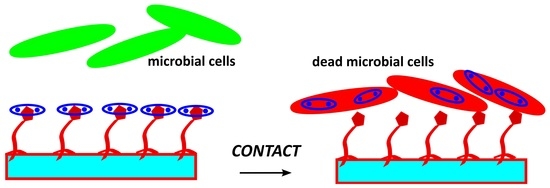
A Supramolecular Approach to Antimicrobial Surfaces
Abstract
:1. Introduction
2. Results and Discussion
2.1. Studies in Solution
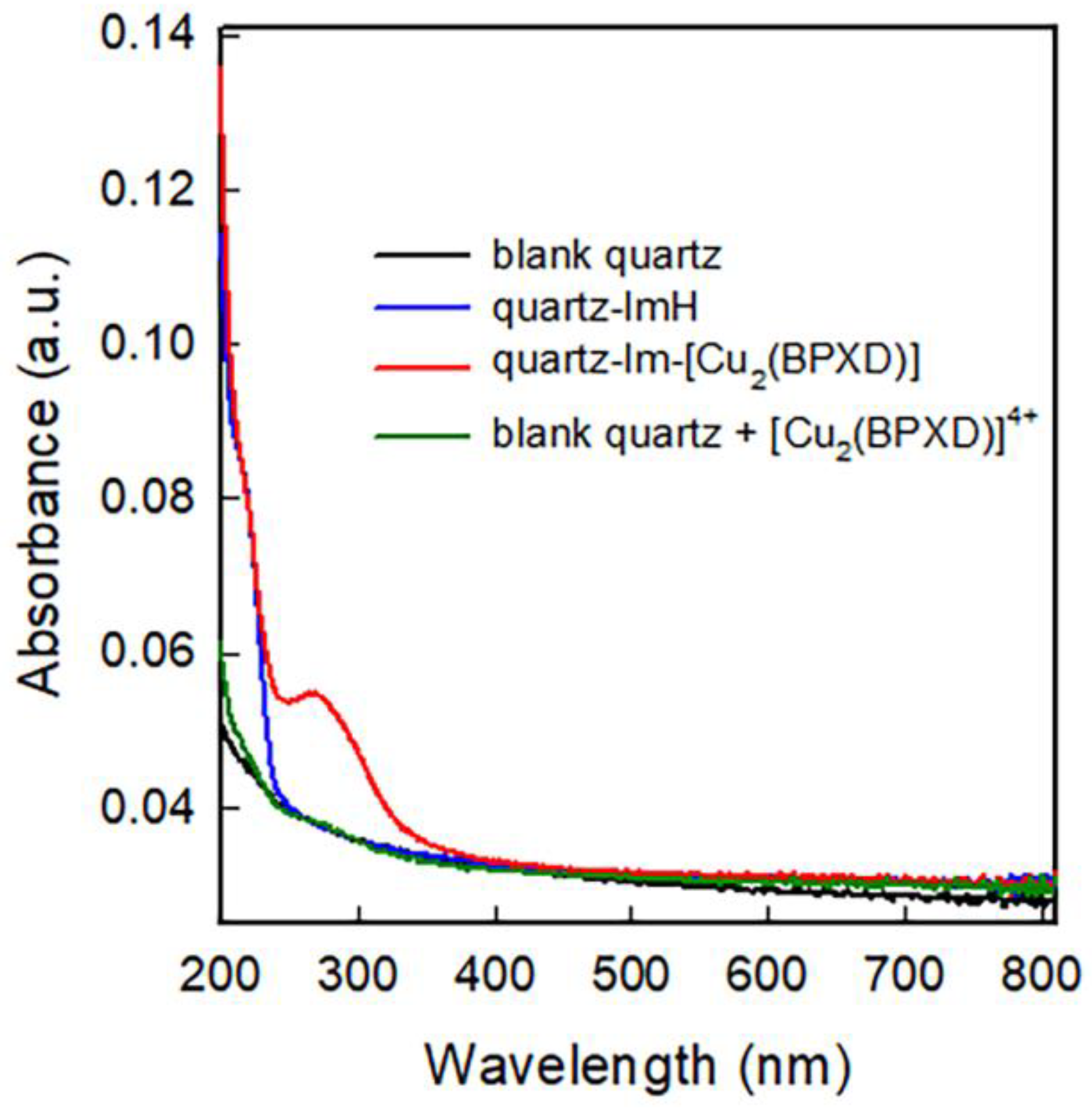
2.2. Bringing the Complex to Glass Surfaces
2.3. Study of Antimicrobial Action
2.4. Cytotoxicity Test
3. Materials and Methods
3.1. Materials and General Procedures
3.2. Potentiometric Titrations
3.3. Glassware, Glass Slides and Quartz Slides Cleaning
3.4. Preparation of Glass-ImH and Quartz-ImH Samples
3.5. Preparation of Glass-Im-[Cu2(BPXD)] and Quartz-Im-[Cu2(BPXD)] Samples
3.6. Determination of Total Cu2+ by ICP
3.7. UV-VIS-NIR Absorption Spectra
3.8. Antibacterial/Antifungal Activity Tests
3.9. Cytotoxicity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| BPXD | 3,6,9,16,19,22-hexaazatricyclo [22.2.2.2(11.14)]triaconta-1(26),11(12),13,24,27, 29-hexaene |
| CM | Culture Media |
| glass-ImH | Glass slides functionalized with grafted imidazole moieties |
| glass-Im-[Cu2(BPXD)] | Glass slides functionalized with imidazolate moieties and copper complexes |
| ICP | Inductively coupled plasma |
| ImH | Imidazole |
| Im− | Imidazolate |
| LMCT | Ligand to Metal Charge Transfer |
| MBC | Minimum Bactericidal Concentration |
| ME | Microbicidal Effect |
| Meff | Maximum Effective Concentration |
| MFC | Minimum Fungicidal Concentration |
| MIC | Minimum Inhibitory Concentration |
| quartz-ImH | Quartz slides functionalized with imidazole moieties |
