Function of Molybdenum Insertases
Abstract
:1. Introduction
2. Catalyzed Reactions of Molybdenum Insertases
3. Characterization of Molybdenum Insertases
3.1. Molecular Characterization of Molybdenum Insertases
3.2. Structural Characterization of Molybdenum Insertases
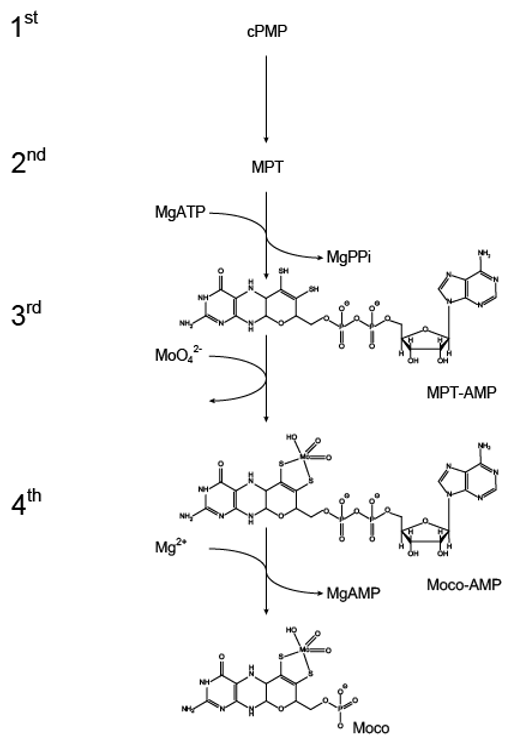
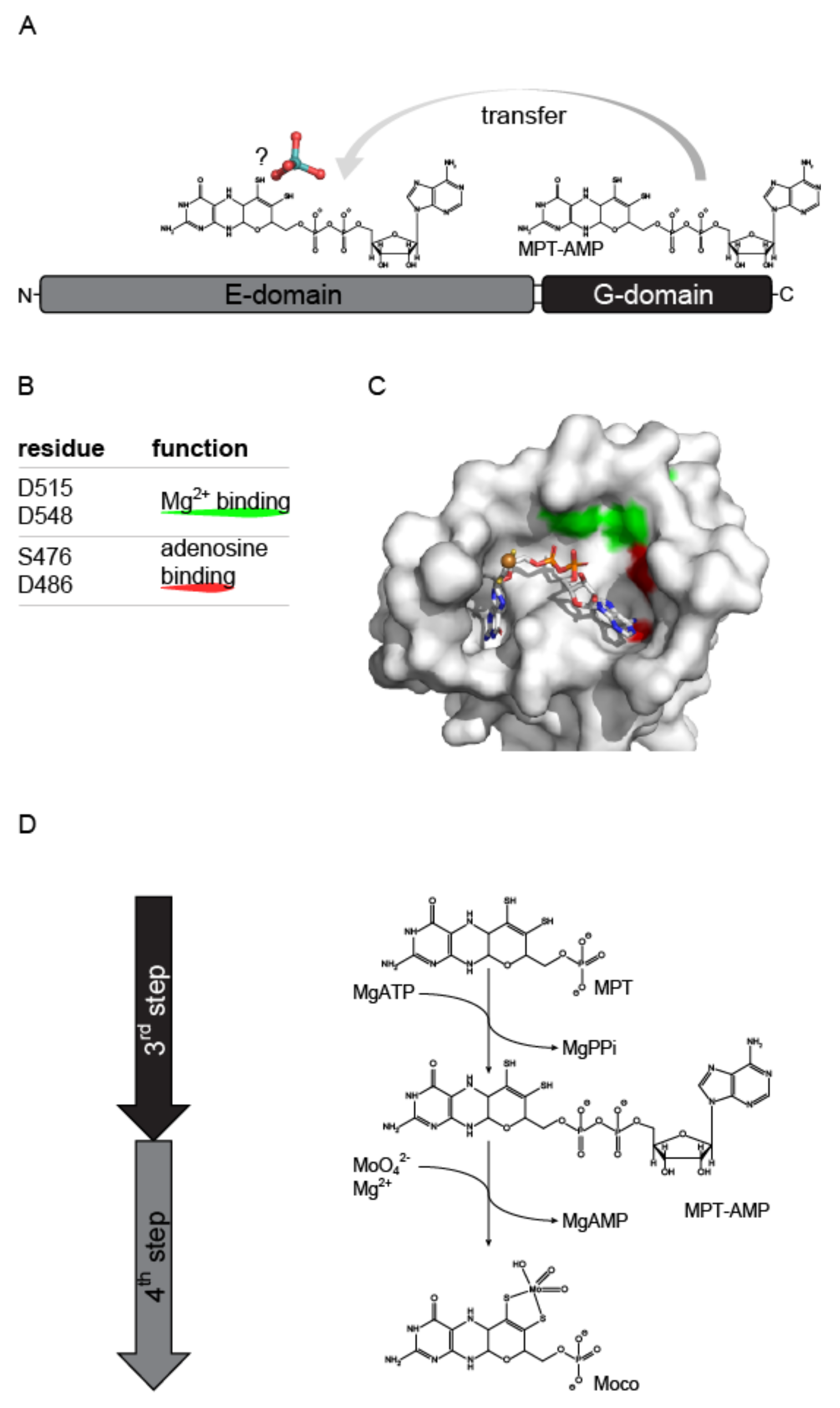
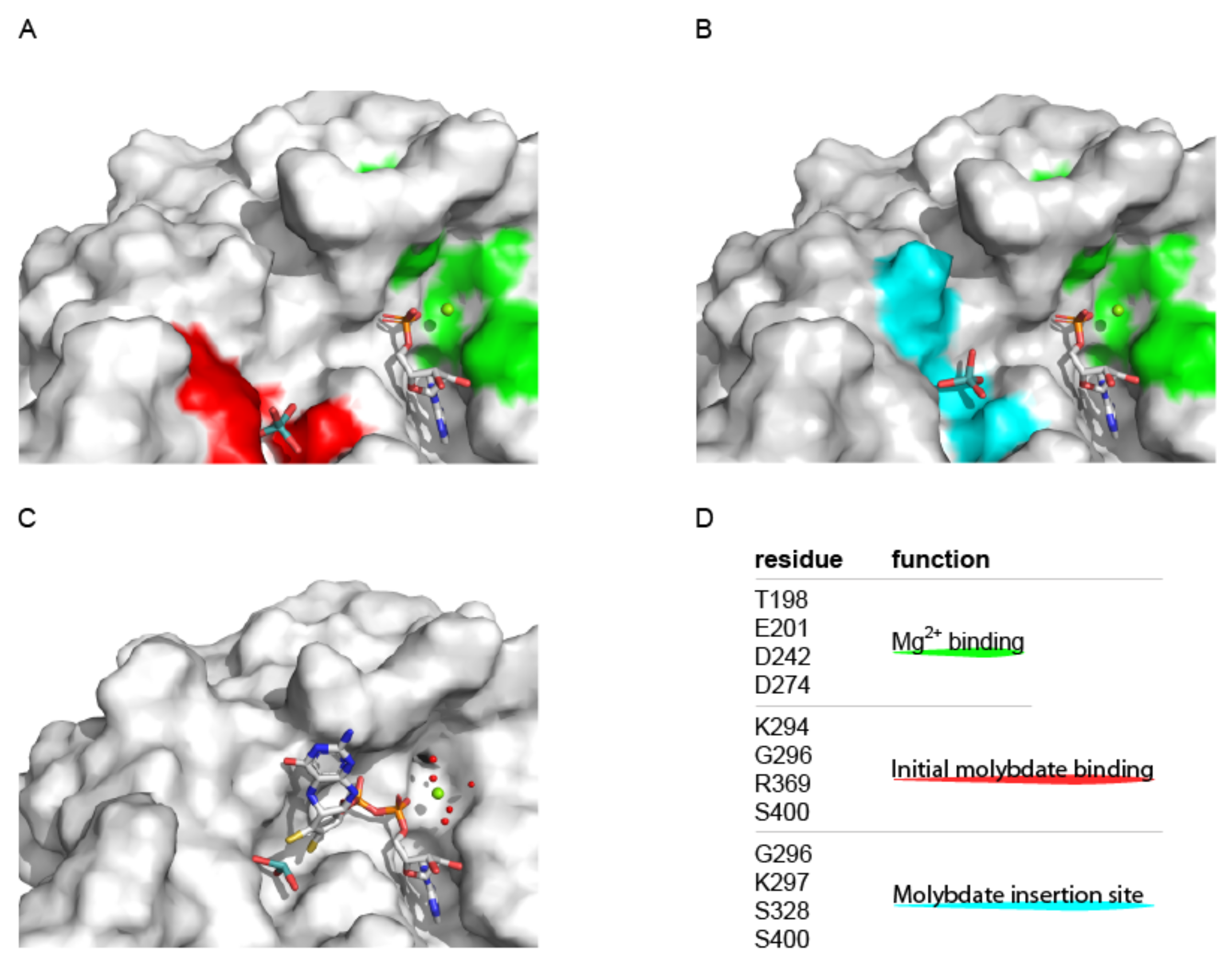
3.2.1. Structural Characterization of the Molybdenum Insertase G-Domain
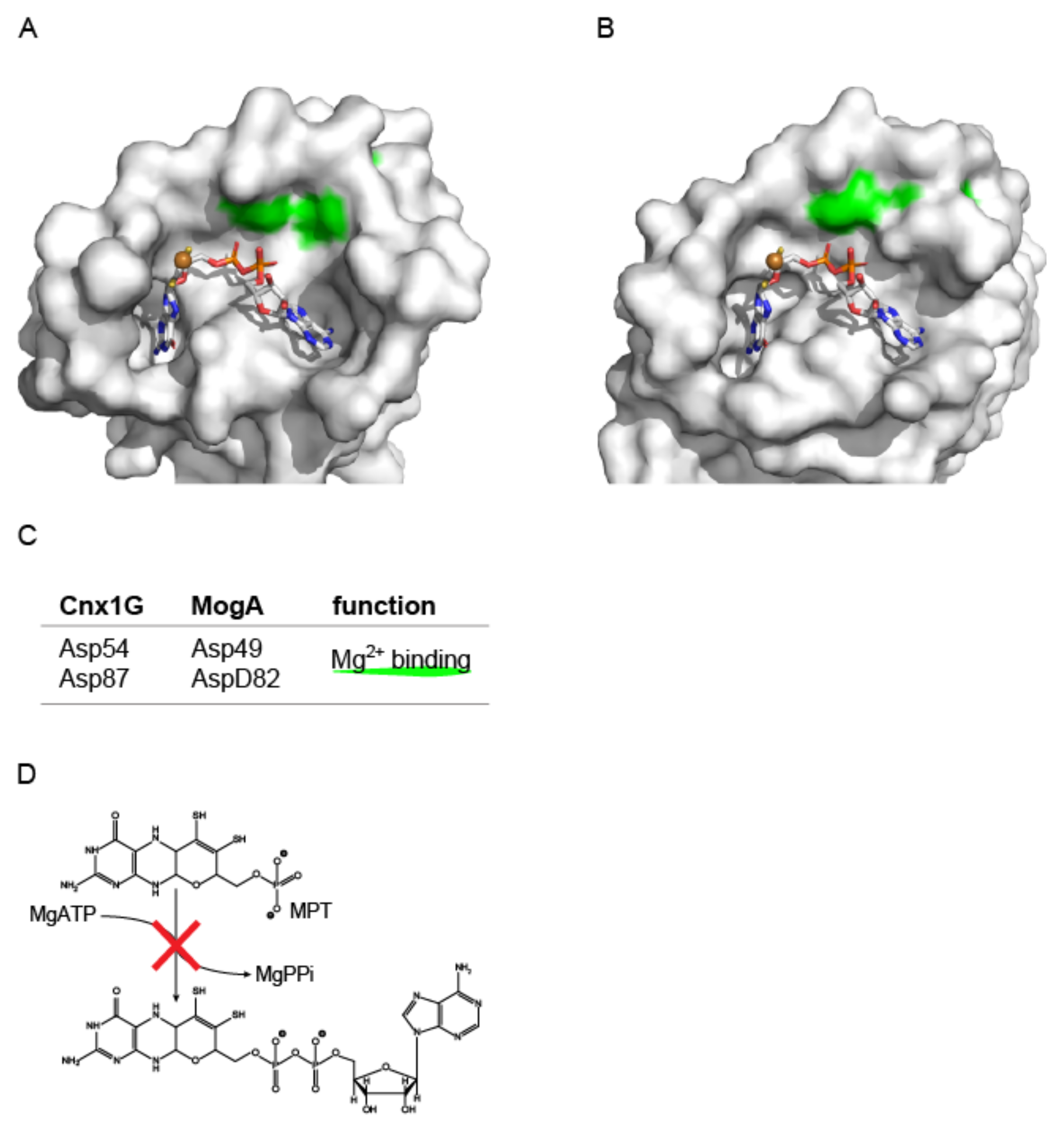
3.2.2. Structural Characterization of the Molybdenum Insertase E-Domain
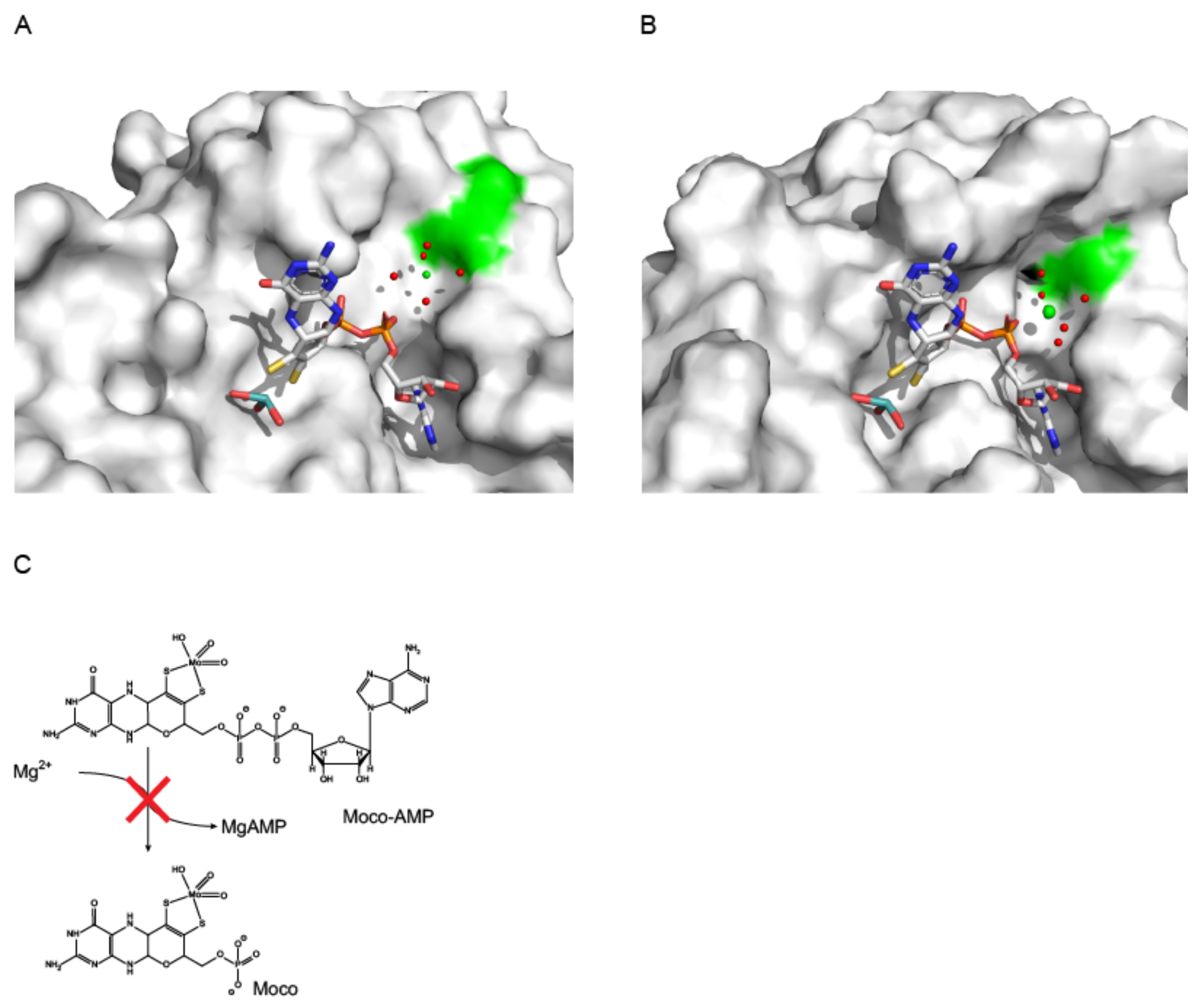
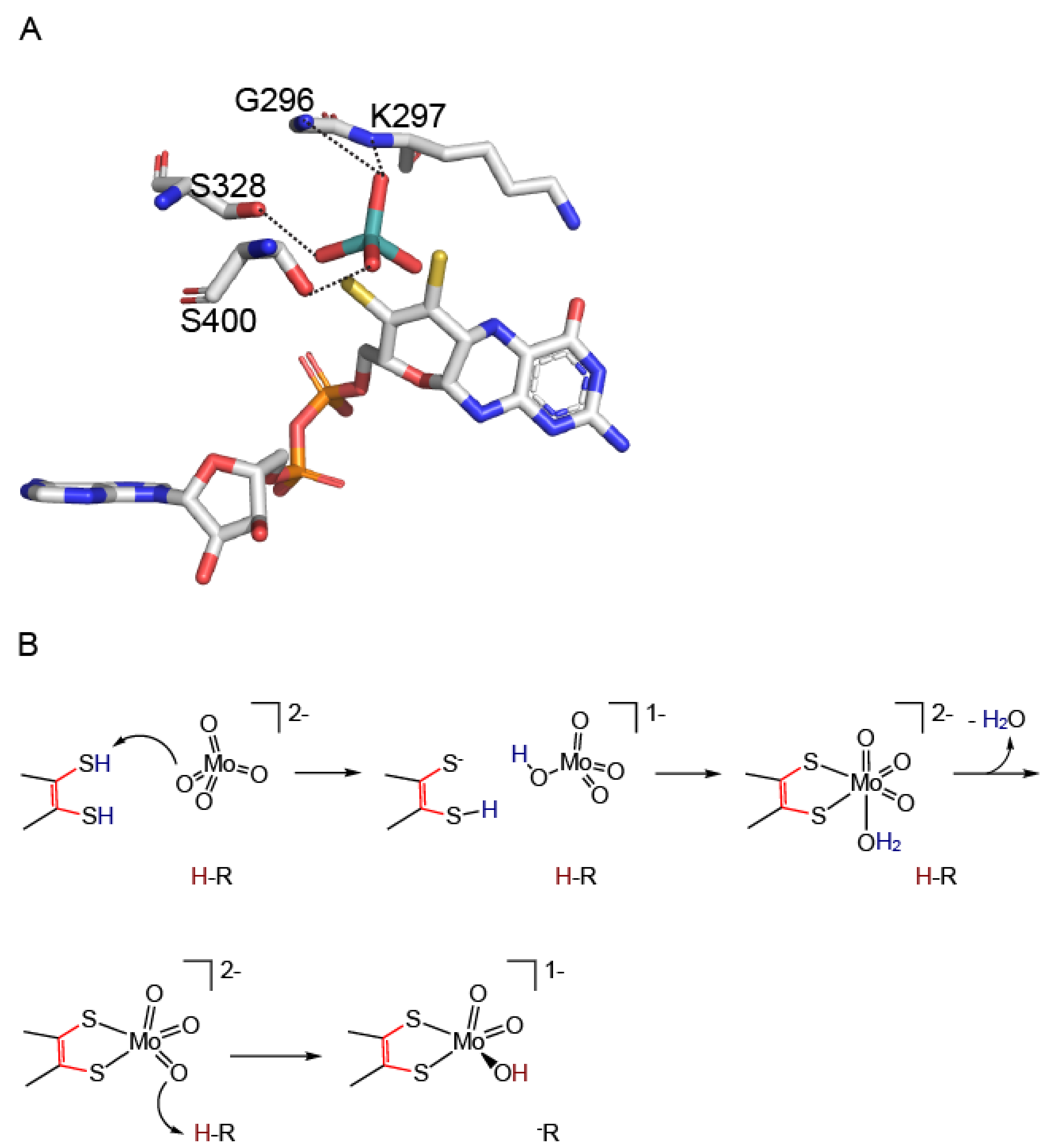
4. Molecular Mechanism of Molybdenum Insertases
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Meulen, H.T. Distribution of Molybdenum. Nature 1932, 130, 966. [Google Scholar] [CrossRef]
- Bortels, H. Molybdän als Katalysator bei der biologischen Stickstoffbindung. Arch. Mikrobiol. 1930, 1, 333–342. [Google Scholar] [CrossRef]
- Hewitt, E. Symptoms of molybdenum deficiency in plants. Soil Sci. Plant Nutr. 1956, 81, 159–172. [Google Scholar] [CrossRef]
- Cove, D.J.; Pateman, J.A. Independently segregating genetic loci concerned with nitrate reductase activity in Aspergillus nidulans. Nature 1963, 198, 262–263. [Google Scholar] [CrossRef]
- Pateman, J.A.; Cove, D.J.; Rever, B.M.; Roberts, D.B. A common cofactor for nitrate reductase and xanthine dehydrogenase wich also regulates the synthesis of nitrate reductase. Nature 1964, 201, 58–60. [Google Scholar] [CrossRef]
