Marine Bacterial Dextranases: Fundamentals and Applications
Abstract
:1. Introduction
2. Classification and Properties of Dextran—Hydrolyzing Enzymes
3. Marine Dextranase
| Species | Enzyme | Isolated from | Concentration Method | Purification Method | Mw (kDa) | Purification (-Fold) | Specific Activity (U/mg) | Ref |
|---|---|---|---|---|---|---|---|---|
| Arthrobacter sp. | Dex410 | Beach mud, fishes, and seaweeds | Ultrafilteration | DEAE-Sepharose | 64 | - | 11.9 | [34] |
| Catenovulum agarivorans MNH15 | - | Sea mud, seaweed, and seawater | Ultrafilteration | - | 110 | - | - | [76] |
| Catenovulum sp. DP03 | Cadex | Sea water | Alcohol and (NH4)2SO4 precipitate | Ion exchange chromatography | 75 | 29.6 | 2309 | [77] |
| Catenovulum sp. DP03 | Cadex2870 | Sea water | Ultrafilteration | Ni-NTA resin | 29.9 | 46.3 | [79] | |
| Arthrobacter oxydans KQ11 | - | - | (NH4)2SO4 precipitate | Ion-exchange chromatography on Q Sepharose Fast Flow | 66.2 | 43.00 | 36.38 | [83] |
| Bacillus aquimaris S5 | BaDex | Shrimps caught | Ultrafilteration | Magnetic bead (His-tag protein purification beads) | 70 | - | - | [84] |
4. Comparison of Dextranases from Marine Bacteria with Those from Other Sources of Microorganisms
5. Effect of Marine Dextranase on Dental Plaque Removal
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Du Boil, P.M.; Wienese, S. Enzymic Reduction of Dextran in Process-Laboratory Evaluation of Dextranases; Citeseer: State College, PA, USA, 2002. [Google Scholar]
- Kaewprapan, K.; Inprakhon, P.; Marie, E.; Durand, A. Enzymatically degradable nanoparticles of dextran esters as potential drug delivery systems. Carbohydr. Polym. 2012, 88, 875–881. [Google Scholar] [CrossRef]
- Zhang, Y.-Q.; Li, R.-H.; Zhang, H.-B.; Wu, M.; Hu, X.-Q. Purification, characterization, and application of a thermostable dextranase from Talaromyces pinophilus. J. Ind. Microbiol. Biotechnol. 2017, 44, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Khalikova, E.; Susi, P.; Korpela, T. Microbial dextran-hydrolyzing enzymes: Fundamentals and applications. Microbiol. Mol. Biol. Rev. 2005, 69, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Salassi, M.E.; Garcia, M.; Breaux, J.B.; No, S.C. Impact of sugarcane delivery schedule on product value at raw sugar factories. J. Agribus. 2004, 22, 61–75. [Google Scholar]
- Brown, C.F.; Inkerman, P.A. Specific method for quantitative measurement of the total dextran content of raw sugar. J. Agric. Food Chem. 1992, 40, 227–233. [Google Scholar] [CrossRef]
- Eggleston, G.; Monge, A.; Montes, B.; Stewart, D. Application of dextranases in sugarcane factory: Overcoming practical problems. Sugar Tech 2009, 11, 135–141. [Google Scholar] [CrossRef]
- Eggleston, G.; Dilks, A.; Blowers, M.; Winters, K. Successful Application of Dextranase in Sugar Beet Factories. In Proceedings of the American Society of Sugarbeet Technologists Meeting, Albuquerque, NM, USA, 2–5 March 2011; pp. 1–16. [Google Scholar]
- Erhardt, F.A.; Jördening, H.-J. Immobilization of dextranase from Chaetomium erraticum. J. Biotechnol. 2007, 131, 440–447. [Google Scholar] [CrossRef]
- Marotta, M.; Martino, A.; De Rosa, A.; Farina, E.; Cartenı, M.; De Rosa, M. Degradation of dental plaque glucans and prevention of glucan formation using commercial enzymes. Process Biochem. 2002, 38, 101–108. [Google Scholar] [CrossRef]
- Patel, S.; Goyal, A. Functional oligosaccharides: Production, properties and applications. World J. Microbiol. Biotechnol. 2011, 27, 1119–1128. [Google Scholar] [CrossRef]
- Bhatia, S.; Bhakri, G.; Arora, M.; Uppal, S.; Batta, S. Dextranase production from Paecilomyces lilacinus and its application for dextran removal from sugarcane juice. Sugar Tech 2010, 12, 133–138. [Google Scholar] [CrossRef]