| quartz-Im-[Cu2(BPXD)] | Quartz slides functionalized with imidazolate moieties and copper complexes |
References
- Campoccia, D.; Montanaro, L.; Arciola, C.R. A review of the clinical implications of anti-infective biomaterials and infection-resistant surfaces. Biomaterials 2013, 34, 8018–8029. [Google Scholar] [CrossRef] [PubMed]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A Brief Recap of Microbial Adhesion and Biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef]
- Pallavicini, P.; Dacarro, G.; Taglietti, A. Self-assembled monolayers of silver nanoparticles: From intrinsic to switchable inorganic antibacterial surfaces. Eur. J. Inorg. Chem. 2018, 2018, 4846–4855. [Google Scholar] [CrossRef]
- Taglietti, A.; Arciola, C.R.; D’Agostino, A.; Dacarro, G.; Montanaro, L.; Campoccia, D.; Cucca, L.; Vercellino, M.; Poggi, A.; Pallavicini, P. Antibiofilm activity of a monolayer of silver nanoparticles anchored to an amino-silanized glass surface. Biomaterials 2014, 35, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Slavin, Y.N.; Asnis, J.; Hafeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Pallavicini, P.; Taglietti, A.; Dacarro, G.; Diaz Fernandez, Y.A.; Galli, M.; Grisoli, P.; Patrini, M.; Santucci De Magistris, G.; Zanoni, R. Self-assembled monolayers of silver nanoparticles firmly grafted on glass surfaces: Low Ag+ release for an efficient antibacterial activity. J. Colloid Interface Sci. 2010, 350, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Pallavicini, P.; Dacarro, G.; Diaz-Fernandez, Y.A.; Taglietti, A. Coordination chemistry of surface-grafted ligands for antibacterial materials. Coord. Chem. Rev. 2014, 275, 37–53. [Google Scholar] [CrossRef]