- Cove, D.J.; Pateman, J.A.; Rever, B.M. Genetic control of nitrate reduction in Aspergillus nidulans. Heredity 1964, 19, 529. [Google Scholar]
- Arst, H.N., Jr.; MacDonald, D.W.; Cove, D.J. Molybdate metabolism in Aspergillus nidulans. I. Mutations affecting nitrate reductase and-or xanthine dehydrogenase. Mol. Genet. Genom. 1970, 108, 129–145. [Google Scholar] [CrossRef]
- Johnson, J.L.; Hainline, B.E.; Rajagopalan, K.V.; Arison, B.H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J. Biol. Chem. 1984, 259, 5414–5422. [Google Scholar] [CrossRef]
- Johnson, J.L.; Rajagopalan, K.V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc. Natl. Acad. Sci. USA 1982, 79, 6856–6860. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.K.; Mukund, S.; Kletzin, A.; Adams, M.W.; Rees, D.C. Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase. Science 1995, 267, 1463–1469. [Google Scholar] [CrossRef]
- Johnson, J.L.; Rajagopalan, K.V.; Mukund, S.; Adams, M.W.W. Identification of molybdopterin as the organic component of the tungsten cofactor in four enzymes from hyperthermophilic archaea. J. Biol. Chem. 1993, 268, 4848–4852. [Google Scholar] [CrossRef]
- Romao, M.J.; Archer, M.; Moura, I.; Moura, J.J.; LeGall, J.; Engh, R.; Schneider, M.; Hof, P.; Huber, R. Crystal structure of the xanthine oxidase-related aldehyde oxido- reductase from D. gigas. Science 1995, 270, 1170–1176. [Google Scholar] [CrossRef]
