L.-J.; Sha, B.-B.; Tian, X.-Y.; Liu, X.-J.; Hu, T.; Chen, P.; Zhang, S.-Y. Design, synthesis and evaluation of novel bis-substituted aromatic amide dithiocarbamate derivatives as colchicine site tubulin polymerization inhibitors with potent anticancer activities. Eur. J. Med. Chem. 2022, 229, 114069. [Google Scholar] [CrossRef]

| Compounds | IC50 (μmol/L) a | ||
|---|---|---|---|
| MGC-803 | HCT-116 | MCF-7 | |
| H1 | 43.2 ± 0.81 b | 35.0 ± 0.93 | 45.7 ± 1.30 |
| H2 | 49.5 ± 0.91 | 23.3 ± 0.80 | 23.3 ± 0.27 |
| H3 | 48.5 ± 2.23 | 44.1 ± 1.35 | 68.9 ± 2.15 |
| H4 | 58.3 ± 1.31 | 23.8 ± 0.61 | 34.7 ± 0.88 |
| H5 | 12.4 ± 0.41 | 13.1 ± 0.42 | 28.1 ± 0.92 |
| H6 | 59.9 ± 2.10 | 68.9 ± 3.87 | >80 |
| H7 | 65.0 ± 3.13 | 25.3 ± 0.24 | 64.5 ± 1.79 |
| H8 | >80 | 34.6 ± 0.87 | 65.4 ± 1.89 |
| H9 | 15.1 ± 0.47 | 15.4 ± 0.32 | >80 |
| H10 | 35.3 ± 1.28 | 32.2 ± 0.52 | 55.9 ± 0.09 |
| H11 | 30.5 ± 2.01 | 30.7 ± 2.34 | 52.1 ± 1.66 |
| H12 | 9.47 ± 0.21 | 9.58 ± 0.48 | 13.1 ± 0.39 |
| H13 | 73.9 ± 0.28 | 43.3 ± 0.83 | 74.3 ± 1.61 |
| H14 | >80 | 48.5 ± 0.54 | 49.8 ± 1.26 |
| H15 | 34.1 ± 1.42 | 22.1 ± 2.94 | 31.9 ± 0.33 |
| H16 | 72.5 ± 3.70 | 23.6 ± 0.56 | 38.8 ± 1.87 |
| H17 | 79.3 ± 1.46 | 46.5 ± 1.08 | 62.5 ± 1.39 |
| H18 | 17.5 ± 0.65 | 18.5 ± 0.51 | 39.1 ± 1.41 |
| 5-Fu | 9.91 ± 0.32 | 18.1 ± 1.68 | 14.8 ± 1.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.-X.; Hu, Y.; Zhang, W.-X.; Tian, X.-Y.; Zhang, S.-Y.; Zhang, Y.; Yuan, S.; Song, J. Design, Synthesis and Biological Evaluation of [1,2,4]Triazolo[1,5-a]pyrimidine Indole Derivatives against Gastric Cancer Cells MGC-803 via the Suppression of ERK Signaling Pathway. Molecules 2022, 27, 4996. https://doi.org/10.3390/molecules27154996
Yu G-X, Hu Y, Zhang W-X, Tian X-Y, Zhang S-Y, Zhang Y, Yuan S, Song J. Design, Synthesis and Biological Evaluation of [1,2,4]Triazolo[1,5-a]pyrimidine Indole Derivatives against Gastric Cancer Cells MGC-803 via the Suppression of ERK Signaling Pathway. Molecules. 2022; 27(15):4996. https://doi.org/10.3390/molecules27154996
Chicago/Turabian StyleYu, Guang-Xi, Ying Hu, Wei-Xin Zhang, Xin-Yi Tian, Sai-Yang Zhang, Yan Zhang, Shuo Yuan, and Jian Song. 2022. "Design, Synthesis and Biological Evaluation of [1,2,4]Triazolo[1,5-a]pyrimidine Indole Derivatives against Gastric Cancer Cells MGC-803 via the Suppression of ERK Signaling Pathway" Molecules 27, no. 15: 4996. https://doi.org/10.3390/molecules27154996