- Taglietti, A.; Fernandez, Y.A.; Amato, E.; Cucca, L.; Dacarro, G.; Grisoli, P.; Necchi, V.; Pallavicini, P.; Pasotti, L.; Patrini, M. Antibacterial activity of glutathione-coated silver nanoparticles against gram positive and gram negative bacteria. Langmuir 2012, 28, 8140–8148. [Google Scholar] [CrossRef]
- Pallavicini, P.; Dacarro, G.; Cucca, L.; Denat, F.; Grisoli, P.; Patrini, M.; Sok, N.; Taglietti, A. A monolayer of a Cu2+-tetraazamacrocyclic complex on glass as the adhesive layer for silver nanoparticles grafting, in the preparation of surface-active antibacterial materials. New J. Chem. 2011, 35, 1198–1201. [Google Scholar] [CrossRef]
- Pallavicini, P.; Dacarro, G.; Grisoli, P.; Mangano, C.; Patrini, M.; Rigoni, F.; Sangaletti, L.; Taglietti, A. Coordination chemistry for antibacterial materials: A monolayer of a Cu2+ 2,2′-bipyridine complex grafted on a glass surface. Dalton Trans. 2013, 42, 4552–4560. [Google Scholar] [CrossRef]
- Dacarro, G.; Cucca, L.; Grisoli, P.; Pallavicini, P.; Patrini, M.; Taglietti, A. Monolayers of polyethilenimine on flat glass: A versatile platform for cations coordination and nanoparticles grafting in the preparation of antibacterial surfaces. Dalton Trans. 2012, 41, 2456–2463. [Google Scholar] [CrossRef]