- Santamaria-Araujo, J.A.; Fischer, B.; Otte, T.; Nimtz, M.; Mendel, R.R.; Wray, V.; Schwarz, G. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J. Biol. Chem. 2004, 279, 15994–15999. [Google Scholar] [CrossRef] [Green Version]
- Probst, C.; Yang, J.; Krausze, J.; Hercher, T.W.; Richers, C.P.; Spatzal, T.; Kc, K.; Giles, L.J.; Rees, D.C.; Mendel, R.R.; et al. Mechanism of molybdate insertion into pterin-based molybdenum cofactors. Nat. Chem. 2021, 13, 758–765. [Google Scholar] [CrossRef]
- Wuebbens, M.M.; Rajagopalan, K.V. Structural characterization of a molybdopterin precursor. J. Biol. Chem. 1993, 268, 13493–13498. [Google Scholar] [CrossRef]
- Santamaria-Araujo, J.A.; Wray, V.; Schwarz, G. Structure and stability of the molybdenum cofactor intermediate cyclic pyranopterin monophosphate. J. Biol. Inorg. Chem. 2012, 17, 113–122. [Google Scholar] [CrossRef]
- Hover, B.M.; Loksztejn, A.; Ribeiro, A.A.; Yokoyama, K. Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 2013, 135, 7019–7032. [Google Scholar] [CrossRef] [Green Version]
- Hover, B.M.; Tonthat, N.K.; Schumacher, M.A.; Yokoyama, K. Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 6347–6352. [Google Scholar] [CrossRef] [Green Version]
- Pitterle, D.M.; Johnson, J.L.; Rajagopalan, K.V. In vitro synthesis of molybdopterin from precursor Z using purified converting factor. Role of protein-bound sulfur in formation of the dithiolene. J. Biol. Chem. 1993, 268, 13506–13509. [Google Scholar] [CrossRef]
