Development and Validation of a Sensitive and Specific LC-MS/MS Method for IWR-1-Endo, a Wnt Signaling Inhibitor: Application to a Cerebral Microdialysis Study
Abstract
:1. Introduction
2. Results and Discussion
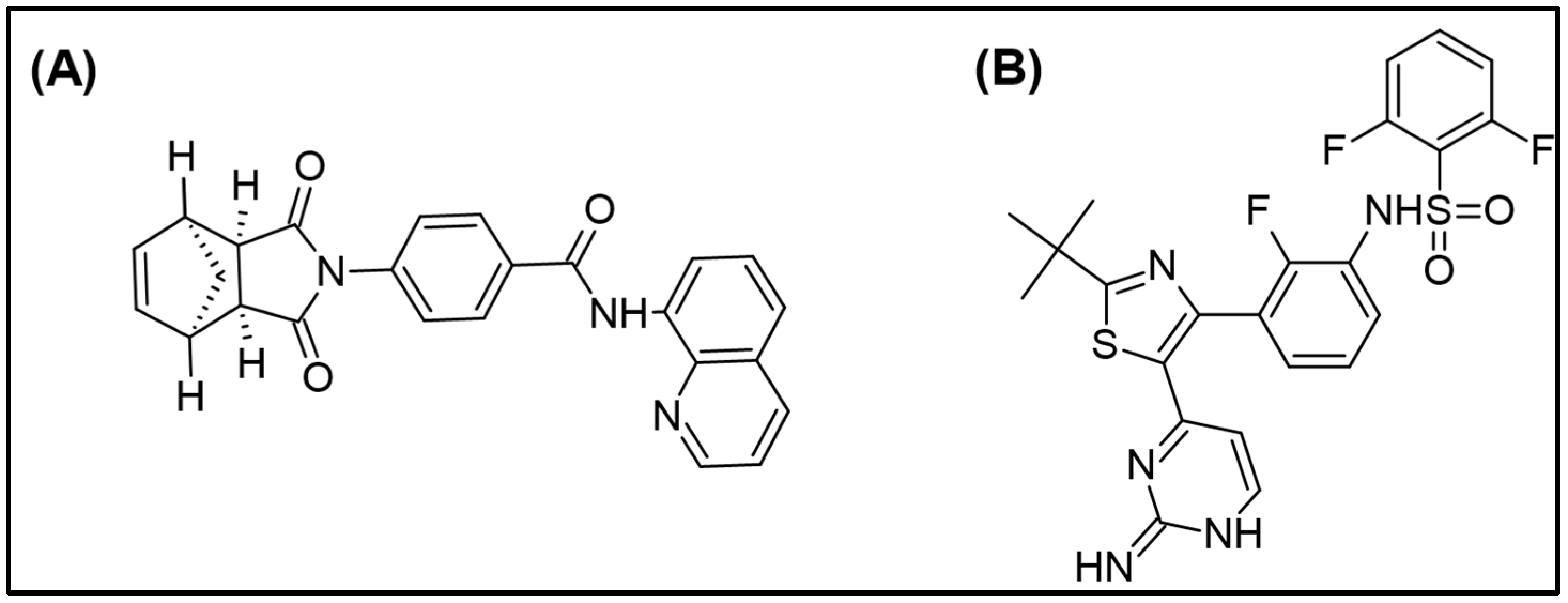
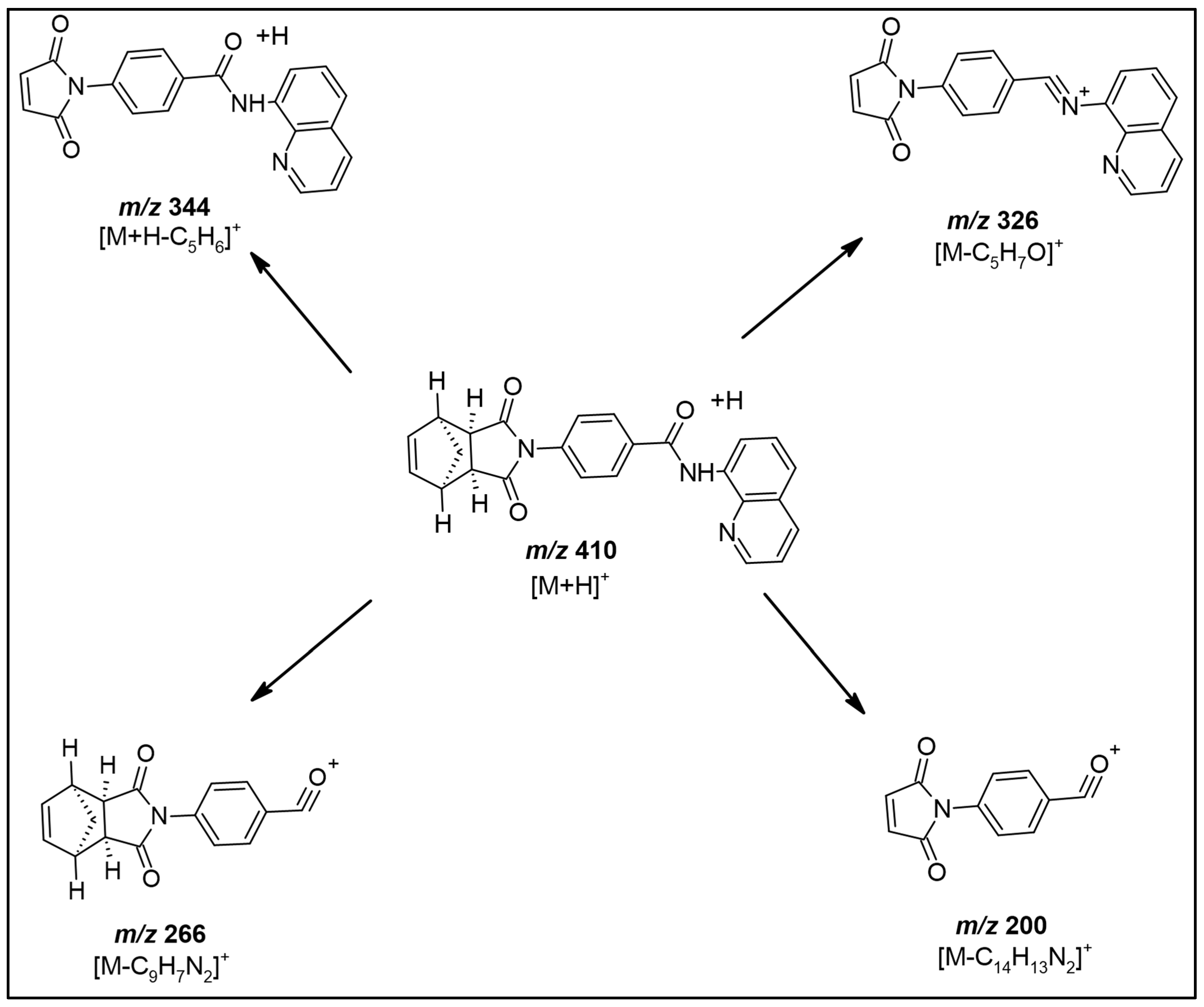
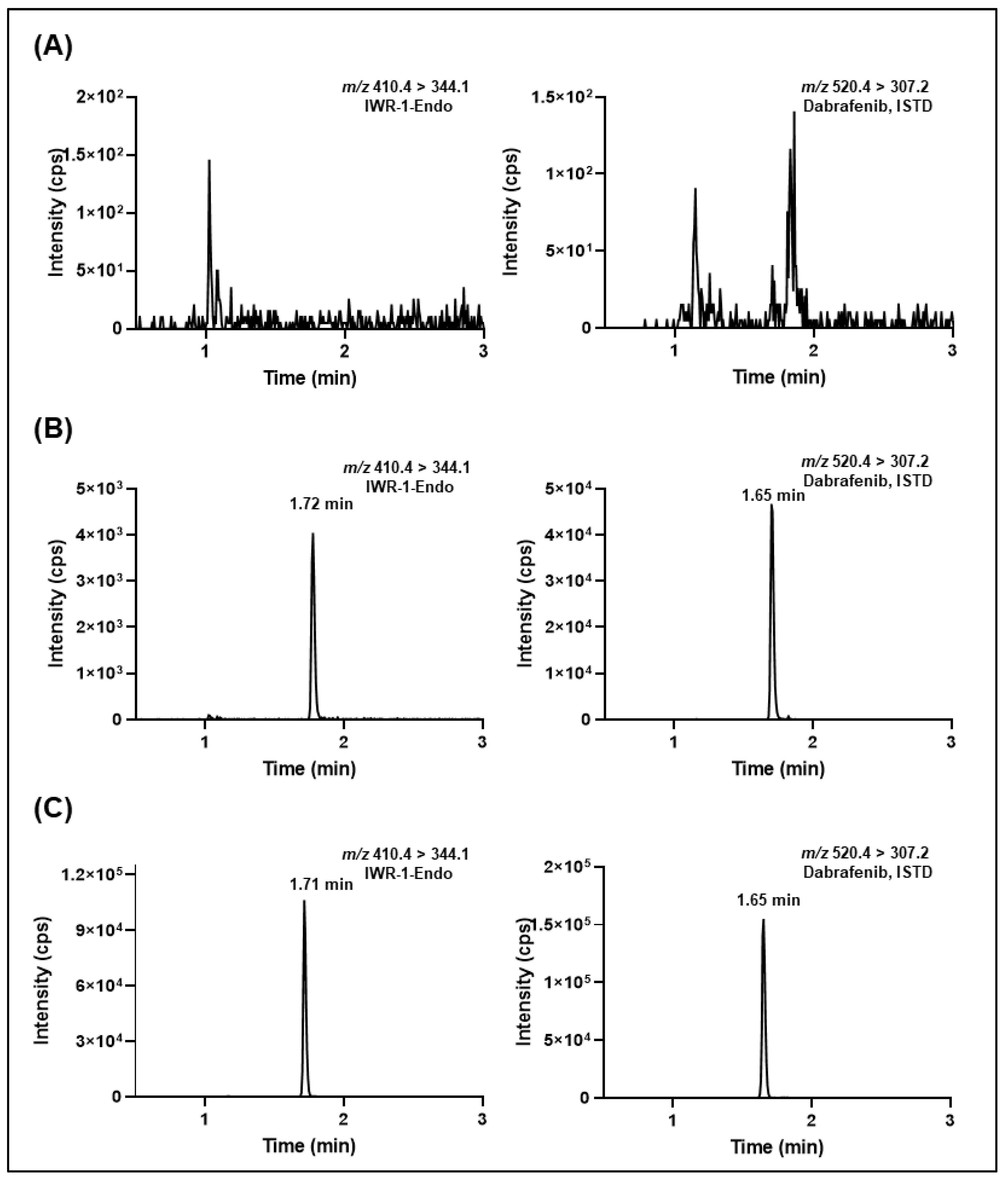
2.1. Optimization of Mass Spectrometric and Chromatographic Conditions
2.2. Optimization of Sample Preparation
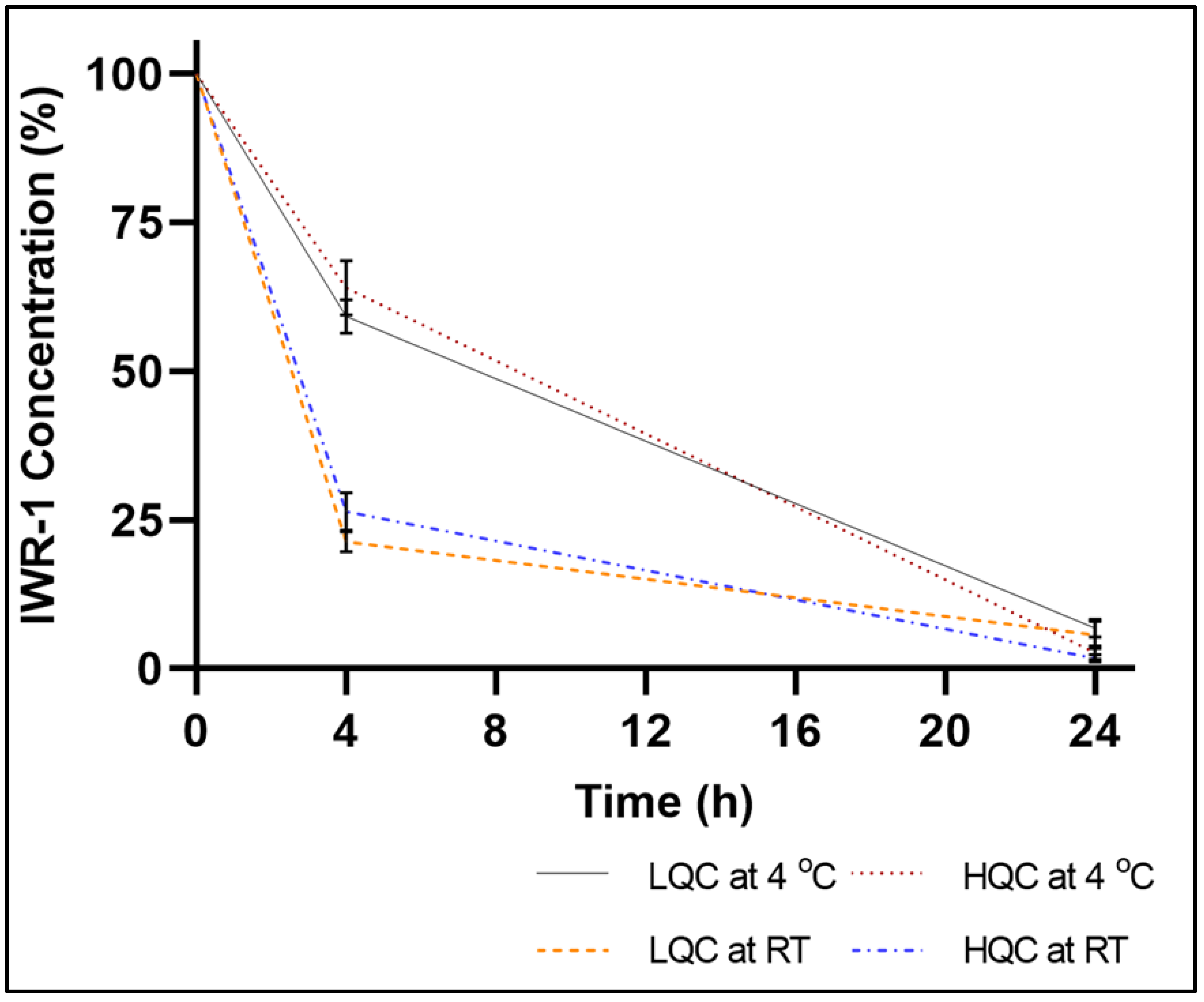
2.3. Stability of IWR-1-Endo in Murine Plasma under Different StorageC
2.4. Addressing IWR-1-Endo Instability in Murine Plasma
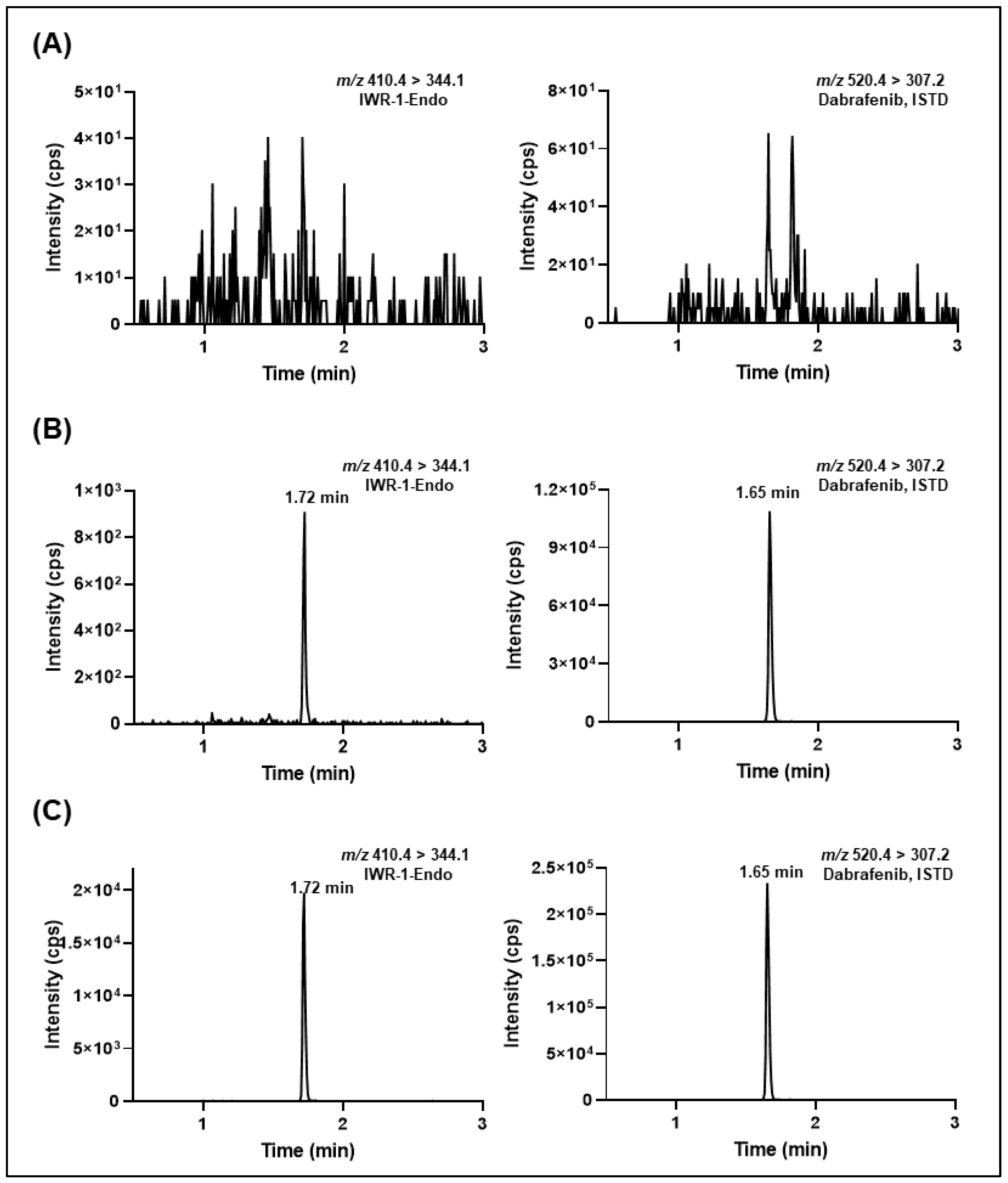
2.5. Validation of the LC-MS/MS Methods
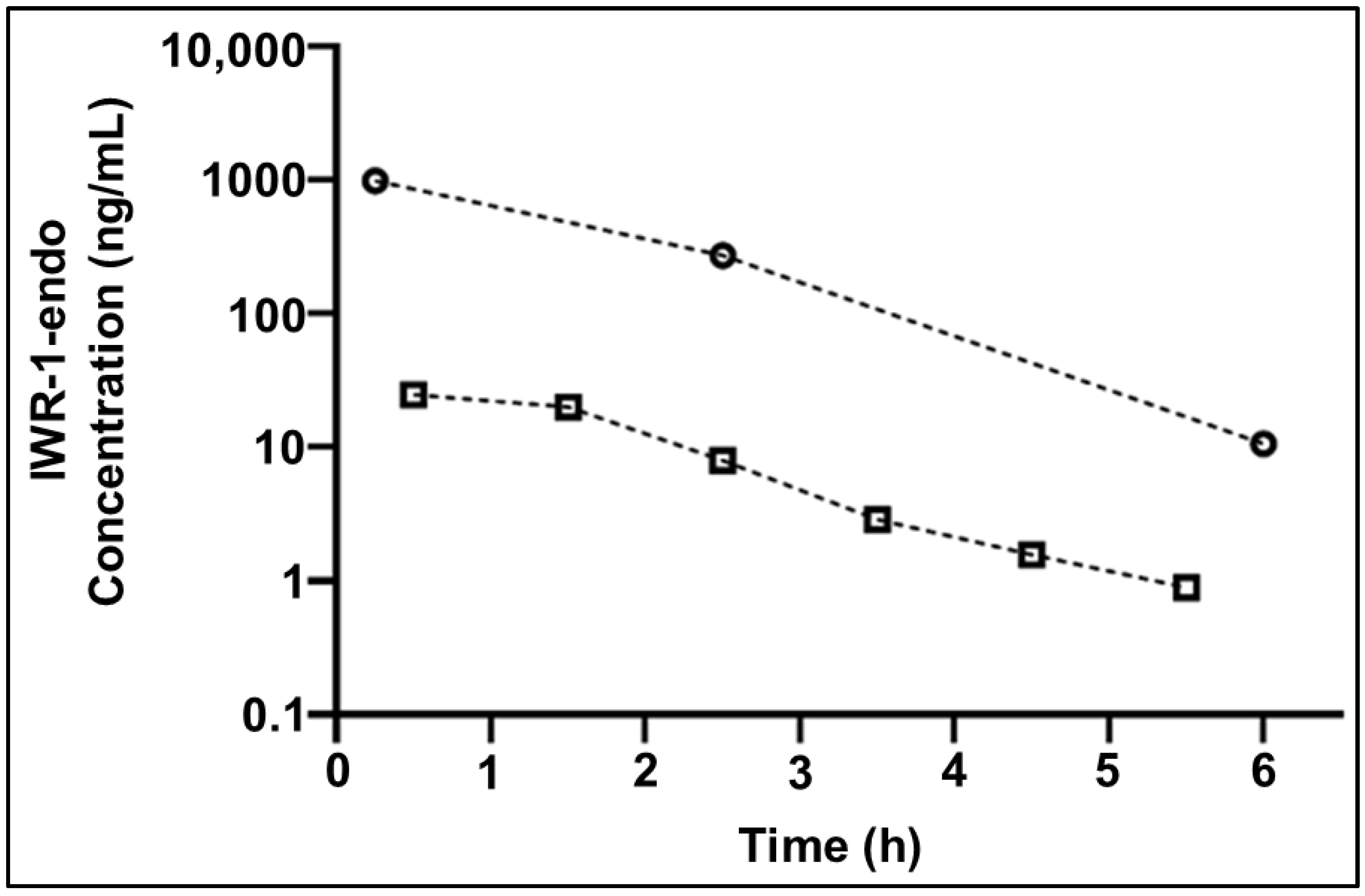
2.6. Application to Cerebral Microdialysis Studies