- Pallavicini, P.; Amendola, V.; Bergamaschi, G.; Cabrini, E.; Dacarro, G.; Rossi, N.; Taglietti, A. A bistren cryptand with a remote thioether function: Cu(ii) complexation in solution and on the surface of gold nanostars. New J. Chem. 2016, 40, 5722–5730. [Google Scholar] [CrossRef]
- Domínguez, M.; Blandez, J.F.; Lozano-Torres, B.; Torre, C.; Licchelli, M.; Mangano, C.; Amendola, V.; Sancenón, F.; Martínez-Máñez, R. A Nanoprobe Based on Gated Mesoporous Silica Nanoparticles for The Selective and Sensitive Detection of Benzene Metabolite t,t-Muconic Acid in Urine. Chem. Eur. J. 2021, 27, 1306–1310. [Google Scholar] [CrossRef]
- Taglietti, A.; Grisoli, P.; Dacarro, G.; Gattesco, A.; Mangano, C.; Pallavicini, P. Grafted monolayers of the neutral Cu (ii) complex of a dioxo-2, 3, 2 ligand: Surfaces with decreased antibacterial action. New J. Chem. 2018, 42, 7595–7598. [Google Scholar] [CrossRef]
- Taglietti, A.; Dacarro, G.; Barbieri, D.; Cucca, L.; Grisoli, P.; Patrini, M.; Arciola, C.R.; Pallavicini, P. High Bactericidal Self-Assembled Nano-Monolayer of Silver Sulfadiazine on Hydroxylated Material Surfaces. Materials 2019, 12, 2761. [Google Scholar] [CrossRef]
- Gao, J.; Reibenspies, J.H.; Martell, A.E. New Macrocyclic Dicopper (II) Complex: Synthesis Structure and Stability. Inorg. Chim. Acta 2002, 335, 125–129. [Google Scholar] [CrossRef]
- Graham, B.; Spiccia, L.; Batten, S.R.; Skelton, B.W.; White, A.H. Polynuclear Nickel (II) and Copper (II) Complexes of Hexaazamacrocycles Incorporating Pairs of Diethylenetriamine Subunits Separated by Aromatic Spacers. Inorg. Chim. Acta 2005, 358, 3983–3994. [Google Scholar] [CrossRef]