- Wuebbens, M.M.; Rajagopalan, K.V. Mechanistic and Mutational Studies of Escherichia coli Molybdopterin Synthase Clarify the Final Step of Molybdopterin Biosynthesis. J. Biol. Chem. 2003, 278, 14523–14532. [Google Scholar] [CrossRef] [Green Version]
- Rudolph, M.J.; Wuebbens, M.M.; Turque, O.; Rajagopalan, K.V.; Schindelin, H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. J. Biol. Chem. 2003, 278, 14514–14522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuper, J.; Llamas, A.; Hecht, H.J.; Mendel, R.R.; Schwarz, G. Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 2004, 430, 803–806. [Google Scholar] [CrossRef]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of adenylated molybdopterin: An essential step for molybdenum insertion. J. Biol. Chem. 2004, 279, 55241–55246. [Google Scholar] [CrossRef] [Green Version]
- Mendel, R.R. The History of the Molybdenum Cofactor—A Personal View. Molecules 2022, 27, 4934. [Google Scholar] [CrossRef]
- Seelmann, C.S.; Willistein, M.; Heider, J.; Boll, M. Tungstoenzymes: Occurrence, Catalytic Diversity and Cofactor Synthesis. Inorganics 2020, 8, 44. [Google Scholar] [CrossRef]
- Krausze, J.; Hercher, T.W.; Zwerschke, D.; Kirk, M.L.; Blankenfeldt, W.; Mendel, R.R.; Kruse, T. The functional principle of eukaryotic molybdenum insertases. Biochem. J. 2018, 475, 1739–1753. [Google Scholar] [CrossRef]