- Igarashi, T.; Morisaki, H.; Goto, N. Molecular characterization of dextranase from Streptococcus rattus. Microbiol. Immunol. 2004, 48, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Naby, M.A.; Ismail, A.-M.S.; Abdel-Fattah, A.M.; Abdel-Fattah, A.F. Preparation and some properties of immobilized Penicillium funiculosum 258 dextranase. Process Biochem. 1999, 34, 391–398. [Google Scholar] [CrossRef]
- Esawy, M.A.; Mansour, S.H.; Ahmed, E.F.; Hassanein, N.M.; El Enshasy, H.A. Characterization of extracellular dextranase from a novel halophilic Bacillus subtilis NRC-B233b a mutagenic honey isolate under solid state fermentation. J. Chem. 2012, 9, 1494–1510. [Google Scholar]
- Khalikova, E.; Susi, P.; Usanov, N.; Korpela, T. Purification and properties of extracellular dextranase from a Bacillus sp. J. Chromatogr. B 2003, 796, 315–326. [Google Scholar] [CrossRef]
- Shahid, F.; Aman, A.; Pervez, S.; Ul Qader, S.A. Degradation of long chain polymer (Dextran) using thermostable dextranase from hydrothermal spring isolate (Bacillus megaterium). Geomicrobiol. J. 2019, 36, 683–693. [Google Scholar] [CrossRef]
- Zohra, R.R.; Aman, A.; Ansari, A.; Haider, M.S.; Qader, S.A.U. Purification, characterization and end product analysis of dextran degrading endodextranase from Bacillus licheniformis KIBGE-IB25. Int. J. Biol. Macromol. 2015, 78, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Deng, T.; Liu, X.; Lai, X.; Feng, Y.; Lyu, M.; Wang, S. Isomalto-oligosaccharides produced by endodextranase Shewanella sp. GZ-7 from sugarcane plants. Nat. Prod. Commun. 2020, 15, 1934578X20953286. [Google Scholar]
- Suzuki, N.; Kishine, N.; Fujimoto, Z.; Sakurai, M.; Momma, M.; Ko, J.-A.; Nam, S.-H.; Kimura, A.; Kim, Y.-M. Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus. J. Biochem. 2016, 159, 331–339. [Google Scholar]
- Lee, J.H.; Nam, S.H.; Park, H.J.; Kim, Y.-M.; Kim, N.; Kim, G.; Seo, E.-S.; Kang, S.-S.; Kim, D. Biochemical characterization of dextranase from Arthrobacter oxydans and its cloning and expression in Escherichia coli. Food Sci. Biotechnol. 2010, 19, 757–762. [Google Scholar] [CrossRef]
- Janson, J. Studies on dextran-degrading enzymes. Isolation and identification of a dextranase-producing strain of Cytophaga johnsonii and studies on the formation of the surface-bound enzyme. Microbiology 1975, 88, 205–208. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Kiso, Y.; Muraki, T.; Kang, M.-S.; Nakai, H.; Saburi, W.; Lang, W.; Kang, H.-K.; Okuyama, M.; Mori, H. Novel dextranase catalyzing cycloisomaltooligosaccharide formation and identification of catalytic amino acids and their functions using chemical rescue approach. J. Biol. Chem. 2012, 287, 19927–19935. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, D.; Miyazaki, T.; Shiwa, Y.; Kimura, K.; Suzuki, S.; Fujita, N.; Yoshikawa, H.; Kimura, A.; Kitamura, S.; Hara, H. A novel intracellular dextranase derived from Paenibacillus sp. 598K with an ability to degrade cycloisomaltooligosaccharides. Appl. Microbiol. Biotechnol. 2019, 103, 6581–6592. [Google Scholar] [CrossRef] [PubMed]
- Hild, E.; Brumbley, S.M.; O’Shea, M.G.; Nevalainen, H.; Bergquist, P.L. A Paenibacillus sp. dextranase mutant pool with improved thermostability and activity. Appl. Microbiol. Biotechnol. 2007, 75, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Pulkownik, A.; Walker, G.J. Purification and substrate specificity of an endo-dextranase of Streptococcus mutans K1-R. Carbohydr. Res. 1977, 54, 237–251. [Google Scholar] [CrossRef]
- Bhatia, S.; Bhakri, G.; Arora, M.; Batta, S.; Uppal, S. Kinetic and thermodynamic properties of partially purified dextranase from Paecilomyces lilacinus and its application in dextran removal from cane juice. Sugar Tech 2016, 18, 204–213. [Google Scholar] [CrossRef]
- Mahmoud, K.; Gibriel, A.; Amin, A.A.; Nessrien, M.; Yassien, N.; El Banna, H.A. Microbial production and characterization of dextranase. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 1095–1113. [Google Scholar]