- Lehn, J.M.; Pine, S.H.; Watanabe, E.; Willard, A.K. Binuclear cryptates. Synthesis and binuclear cation inclusion complexes of bis-tren macrobicyclic ligands. J. Am. Chem. Soc. 1977, 99, 6766–6768. [Google Scholar] [CrossRef]
- Fabbrizzi, L.; Marcotte, N.; Stomeo, F.; Taglietti, A. Pyrophosphate detection in water by fluorescence competition assays: Inducing selectivity through the choice of the indicator. Angew. Chem. Int. Ed. 2002, 41, 3811–3814. [Google Scholar] [CrossRef]
- Hortalá, M.A.; Fabbrizzi, L.; Marcotte, N.; Stomeo, F.; Taglietti, A. Designing the Selectivity of the Fluorescent Detection of Amino Acids: A Chemosensing Ensemble for Histidine. J. Am. Chem. Soc. 2003, 125, 20–21. [Google Scholar] [CrossRef]
- Fabbrizzi, L.; Pallavicini, P.; Parodi, L.; Perotti, A.; Taglietti, A. Molecular recognition of the imidazole residue by a dicopper (II) complex with a bisdien macrocycle bearing two pendant arms. J. Chem. Soc. Chem. Commun. 1995, 23, 2439–2440. [Google Scholar] [CrossRef]
- Fabbrizzi, L.; Foti, F.; Patroni, S.; Pallavicini, P.; Taglietti, A. A Sleeping Host Awoken by Its Guest: Recognition and Sensing of Imidazole-Containing Molecules Based on Double Cu2+ Translocation inside a Polyaza Macrocycle. Angew. Chem. Int. Ed. 2004, 43, 5073–5077. [Google Scholar] [CrossRef]