- Llamas, A.; Otte, T.; Multhaup, G.; Mendel, R.R.; Schwarz, G. The Mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. A novel route toward metal cofactor assembly. J. Biol. Chem. 2006, 281, 18343–18350. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, K.V.; Johnson, J.L. The pterin molybdenum cofactors. J. Biol. Chem. 1992, 267, 10199–10202. [Google Scholar] [CrossRef]
- Hasona, A.; Ray, R.M.; Shanmugam, K.T. Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J. Bacteriol. 1998, 180, 1466–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 2008, 379, 881–899. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Gladyshev, V.N. Comparative genomics of trace elements: Emerging dynamic view of trace element utilization and function. Chem. Rev. 2009, 109, 4828–4861. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G.; Schulze, J.; Bittner, F.; Eilers, T.; Kuper, J.; Bollmann, G.; Nerlich, A.; Brinkmann, H.; Mendel, R.R. The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 2000, 12, 2455–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufholdt, D.; Baillie, C.K.; Meinen, R.; Mendel, R.R.; Hansch, R. The Molybdenum Cofactor Biosynthesis Network: In vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 2017, 8, 1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, T. Eukaryotic Molybdenum Insertases. In Encyclopedia of Inorganic and Bioinorganic Chemistry; Scott, R.A., Ed.; Wiley: Hoboken, NJ, USA, 2020. [Google Scholar]