- Huang, R.; Zhong, L.; Xie, F.; Wei, L.; Gan, L.; Wang, X.; Liao, A. Purification, characterization and degradation performance of a novel dextranase from Penicillium cyclopium CICC-4022. Int. J. Mol. Sci. 2019, 20, 1360. [Google Scholar] [CrossRef]
- Wu, D.-T.; Zhang, H.-B.; Huang, L.-J.; Hu, X.-Q. Purification and characterization of extracellular dextranase from a novel producer, Hypocrea lixii F1002, and its use in oligodextran production. Process Biochem. 2011, 46, 1942–1950. [Google Scholar] [CrossRef]
- Sufiate, B.L.; Soares, F.E.; Gouveia, A.S.; Moreira, S.S.; Cardoso, E.F.; Tavares, G.P.; Braga, F.R.; Araújo, J.V.; Queiroz, J.H. Statistical tools application on dextranase production from Pochonia chlamydosporia (VC4) and its application on dextran removal from sugarcane juice. An. Da Acad. Bras. De Ciênc. 2018, 90, 461–470. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.; Ibarra-Junquera, V.; Escalante-Minakata, P.; Ornelas-Paz, J.D.J.; Osuna-Castro, J.; González-Potes, A. Kinetics and thermodynamic of the purified dextranase from Chaetomium erraticum. J. Mol. Catal. B Enzym. 2015, 122, 80–86. [Google Scholar] [CrossRef]
- Netsopa, S.; Niamsanit, S.; Araki, T.; Kongkeitkajorn, M.B.; Milintawisamai, N. Purification and characterization including dextran hydrolysis of dextranase from Aspergillus allahabadii X26. Sugar Tech 2019, 21, 329–340. [Google Scholar] [CrossRef]
- Jiao, Y.-L.; Wang, S.-J.; Lv, M.-S.; Jiao, B.-H.; Li, W.-J.; Fang, Y.-W.; Liu, S. Characterization of a marine-derived dextranase and its application to the prevention of dental caries. J. Ind. Microbiol. Biotechnol. 2014, 41, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Dong, D.; Tian, X.; Zu, H.; Tian, X.; Lyu, M.; Wang, S. Alkalic dextranase produced by marine bacterium Cellulosimicrobium sp. PX02 and its application. J. Basic Microbiol. 2021, 61, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Wang, S.; Lü, M.; Wang, X.; Fang, Y.; Jiao, Y.; Hu, J. Optimization of four types of antimicrobial agents to increase the inhibitory ability of marine Arthrobacter oxydans KQ11 dextranase mouthwash. Chin. J. Oceanol. Limnol. 2016, 34, 354–366. [Google Scholar] [CrossRef]
- Ren, W.; Liu, L.; Gu, L.; Yan, W.; Feng, Y.L.; Dong, D.; Wang, S.; Lyu, M.; Wang, C. Crystal structure of GH49 dextranase from Arthrobacter oxidans KQ11: Identification of catalytic base and improvement of thermostability using semirational design based on B-factors. J. Agric. Food Chem. 2019, 67, 4355–4366. [Google Scholar] [CrossRef]
- Abdelwahed, N.A.; Ahmed, E.F.; El-Gammal, E.W.; Hawas, U.W. Application of statistical design for the optimization of dextranase production by a novel fungus isolated from Red Sea sponge. 3 Biotech 2014, 4, 533–544. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Kim, D. Characterization of novel thermostable dextranase from Thermotoga lettingae TMO. Appl. Microbiol. Biotechnol. 2010, 85, 581–587. [Google Scholar] [CrossRef]
- Park, T.-S.; Jeong, H.-J.; Ko, J.-A.; Ryu, Y.-B.; Park, S.-J.; Kim, D.-M.; Kim, Y.-M.; Lee, W.-S. Biochemical characterization of thermophilic dextranase from a thermophilic bacterium, Thermoanaerobacter pseudethanolicus. J. Microbiol. Biotechnol. 2012, 22, 637–641. [Google Scholar] [CrossRef]
- Papaleo, E.; Tiberti, M.; Invernizzi, G.; Pasi, M.; Ranzani, V. Molecular determinants of enzyme cold adaptation: Comparative structural and computational studies of cold-and warm-adapted enzymes. Curr. Protein Pept. Sci. 2011, 12, 657–683. [Google Scholar] [CrossRef]
- Cieśliński, H.; Kur, J.; Białkowska, A.; Baran, I.; Makowski, K.; Turkiewicz, M. Cloning, expression, and purification of a recombinant cold-adapted β-galactosidase from Antarctic bacterium Pseudoalteromonas sp. 22b. Protein Expr. Purif. 2005, 39, 27–34. [Google Scholar] [CrossRef]
- Majeed, A.; Grobler, S.R.; Moola, M.H. The pH of various tooth-whitening products on the South African market: Scientific. S. Afr. Dent. J. 2011, 66, 278–281. [Google Scholar]