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Preparation of Stock and Working Solutions
3.3. Calibration Standards and Quality Controls
3.4. Plasma and Microdialysate Sample Preparation
3.5. Chromatographic Conditions
3.6. Mass Spectrometry Conditions
3.7. Method Validation Procedures
3.7.1. Linearity
3.7.2. Selectivity and Sensitivity
3.7.3. Precision and Accuracy
3.7.4. Matrix Effect and Recovery
3.7.5. Stability
3.7.6. Carry-Over
3.7.7. Dilution Integrity
3.8. Method Application to Cerebral Microdialysis Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Primers 2019, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Morfouace, M.; Cheepala, S.; Jackson, S.; Fukuda, Y.; Patel, Y.T.; Fatima, S.; Kawauchi, D.; Shelat, A.A.; Stewart, C.F.; Sorrentino, B.P.; et al. ABCG2 Transporter Expression Impacts Group 3 Medulloblastoma Response to Chemotherapy. Cancer Res. 2015, 75, 3879–3889. [Google Scholar] [CrossRef]
- Correa, S.; Binato, R.; Du Rocher, B.; Castelo-Branco, M.T.; Pizzatti, L.; Abdelhay, E. Wnt/beta-catenin pathway regulates ABCB1 transcription in chronic myeloid leukemia. BMC Cancer 2012, 12, 303. [Google Scholar] [CrossRef]
- Hung, T.H.; Hsu, S.C.; Cheng, C.Y.; Choo, K.B.; Tseng, C.P.; Chen, T.C.; Lan, Y.W.; Huang, T.T.; Lai, H.C.; Chen, C.M.; et al. Wnt5A regulates ABCB1 expression in multidrug-resistant cancer cells through activation of the non-canonical PKA/beta-catenin pathway. Oncotarget 2014, 5, 12273–12290. [Google Scholar] [CrossRef]
- Gustafson, C.T.; Mamo, T.; Maran, A.; Yaszemski, M.J. Efflux inhibition by IWR-1-endo confers sensitivity to doxorubicin effects in osteosarcoma cells. Biochem. Pharmacol. 