- Stallmeyer, B.; Nerlich, A.; Schiemann, J.; Brinkmann, H.; Mendel, R.R. Molybdenum co-factor biosynthesis: The Arabidopsis thaliana cDNA cnx1 encodes a multifunctional two-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant J. 1995, 8, 751–762. [Google Scholar] [CrossRef]
- Llamas, A.; Tejada-Jimenez, M.; Gonzalez-Ballester, D.; Higuera, J.J.; Schwarz, G.; Galvan, A.; Fernandez, E. Chlamydomonas reinhardtii CNX1E reconstitutes molybdenum cofactor biosynthesis in Escherichia coli mutants. Eukaryot. Cell 2007, 6, 1063–1067. [Google Scholar] [CrossRef] [Green Version]
- Krausze, J.; Probst, C.; Curth, U.; Reichelt, J.; Saha, S.; Schafflick, D.; Heinz, D.W.; Mendel, R.R.; Kruse, T. Dimerization of the plant molybdenum insertase Cnx1E is required for synthesis of the molybdenum cofactor. Biochem. J. 2017, 474, 163–178. [Google Scholar] [CrossRef]
- Schwarz, G.; Boxer, D.H.; Mendel, R.R. Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 1997, 272, 26811–26814. [Google Scholar] [CrossRef] [Green Version]
- Stewart, V.; MacGregor, C.H. Nitrate reductase in Escherichia coli K-12: Involvement of chlC, chlE, and chlG loci. J. Bacteriol. 1982, 151, 788–799. [Google Scholar] [CrossRef] [Green Version]
- Kuper, J.; Palmer, T.; Mendel, R.R.; Schwarz, G. Mutations in the molybdenum cofactor biosynthetic protein Cnx1G from Arabidopsis thaliana define functions for molybdopterin binding, molybdenum insertion, and molybdenum cofactor stabilization. Proc. Natl. Acad. Sci. USA 2000, 97, 6475–6480. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.T.; Wuebbens, M.M.; Rajagopalan, K.V.; Schindelin, H. Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli. J. Biol. Chem. 2000, 275, 1814–1822. [Google Scholar] [CrossRef] [Green Version]
- Kuper, J.; Winking, J.; Hecht, H.J.; Mendel, R.R.; Schwarz, G. The active site of the molybdenum cofactor biosynthetic protein domain Cnx1G. Arch. Biochem. Biophys. 2003, 411, 36–46. [Google Scholar] [CrossRef]
- Schwarz, G.; Schrader, N.; Mendel, R.R.; Hecht, H.J.; Schindelin, H. Crystal structures of human gephyrin and plant Cnx1 G domains: Comparative analysis and functional implications. J. Mol. Biol. 2001, 312, 405–418. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, V.S., LLC. Available online: https://pymol.org/2/ (accessed on 6 June 2012).
- Dai, X.; Hayashi, K.; Nozaki, H.; Cheng, Y.; Zhao, Y. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3129–3134. [Google Scholar] [CrossRef] [Green Version]
- Kasaragod, V.B.; Schindelin, H. Structural Framework for Metal Incorporation during Molybdenum Cofactor Biosynthesis. Structure 2016, 24, 782–788. [Google Scholar] [CrossRef]
- Hercher, T.W.; Krausze, J.; Hoffmeister, S.; Zwerschke, D.; Lindel, T.; Blankenfeldt, W.; Mendel, R.R.; Kruse, T. Insights into the Cnx1E catalyzed MPT-AMP hydrolysis. Biosci. Rep. 2020, 40, BSR20191806. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.; Nichols, J.; Rajagopalan, K.V.; Schindelin, H. The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin. Structure 2001, 9, 299–310. [Google Scholar] [CrossRef] [Green Version]