- Thitaram, S.; Chung, C.-H.; Day, D.; Hinton, A., Jr.; Bailey, J.; Siragusa, G. Isomaltooligosaccharide increases cecal Bifidobacterium population in young broiler chickens. Poult. Sci. 2005, 84, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.M.; Andersson, R.; Ståhlberg, J.; Kenne, L.; Jones, T.A. Dextranase from Penicillium minioluteum: Reaction course, crystal structure, and product complex. Structure 2003, 11, 1111–1121. [Google Scholar] [CrossRef]
- Hayacibara, M.F.; Koo, H.; Smith, A.M.V.; Kopec, L.K.; Scott-Anne, K.; Cury, J.A.; Bowen, W.H. The influence of mutanase and dextranase on the production and structure of glucans synthesized by streptococcal glucosyltransferases. Carbohydr. Res. 2004, 339, 2127–2137. [Google Scholar] [CrossRef]
- Kim, D.; Day, D.F. A new process for the production of clinical dextran by mixed-culture fermentation of Lipomyces starkeyi and Leuconostoc mesenteroides. Enzym. Microb. Technol. 1994, 16, 844–848. [Google Scholar] [CrossRef]
- Simonson, M.R. Designing Instruction for Attitudinal Outcomes. J. Instr. Dev. 1979, 2, 15–19. [Google Scholar] [CrossRef]
- Aoki, H.; Yopi, S.Y. Molecular cloning and heterologous expression of the isopullulanase gene from Aspergillus niger A.T.C.C. 9642. Biochem. J. 1997, 323, 757–764. [Google Scholar] [CrossRef]
- Larsson, C.L.; Rönnlund, U.; Johansson, G.; Dahlgren, L. Veganism as status passage: The process of becoming a vegan among youths in Sweden. Appetite 2003, 41, 61–67. [Google Scholar] [CrossRef]
- García, B.; Rodríguez, E. Carbon source regulation of a dextranase gene from the filamentous fungus Penicillium minioluteum. Curr. Genet. 2000, 37, 396–402. [Google Scholar] [CrossRef]
- García, B.; Castellanos, A.; Menéndez, J.; Pons, T. Molecular cloning of an α-glucosidase-like gene from Penicillium minioluteum and structure prediction of its gene product. Biochem. Biophys. Res. Commun. 2001, 281, 151–158. [Google Scholar] [CrossRef]
- Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes; Academic Press: Cambridge, MA, USA, 1965. [Google Scholar]
- Oguma, T.; Kawamoto, H. Production of cyclodextran and its application. Trends Glycosci. Glycotechnol. 2003, 15, 91–99. [Google Scholar] [CrossRef]
- Jensen, B.; Olsen, J. Extracellular α-glucosidase with dextran-hydrolyzing activity from the thermophilic fungus, Thermomyces lanuginosus. Curr. Microbiol. 1996, 33, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G.J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 2000, 124, 1515–1519. [Google Scholar] [CrossRef]
- Aoki, H.; Sakano, Y. A classification of dextran-hydrolysing enzymes based on amino-acid-sequence similarities. Biochem. J. 1997, 323, 859. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Henrissat, B. Glycoside hydrolases and glycosyltransferases: Families and functional modules. Curr. Opin. Struct. Biol. 2001, 11, 593–600. [Google Scholar] [CrossRef]
- Lovelock, J.E.; Rapley, C.G. Ocean pipes could help the Earth to cure itself. Nature 2007, 449, 403. [Google Scholar] [CrossRef]
- Haefner, B. Drugs from the deep: Marine natural products as drug candidates. Drug Discov. Today 2003, 8, 536–544. [Google Scholar] [CrossRef]
- Barzkar, N.; Fariman, G.A.; Taheri, A. Proximate composition and mineral contents in the body wall of two species of sea cucumber from Oman Sea. Environ. Sci. Pollut. Res. 2017, 24, 18907–18911. [Google Scholar] [CrossRef]
- Jahromi, S.T.; Pourmozaffar, S.; Jahanbakhshi, A.; Rameshi, H.; Gozari, M.; Khodadadi, M.; Sohrabipour, J.; Behzadi, S.; Barzkar, N.; Nahavandi, R. Corrigendum to “Effect of different levels of dietary Sargassum cristaefolium on growth performance, hematological parameters, histological structure of hepatopancreas and intestinal microbiota of Litopenaeus vannamei”. Aquaculture 2021, 535, 736376. [Google Scholar] [CrossRef]