2018, 150, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Dodge, M.E.; Tang, W.; Lu, J.; Ma, Z.; Fan, C.W.; Wei, S.; Hao, W.; Kilgore, J.; Williams, N.S.; et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 2009, 5, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Gose, T.; Shafi, T.; Fukuda, Y.; Das, S.; Wang, Y.; Allcock, A.; Gavan McHarg, A.; Lynch, J.; Chen, T.; Tamai, I.; et al. ABCG2 requires a single aromatic amino acid to “clamp” substrates and inhibitors into the binding pocket. FASEB J. 2020, 34, 4890–4903. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Davis, A.; Campagne, O.; Schuetz, J.D.; Stewart, C.F. Development and validation of an LC-MS/MS method to quantify the bromodomain and extra-terminal (BET) inhibitor JQ1 in mouse plasma and brain microdialysate: Application to cerebral microdialysis study. J. Pharm. Biomed. Anal. 2021, 204, 114274. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, J.; Tse, F.L. Strategies in quantitative LC-MS/MS analysis of unstable small molecules in biological matrices. Biomed. Chromatogr. BMC 2011, 25, 258–277. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, C.J.; Hage, D.S. Factors affecting the stability of drugs and drug metabolites in biological matrices. Bioanalysis 2009, 1, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hsieh, Y. Stabilizing drug molecules in biological samples. Ther. Drug Monit. 2005, 27, 617–624. [Google Scholar] [CrossRef]
- Hartman, D.A. Determination of the stability of drugs in plasma. Curr. Protoc. Pharmacol. 2002, 19, 7.6.1–7.6.8. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, G.; Uges, D.R.; Franke, J.P. pH adjustment of human blood plasma prior to bioanalytical sample preparation. J. Pharm. Biomed. Anal. 2008, 47, 126–133. [Google Scholar] [CrossRef] [PubMed]
- US DHHS; FDA; CDER; CVM. Guidance for Industry, Bioanalytical Method Validation; US Department of Health and Human Services; Food and Drug Administration; Center for Drug Evaluation and Research; Center for Veterinary Medicine: Washington, DC, USA, 2018. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioanalytical-method-validation-guidance-industry (accessed on 1 June 2020).
- Guideline on Bioanalytical Method Validation, European Medicines Agency. February 2012. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf (accessed on 1 June 2020).