- Schrag, J.D.; Huang, W.; Sivaraman, J.; Smith, C.; Plamondon, J.; Larocque, R.; Matte, A.; Cygler, M. The crystal structure of Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway. J. Mol. Biol. 2001, 310, 419–431. [Google Scholar] [CrossRef]
- Sola, M.; Kneussel, M.; Heck, I.S.; Betz, H.; Weissenhorn, W. X-ray crystal structure of the trimeric N-terminal domain of gephyrin. J. Biol. Chem. 2001, 276, 25294–25301. [Google Scholar] [CrossRef] [Green Version]
- Sola, M.; Bavro, V.N.; Timmins, J.; Franz, T.; Ricard-Blum, S.; Schoehn, G.; Ruigrok, R.W.; Paarmann, I.; Saiyed, T.; O’Sullivan, G.A.; et al. Structural basis of dynamic glycine receptor clustering by gephyrin. EMBO J. 2004, 23, 2510–2519. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.D.; Xiang, S.; Schindelin, H.; Rajagopalan, K.V. Mutational analysis of Escherichia coli MoeA: Two functional activities map to the active site cleft. Biochemistry 2007, 46, 78–86. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruse, T. Function of Molybdenum Insertases. Molecules 2022, 27, 5372. https://doi.org/10.3390/molecules27175372
Kruse T. Function of Molybdenum Insertases. Molecules. 2022; 27(17):5372. https://doi.org/10.3390/molecules27175372
Chicago/Turabian StyleKruse, Tobias. 2022. "Function of Molybdenum Insertases" Molecules 27, no. 17: 5372. https://doi.org/10.3390/molecules27175372