- Barzkar, N. Marine microbial alkaline protease: An efficient and essential tool for various industrial applications. Int. J. Biol. Macromol. 2020, 161, 1216–1229. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Homaei, A.; Hemmati, R.; Patel, S. Thermostable marine microbial proteases for industrial applications: Scopes and risks. Extremophiles 2018, 22, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Jahromi, S.T.; Poorsaheli, H.B.; Vianello, F. Metabolites from marine microorganisms, micro, and macroalgae: Immense scope for pharmacology. Mar. Drugs 2019, 17, 464. [Google Scholar] [CrossRef]
- Barzkar, N.; Jahromi, S.T.; Vianello, F. Marine Microbial Fibrinolytic Enzymes: An Overview of Source, Production, Biochemical Properties and Thrombolytic Activity. Mar. Drugs 2022, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Khan, Z.; Jahromi, S.T.; Pourmozaffar, S.; Gozari, M.; Nahavandi, R. A critical review on marine serine protease and its inhibitors: A new wave of drugs? Int. J. Biol. Macromol. 2021, 170, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Sohail, M. An overview on marine cellulolytic enzymes and their potential applications. Appl. Microbiol. Biotechnol. 2020, 104, 6873–6892. [Google Scholar] [CrossRef]
- Barzkar, N.; Sohail, M.; Jahromi, S.T.; Gozari, M.; Poormozaffar, S.; Nahavandi, R.; Hafezieh, M. Marine bacterial esterases: Emerging biocatalysts for industrial applications. Appl. Biochem. Biotechnol. 2021, 193, 1187–1214. [Google Scholar] [CrossRef]
- Barzkar, N.; Sohail, M.; Jahromi, S.T.; Nahavandi, R.; Khodadadi, M. Marine microbial L-glutaminase: From pharmaceutical to food industry. Appl. Microbiol. Biotechnol. 2021, 105, 4453–4466. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.A.; Belaich, J.-P.; Shoham, Y.; Lamed, R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef]
- Jahromi, S.T.; Barzkar, N. Marine bacterial chitinase as sources of energy, eco-friendly agent, and industrial biocatalyst. Int. J. Biol. Macromol. 2018, 120, 2147–2154. [Google Scholar] [CrossRef]
- Jahromi, S.T.; Barzkar, N. Future direction in marine bacterial agarases for industrial applications. Appl. Microbiol. Biotechnol. 2018, 102, 6847–6863. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Sheng, R.; Sohail, M.; Jahromi, S.T.; Babich, O.; Sukhikh, S.; Nahavandi, R. Alginate Lyases from Marine Bacteria: An Enzyme Ocean for Sustainable Future. Molecules 2022, 27, 3375. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Liu, X.; Liu, X.; Deng, T.; Feng, Y.; Tian, X.; Lyu, M.; Wang, S. The marine Catenovulum agarivorans MNH15 and dextranase: Removing dental plaque. Mar. Drugs 2019, 17, 592. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Cai, R.; Yan, W.; Lyu, M.; Fang, Y.; Wang, S. Purification and characterization of a biofilm-degradable dextranase from a marine bacterium. Mar. Drugs 2018, 16, 51. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Lu, M.; Fang, Y.; Jiao, Y.; Zhu, Q.; Liu, Z.; Wang, S. Screening, production, and characterization of dextranase from Catenovulum sp. Ann. Microbiol. 2014, 64, 147–155. [Google Scholar] [CrossRef]
- Deng, T.; Feng, Y.; Xu, L.; Tian, X.; Lai, X.; Lyu, M.; Wang, S. Expression, purification and characterization of a cold-adapted dextranase from marine bacteria and its ability to remove dental plaque. Protein Expr. Purif. 2020, 174, 105678. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ren, W.; Ly, M.; Li, H.; Wang, S. Characterization of an alkaline GH49 dextranase from marine bacterium Arthrobacter oxydans KQ11 and its application in the preparation of isomalto-oligosaccharide. Mar. Drugs 2019, 17, 479. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, H.; Lu, M.; Fang, Y.; Jiao, Y.; Li, W.; Zhao, G.; Wang, S. Dextranase from Arthrobacter oxydans KQ11-1 inhibits biofilm formation by polysaccharide hydrolysis. Biofouling 2016, 32, 1223–1233. [Google Scholar] [CrossRef]