- Goswami, D.; Khuroo, A.; Gurule, S.; Modhave, Y.; Monif, T. Controlled ex-vivo plasma hydrolysis of valaciclovir to acyclovir demonstration using tandem mass spectrometry. Biomed. Chromatogr. BMC 2011, 25, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, S.; Menon, S.; Shailajan, S. A rapid HPLC-ESI-MS/MS method for determination of beta-asarone, a potential anti-epileptic agent, in plasma after oral administration of Acorus calamus extract to rats. Biomed. Chromatogr. BMC 2013, 27, 318–326. [Google Scholar] [CrossRef]
- Jacus, M.O.; Throm, S.L.; Turner, D.C.; Patel, Y.T.; Freeman, B.B., 3rd; Morfouace, M.; Boulos, N.; Stewart, C.F. Deriving therapies for children with primary CNS tumors using pharmacokinetic modeling and simulation of cerebral microdialysis data. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2014, 57, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Elmeliegy, M.A.; Carcaboso, A.M.; Tagen, M.; Bai, F.; Stewart, C.F. Role of ATP-binding cassette and solute carrier transporters in erlotinib CNS penetration and intracellular accumulation. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Parameter | Sample Matrix | |
|---|---|---|
| Murine Plasma | Microdialysates | |
| Calibration Range (ng/mL) | 5–1000 | 0.5–500 |
| LLOQ (ng/mL) | 5 | 0.5 |
| LOD (ng/mL) | 0.5 | 0.2 |
| Intercept (a) | −1.50 × 10−3 | 1.70 × 10−4 |
| Sa | 1.20 × 10−3 | 1.17 × 10−4 |
| Slope (b) | 3.90 × 10−3 | 1.33 × 10−2 |
| Sb | 0.20 × 10−3 | 0.07 × 10−2 |
| Correlation Coefficient (r2) | 0.9989 | 0.9990 |
| P & A Batch Run | Replicates | Murine Plasma | |||
|---|---|---|---|---|---|
| Nominal Conc. | Calculated conc. | Accuracy | Precision | ||
| (n) | (ng/mL) | (ng/mL) | (% R.E.) | (% R. S. D.) | |
| Within-Run 1 | 6 | 5 | 5.30 | 6.07 | 5.04 |
| 6 | 15 | 15.06 | 0.38 | 4.06 | |
| 6 | 300 | 312.81 | 4.27 | 2.89 | |
| 6 | 850 | 895.12 | 5.31 | 1.74 | |
| Within-Run 2 | 6 | 5 | 5.09 | 1.75 | 10.10 |
| 6 | 15 | 14.87 | −0.86 | 5.87 | |
| 6 | 300 | 306.98 | 2.33 | 2.29 | |
| 6 | 850 | 908.24 | 6.85 | 6.80 | |
| Within-Run 3 | 6 | 5 | 5.46 | 9.26 | 6.58 |
| 6 | 15 | 15.65 | 4.31 | 4.50 | |
| 6 | 300 | 324.06 | 8.02 | 3.30 | |
| 6 | 850 | 924.98 | 8.82 | 1.03 | |
| Average of Runs 1, 2, and 3 (Between-Runs) | 18 | 5 | 5.28 | 5.69 | 7.61 |
| 18 | 15 | 15.19 | 1.28 | 5.08 | |
| 18 | 300 | 314.62 | 4.87 | 3.56 | |
| 18 | 850 | 909.45 | 6.99 | 4.08 | |
| P & A Batch Run | Replicates | Microdialysate | |||
|---|---|---|---|---|---|
| Nominal Conc. | Calculated conc. | Accuracy | Precision | ||
| (n) | (ng/mL) | (ng/mL) | (% R.E.) | (% R. S. D.) | |
| Within-Run 1 | 6 | 0.5 | 0.46 | −8.86 | 3.33 |
| 6 | 1.5 | 1.39 | −7.25 | 5.25 | |
| 6 | 150 | 141.88 | −5.42 | 2.25 | |
| 6 | 400 | 372.08 | −6.98 | 4.66 | |
| Within-Run 2 | 6 | 0.5 | 0.49 | −1.16 | 6.95 |
| 6 | 1.5 | 1.49 | −0.53 | 7.31 | |
| 6 | 150 | 148.13 | −1.24 | 4.11 | |
| 6 | 400 | 399.15 | −0.21 | 3.29 | |
| Within-Run 3 | 6 | 0.5 | 0.49 | −2.36 | 3.68 |
| 6 | 1.5 | 1.44 | −3.69 | 6.11 | |
| 6 | 150 | 144.89 | −3.41 | 3.29 | |
| 6 | 400 | 395.57 | −1.11 | 3.32 | |
| Average of Runs 1, 2, and 3 (Between-Runs) | 18 | 0.5 | 0.48 | −4.13 | 5.95 |
| 18 | 1.5 | 1.44 | −3.82 | 6.64 | |
| 18 | 150 | 144.97 | −3.36 | 3.62 | |
| 18 | 400 | 388.94 | −2.77 | 4.76 | |