- Wang, X.; Lu, M.; Wang, S.; Fang, Y.; Wang, D.; Ren, W.; Zhao, G. The atmospheric and room-temperature plasma (ARTP) method on the dextranase activity and structure. Int. J. Biol. Macromol. 2014, 70, 284–291. [Google Scholar] [CrossRef]
- Wang, D.; Lu, M.; Wang, S.; Jiao, Y.; Li, W.; Zhu, Q.; Liu, Z. Purification and characterization of a novel marine Arthrobacter oxydans KQ11 dextranase. Carbohydr. Polym. 2014, 106, 71–76. [Google Scholar] [CrossRef]
- Dong, D.; Wang, X.; Deng, T.; Ning, Z.; Tian, X.; Zu, H.; Ding, Y.; Wang, C.; Wang, S.; Lyu, M. A novel dextranase gene from the marine bacterium Bacillus aquimaris S5 and its expression and characteristics. FEMS Microbiol. Lett. 2021, 368, fnab007. [Google Scholar] [CrossRef] [PubMed]
- Lawman, P.; Bleiweis, A. Molecular cloning of the extracellular endodextranase of Streptococcus salivarius. J. Bacteriol. 1991, 173, 7423–7428. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Li, P.; Dong, X.; Lan, Y.; Xu, L.; Wei, Z.; Wang, S. Purification, Characterization, and Hydrolysate Analysis of Dextranase From Arthrobacter oxydans G6-4B. Front. Bioeng. Biotechnol. 2022, 9, 813079. [Google Scholar] [CrossRef] [PubMed]
- Sufiate, B.L.; Soares, F.E.D.F.; Moreira, S.S.; Gouveia, A.D.S.; Cardoso, E.F.; Braga, F.R. In Vitro and In Silico Characterization of a Novel Dextranase from Pochonia Chlamydosporia. Biotech 2018, 8, 167. [Google Scholar] [CrossRef]
- Lee, S.Y.; Khoiroh, I.; Ooi, C.W.; Ling, T.C.; Show, P.L. Recent Advances in Protein Extraction Using Ionic Liquid-Based Aqueous Two-Phase Systems. Sep. Purif. Rev. 2017, 46, 291–304. [Google Scholar] [CrossRef]
- Phong, W.N.; Show, P.L.; Le, C.F.; Tao, Y.; Chang, J.-S.; Ling, T.C. Improving Cell Disruption Efficiency to Facilitate Protein Release from Microalgae Using Chemical and Mechanical Integrated Method. Biochem. Eng. J. 2018, 135, 83–90. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, L.; Wei, X.; Li, K.; Liu, J. Food-Grade Expression and Characterization of a Dextranase from Chaetomium Gracile Suitable for Sugarcane Juice Clarification. Chem. Biodivers. 2021, 18, e2000797. [Google Scholar] [CrossRef]
- Huang, S.-X.; Hou, D.-Z.; Qi, P.-X.; Wang, Q.; Chen, H.-L.; Ci, L.-Y. Enzymatic Synthesis of Non-Digestible Oligosaccharide Catalyzed by Dextransucrase and Dextranase from Maltose Acceptor Reaction. Biochem. Biophys. Res. Commun. 2020, 523, 651–657. [Google Scholar] [CrossRef]
- Kubo, S.; Kubota, H.; Ohnishi, Y.; Morita, T.; Matsuya, T.; Matsushiro, A. Expression and secretion of an Arthrobacter dextranase in the oral bacterium Streptococcus gordonii. Infect. Immun. 1993, 61, 4375–4381. [Google Scholar] [CrossRef]
- Juntarachot, N.; Kantachote, D.; Peerajan, S.; Sirilun, S.; Chaiyasut, C. Optimization of fungal dextranase production and its antibiofilm activity, encapsulation and stability in toothpaste. Molecules 2020, 25, 4784. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Song, X.; Cai, J. Improving the stability and reusability of dextranase by immobilization on polyethylenimine modified magnetic particles. New J. Chem. 2018, 42, 8391–8399. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Abd AL-MAnhEL, A.J. Production of exopolysaccharide from local fungal isolate. Curr. Res. Nutr. Food Sci. J. 2017, 5, 338–346. [Google Scholar] [CrossRef]
- Yano, A.; Kikuchi, S.; Yamashita, Y.; Sakamoto, Y.; Nakagawa, Y.; Yoshida, Y. The inhibitory effects of mushroom extracts on sucrose-dependent oral biofilm formation. Appl. Microbiol. Biotechnol. 2010, 86, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Pittrof, S.L.; Kaufhold, L.; Fischer, A.; Wefers, D. Products Released from Structurally Different Dextrans by Bacterial and Fungal Dextranases. Foods 2021, 10, 244. [Google Scholar] [CrossRef]
- Tsutsumi, K.; Gozu, Y.; Nishikawa, A.; Tonozuka, T. Structural insights into polysaccharide recognition by Flavobacterium johnsoniae dextranase, a member of glycoside hydrolase family 31. FEBS J. 2020, 287, 1195–1207. [Google Scholar] [CrossRef]
- Karpova, N.; Mogilevskaya, I.; Semenova, O.Y.; Kolotova, O.; Suprunyuk, A.; Zibarov, E.; Kravchenko, T. Advantages of Using the Dextranase Enzyme Based on Saccharomyces Cerevisiae in Beet Sugar Production. Int. Res. J. 2021, 6, 33–38. (In Russion) [Google Scholar]
- Karpova, N.N.; Mogilevskaya, I.V. Selection of culture medium components for S. cerevisiae strain Y-1531-dextranase producer. In Proceedings of the Annual Youth Scientific Conference, South of Russia: Challenges of Time, Discoveries, Prospects: Materials of the Conf, Rostov-on-Don, Russia, 20 April 2021. [Google Scholar]
- Gres, N.A.; Skalny, A.V. Bioelemental Status of the Population of Belarus: Ecological, Physiological and Pathological Aspects; Harvest: Minsk, Belarus, 2011. [Google Scholar]
- Volgin, M.; Mayer-Lukel, H.A. Kalbassa, Etiology, pathogenesis and treatment methods. Dent. Art. 2006, 3, 59–63. [Google Scholar]
- Tsimbalistov, A.; Voytyatskaya, I. Pikhur OL P Increased abrasion of hard dental tissues. Clin. Pict. Morphol. Cryst. Struct. Klin. Stomatol. 2005, 2, 12–14. [Google Scholar]
- Ameen, F.; AlNadhari, S.; Al-Homaidan, A.A. Marine microorganisms as an untapped source of bioactive compounds. Saudi J. Biol. Sci. 2021, 28, 224–231. [Google Scholar] [CrossRef]
- Fedorov, Y.; Drojjina, V.; Matelo, S.; Tumanova, S. Klinicheskie vozmojnosti primeneniya sovremennyih remineralizuyuschih sostavov u vzroslyih [Clinical possibilities of application of modern remineralizing compositions in adults]. Clin. Stomatol. 2008, 3, 32–34. [Google Scholar]
- Kobiyasova, I.V. Objective assessment method enamel mineral maturity and efficiency the effect of the drug “Calcium-D3 Nycomed” on the rate of maturation of hard tissues of permanent teeth in adolescents. Dent. Forum 2005, 3, 37–42. [Google Scholar]
- Matviychuk, O.Y. Occlusal damage yak one of the primary reasons for vindication of non-carious cervical lesions. Bull. Dent. 2005, 1, 32–34. [Google Scholar]
- Ahmedbeyli, R.M. Modern data on the mineral composition, structure and properties of hard dental tissues. BioMed 2016, 2, 22–27. [Google Scholar]
- Gurenlian, J.R. The role of dental plaque biofilm in oral health. Am. Dent. Hyg. Assoc. 2007, 81, 116. [Google Scholar]
- Marsh, P.D. Microbiology of dental plaque biofilms and their role in oral health and caries. Dent. Clin. 2010, 54, 441–454. [Google Scholar] [CrossRef]
- Jenkinson, H.F.; Lamont, R.J. Oral microbial communities in sickness and in health. Trends Microbiol. 2005, 13, 589–595. [Google Scholar] [CrossRef]
- Beighton, D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Commun. Dent. Oral Epidemiol. 2005, 33, 248–255. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J., Jr. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar] [CrossRef]
- Salam, M.A.; Matsumoto, N.; Matin, K.; Tsuha, Y.; Nakao, R.; Hanada, N.; Senpuku, H. Establishment of an animal model using recombinant NOD. B10. D2 mice to study initial adhesion of oral streptococci. Clin. Vaccine Immunol. 2004, 11, 379–386. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, S.H.; Park, J.Y.; Jin, X.J.; Oh, D.K.; Soo Kang, S.; Kim, D. Cloning and characterization of a dextranase gene from Lipomyces starkeyi and its expression in Saccharomyces cerevisiae. Yeast 2005, 22, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.; Spinell, D.; Stoudt, T. Enzymatic removal of artificial plaques. Arch. Oral Biol. 1968, 13, 125–128. [Google Scholar] [CrossRef]
- Gibbons, R.; Keyes, P. Inhibition of insoluble dextran synthesis, plaque formation and dental caries in hamsters by low molecular weight dextran. Arch. Oral Biol. 1969, 14, 721–724. [Google Scholar] [CrossRef]
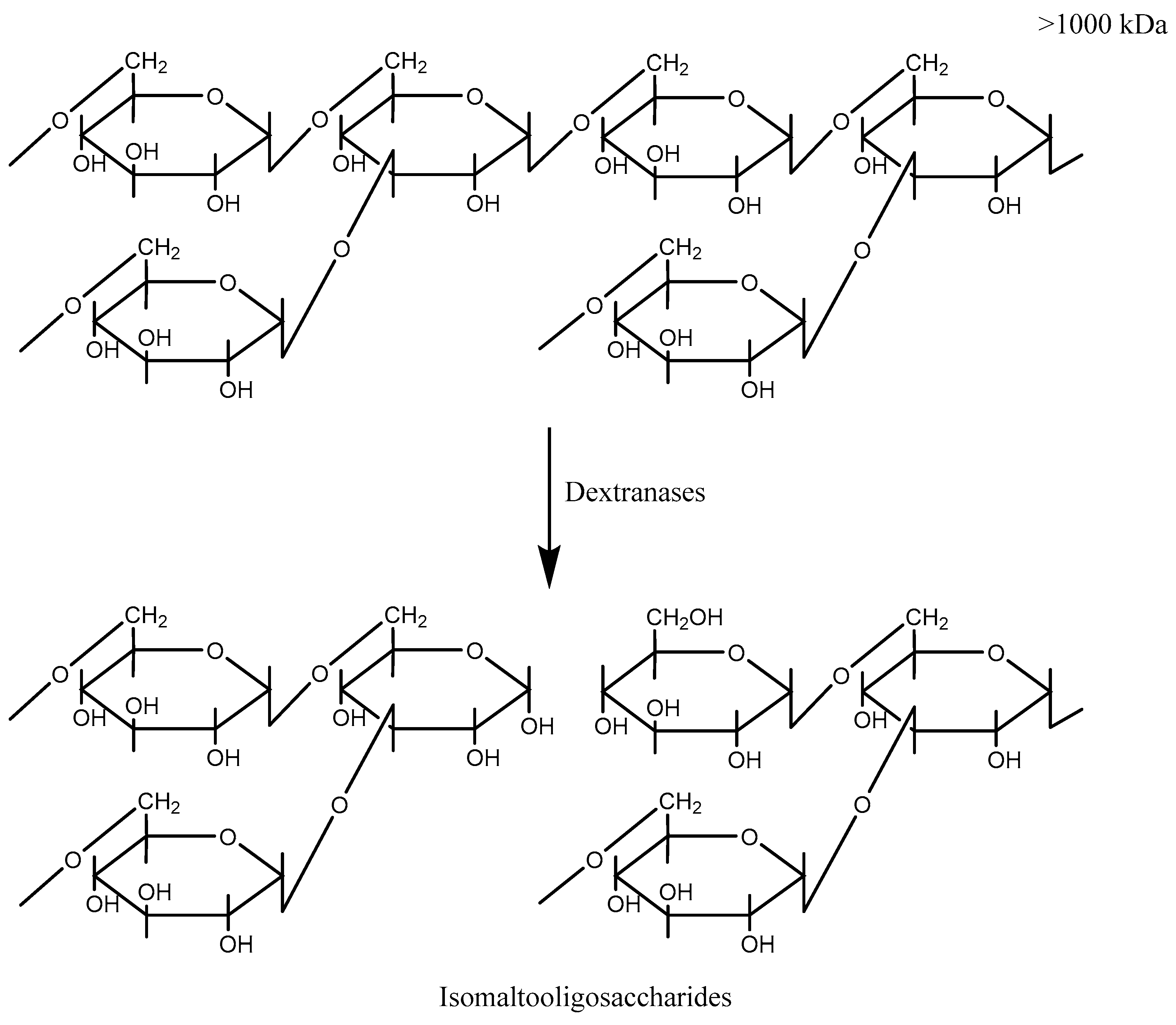
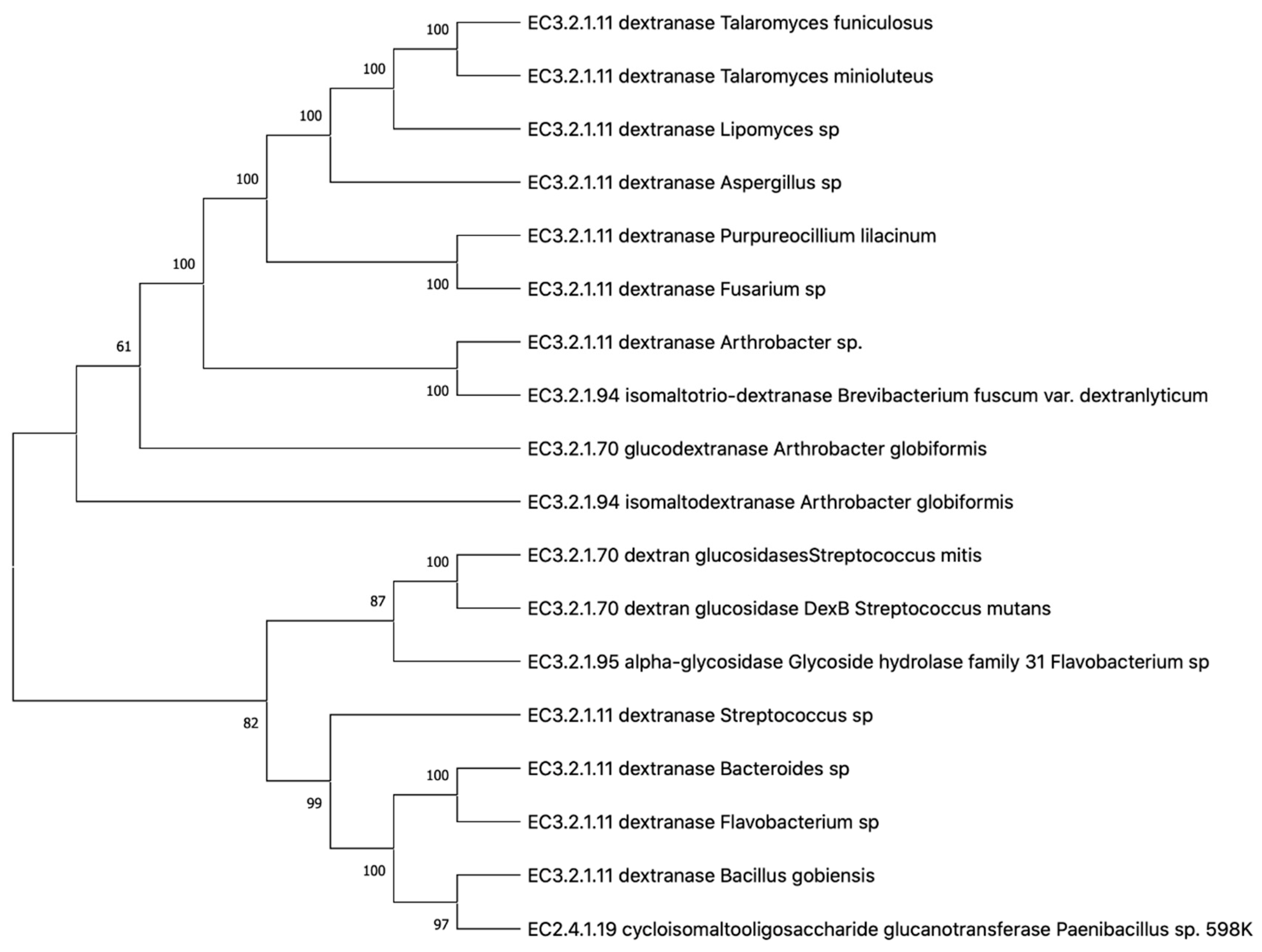
| Species | Enzyme | Enzyme Family | Enzymatic Digestion Products | pH (Opt) | Temp (Opt) | Stable at pH | Stable at Temp | Activator | Inhibitor | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| Arthrobacter sp. | Dex410 | Endoglycosidase | isomaltotriose, isomaltoteraose, and isomaltopentaose | 5.5 | 45 | - | - | - | - | [34] |