| Sample Matrix | Compound | Nominal Concentration (ng/mL) | Matrix Effect | Recovery | ||
|---|---|---|---|---|---|---|
| Mean Calculated MF value | % R.S.D. | % Mean Recovery | % R.S.D. | |||
| Mouse Plasma CD1 | IWR-1 | 15 | 1.02 | 2.55 | 97.16 | 1.57 |
| 850 | 1.00 | 1.07 | 97.92 | 1.94 | ||
| ISTD | 500 | 0.97 | 1.07 | 102.45 | 1.52 | |
| Mouse plasma CD1 Nude | IWR-1 | 15 | 0.99 | 2.51 | 100.67 | 2.40 |
| 850 | 1.00 | 1.81 | 97.34 | 3.23 | ||
| ISTD | 500 | 0.95 | 0.64 | 104.58 | 2.24 | |
| Microdialysate | IWR-1 | 1.5 | 1.05 | 9.73 | 86.56 | 10.00 |
| 150 | 1.13 | 1.64 | 90.96 | 3.85 | ||
| ISTD | 500 | 0.95 | 3.85 | 94.94 | 4.78 | |
| Sample Matrix | Stability Study | Nominal Concentration (ng/mL) | Mean ± S.D. Calculated Concentration (ng/mL) | Precision (% R.S.D.) | Accuracy (% R.E.) | % Mean Deviation |
|---|---|---|---|---|---|---|
| Murine Plasma | Process a | 15 | 16.33 ± 0.63 | 3.87 | 8.88 | 9.35 |
| 850 | 916.82 ± 10.54 | 1.15 | 7.86 | 12.40 | ||
| Bench-Top RT b | 15 | 15.87 ± 0.45 | 2.84 | 5.82 | 6.27 | |
| 850 | 915.08 ± 14.18 | 1.55 | 7.66 | 12.18 | ||
| Bench-Top 4 °C c | 15 | 15.20 ± 0.24 | 1.55 | 1.34 | 1.78 | |
| 850 | 915.69 ± 16.12 | 1.76 | 7.73 | 12.26 | ||
| Freeze–Thaw d | 15 | 16.01 ± 0.88 | 5.48 | 6.70 | 7.16 | |
| 850 | 917.18 ± 30.24 | 3.30 | 7.90 | 12.44 | ||
| Long-Term e | 15 | 15.93 ± 0.21 | 1.35 | 6.17 | −5.65 | |
| 850 | 938.89 ± 15.84 | 1.77 | 10.46 | −0.33 | ||
| Microdialysate | Process a | 1.5 | 1.56 ± 0.042 | 2.68 | 4.16 | 7.79 |
| 400 | 429.35 ± 7.00 | 1.63 | 7.34 | 6.64 | ||
| Bench-Top RT b | 1.5 | 1.60 ± 0.05 | 3.02 | 6.35 | 9.35 | |
| 400 | 434.08 ± 17.01 | 3.92 | 8.52 | 7.61 | ||
| Bench-Top 4 °C c | 1.5 | 1.60 ± 0.08 | 4.86 | 6.40 | 11.50 | |
| 400 | 415.84 ± 6.85 | 1.65 | 3.96 | 3.09 | ||
| Freeze–Thaw d | 1.5 | 1.54 ± 0.07 | 4.56 | 2.34 | 5.91 | |
| 400 | 424.79 ± 9.18 | 2.16 | 6.20 | 5.31 | ||
| Long-Term f | 1.5 | 1.39 ± 0.008 | 0.60 | −7.63 | −4.41 | |
| 400 | 390.35 ± 21.53 | 5.52 | −2.41 | −3.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nair, S.; Davis, A.; Campagne, O.; Schuetz, J.D.; Stewart, C.F. Development and Validation of a Sensitive and Specific LC-MS/MS Method for IWR-1-Endo, a Wnt Signaling Inhibitor: Application to a Cerebral Microdialysis Study. Molecules 2022, 27, 5448. https://doi.org/10.3390/molecules27175448
Nair S, Davis A, Campagne O, Schuetz JD, Stewart CF. Development and Validation of a Sensitive and Specific LC-MS/MS Method for IWR-1-Endo, a Wnt Signaling Inhibitor: Application to a Cerebral Microdialysis Study. Molecules. 2022; 27(17):5448. https://doi.org/10.3390/molecules27175448
Chicago/Turabian StyleNair, Sreenath, Abigail Davis, Olivia Campagne, John D. Schuetz, and Clinton F. Stewart. 2022. "Development and Validation of a Sensitive and Specific LC-MS/MS Method for IWR-1-Endo, a Wnt Signaling Inhibitor: Application to a Cerebral Microdialysis Study" Molecules 27, no. 17: 5448. https://doi.org/10.3390/molecules27175448
APA StyleNair, S., Davis, A., Campagne, O., Schuetz, J. D., & Stewart, C. F. (2022). Development and Validation of a Sensitive and Specific LC-MS/MS Method for IWR-1-Endo, a Wnt Signaling Inhibitor: Application to a Cerebral Microdialysis Study. Molecules, 27(17), 5448. https://doi.org/10.3390/molecules27175448