| Cellulosimicrobium sp. PX02 | - | - | isomaltotriose, maltopentaose, and isomaltooligosaccharide | 7.5 | 40 | 7.0–9.0 | up to 45 | - | - | [35] |
| Catenovulum agarivorans MNH15 | - | α-1,6-glucosidic bonds | glucose, maltose, and maltoheptaose | 8.0 | 40 | 5.0–9.0 | 30 °C | Sr2+ | NH4+, Co2+, Cu2+, Li+ | [76] |
| Catenovulum sp. | Cadex | α-1,6 glycosidic bond | Isomaltoogligosaccharides | 8.0 | 40 | 5.0- 11.0 | under 30 °C | Mn2+, Sr2+ | Cu2+, Fe3+, Zn2+, Cd2+, Ni2+, Co2+ | [77] |
| Catenovulum sp. DP03 | - | - | - | 8.0 | 40 | 6.0–11.0 | 30 | - | - | [78] |
| Catenovulum sp. DP03 | Cadex2870 | - | maltose, maltotetraose, maltopentose, maltoheptaose and higher molecular weight maltooligosaccharides | 8 | 45 | 60% activity at pH 5–9 for 1 h | 10% catalytic activity at 0 °C | - | - | [79] |
| Arthrobacter oxydans KQ11-1 | - | - | glucose, maltose, maltotriose, and maltotetraose | 6.5 | 60 | - | - | - | - | [81] |
| Arthrobacter oxydans KQ11 | - | - | - | 7.0 | 50 | - | >60% activity at 60 °C for 1 h | Co2+, Ca2+, xylitol, alcohol | Ni2+, Fe3+, 0.05% SDS | [83] |
| Bacillus aquimaris S5 | BaDex | 66 | - | 6.0 | 40 | - | 80% after incubation at 10–30 °C for 3 h | - | - | [84] |
| Bacterial Strain | Dextranase Gene | Primer | Host Cell | Vector Plasmid | GeneBank Accession Number | Ref |
|---|---|---|---|---|---|---|
| Arthrobacter sp. | Dex410 | 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) | - | - | JX481352 | [34] |
| Arthrobacter oxidans KQ11 | Aodex | - | E. coli DH5α and E. coli BL21 (DE3) | pCold III-KQ | KJ571608 | [37] |
| Catenovulum agarivorans MNH15 | - | 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) | E. coli | pMD19-T | - | [76] |
| Catenovulum sp. | Cadex | - | - | pMD19-T | - | [77] |
| Catenovulum sp. DP03 | Cadex2870 | F (5′-GAAGATCTGGGCTGCTCAAGCAGCAGCTCGT-3′) and R (5′-ATAAGAATGCGGCCGCAATTTCGA TTTTTGTAATTTGATA-3′) | E. coli BL21(DE3) | pET29a | - | [79] |
| Arthrobacter oxydans KQ11 | DexKQ | KQ-28aF: GGGAATTCCATATGAAGCATTACCTCCGTCTA; KQ-28aR: CCCAAGCTTCC-ACGCGTTCCAGTTATCCCA | E. coli BL21(DE3) | pET28a | AHZ97853.1 | [80] |
| Arthrobacter oxydans KQ11-1 | - | 5′-CGCGGATCCCAGGAGCCCCGCTGCGACAGA-3′ (BamHI site is underlined) and (5′-CCCAAGCTTCCACGCGTTCCAGTTATCCA-3′ (HindIII site is underlined) | E. coli DH5α | pET-28a-(+) | D00834.1 | [81] |
| Arthrobacter KQ11 | - | 5′-CGCGGATCCCAGGAGCCCCGCTGCGACAGA-3′ and 5′-CCCAAGCTTCCACGCGTTCCA TTATCCA-3′ | E. coli DH 5α | PMD-19 | KJ571608 | [82] |
| Bacillus aquimaris S5 | BaDex | F (5′-CGCGAGCTCATGGGGAAAAAGAA-3′) and R (5′-CCGCTCGAGTTTATAGTCGATCACGACC-3′) | E. coli BL21(DE3) | pET29a | - | [84] |
| Dextranase Source | Advantages | Disadvantages |
|---|---|---|
| Mold fungi | high enzymatic activity | some mold fungi can be poisonous; fungal spores are volatile and can contaminate production facilities |
| Bacteria | fast cultivation; high enzymatic activity | development of a complex isolation method is required |
| Yeast | there is no bacterial DNA in the preparations produced by cultivating yeast | long cultivation period |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzkar, N.; Babich, O.; Das, R.; Sukhikh, S.; Tamadoni Jahromi, S.; Sohail, M. Marine Bacterial Dextranases: Fundamentals and Applications. Molecules 2022, 27, 5533. https://doi.org/10.3390/molecules27175533
Barzkar N, Babich O, Das R, Sukhikh S, Tamadoni Jahromi S, Sohail M. Marine Bacterial Dextranases: Fundamentals and Applications. Molecules. 2022; 27(17):5533. https://doi.org/10.3390/molecules27175533
Chicago/Turabian StyleBarzkar, Noora, Olga Babich, Rakesh Das, Stanislav Sukhikh, Saeid Tamadoni Jahromi, and Muhammad Sohail. 2022. "Marine Bacterial Dextranases: Fundamentals and Applications" Molecules 27, no. 17: 5533. https://doi.org/10.3390/molecules27175533