- Walba, H.; Isensee, W. Acidity Constants of Some Arylimidazoles and Their Cations. J. Org. Chem. 1961, 26, 2789–2791. [Google Scholar] [CrossRef]
- Basallote, M.G.; Durán, J.; Fernández-Trujillo, J.; Máñez, M.A.; Quirós, M.; Salas, J.M. Equilibrium studies on the protonation and Cu(II) complexation by an hexaaza macrocycle containing p-xylyl spacers. The crystal structure of the hexaprotonated ligand and the kinetics of decomposition of the Cu(II) complexes. Polyhedron 2001, 20, 297–305. [Google Scholar] [CrossRef]
- Licsandru, E.; Petit, E.; Moldovan, S.; Ersen, O.; Barboiu, M. Biomimetic Autocatalytic Synthesis of Organized Silica Hybrids. Eur. J. Inorg. Chem. 2015, 22, 3637–3641. [Google Scholar] [CrossRef]
- Gargioni, C.; Borzenkov, M.; D’Alfonso, L.; Sperandeo, P.; Polissi, A.; Cucca, L.; Dacarro, G.; Grisoli, P.; Pallavicini, P.; D’Agostino, A.; et al. Self-Assembled Monolayers of Copper Sulfide Nanoparticles on Glass as Antibacterial Coatings. Nanomaterials 2020, 10, 352. [Google Scholar] [CrossRef] [PubMed]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- NCCLS. Methods for Determining Bactericidal Activity on Antimicrobial Agents. Approved Guideline; NCCLS: Wayne, PA, USA, 1999; Volume 19. [Google Scholar]
- NCCLS. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard M7-A6, 6th ed.; NCCLS: Wayne, PA, USA, 2003. [Google Scholar]

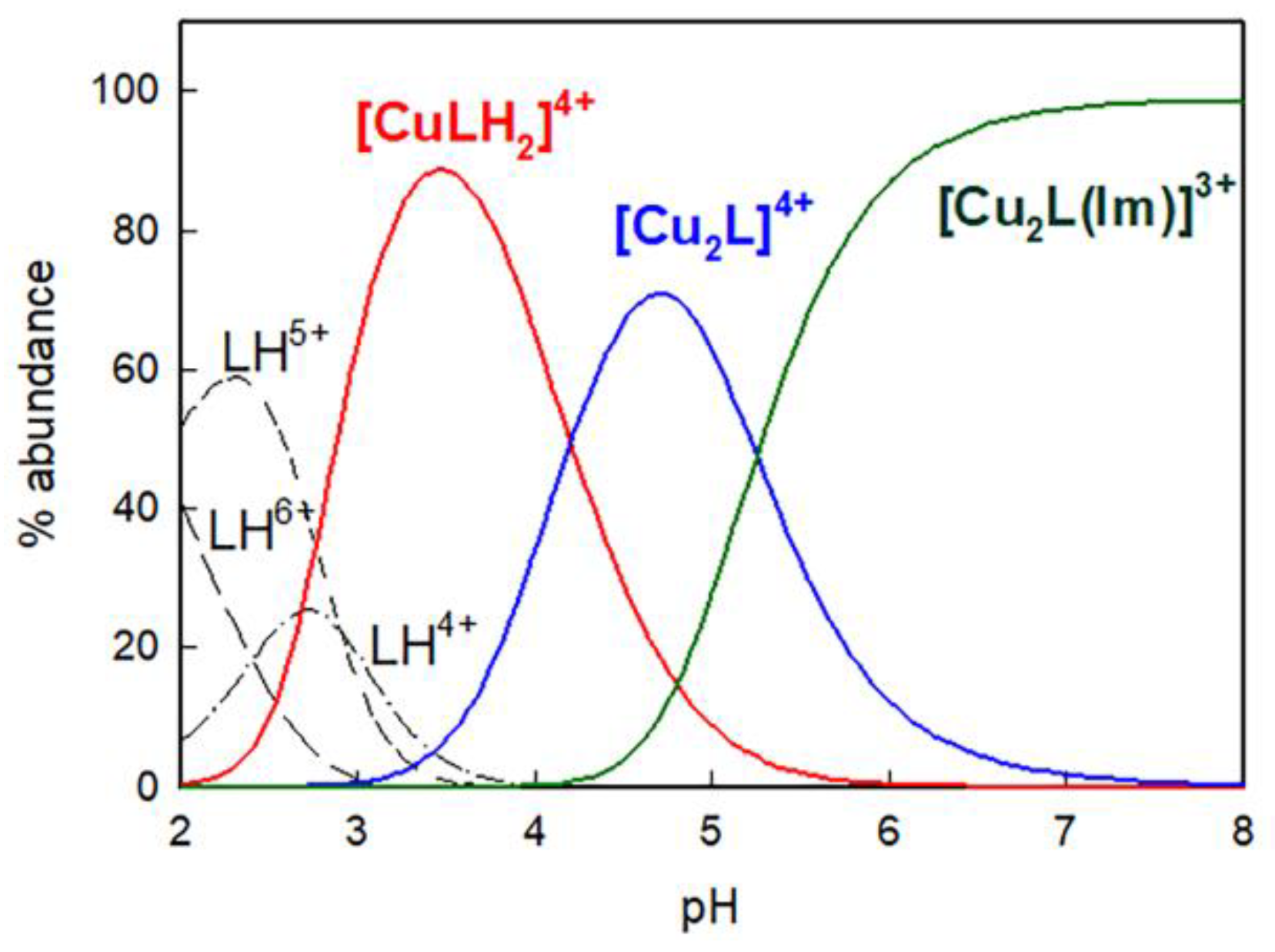
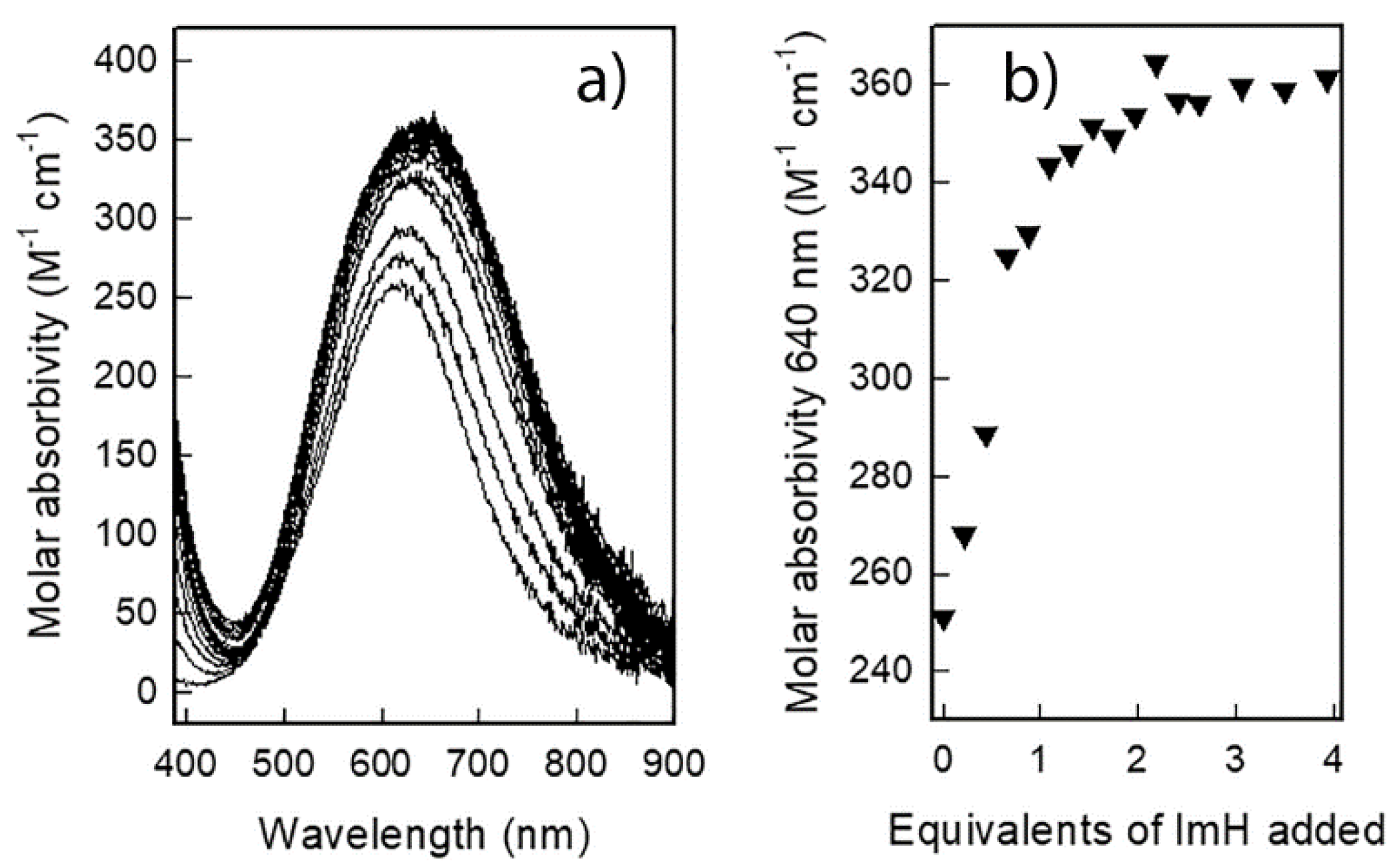


| Equilibrium | logβ |
|---|---|
| L + H+ ↔ LH+ | 9.64 (3) |
| L + 2H+ ↔ LH22+ | 18.22 (4) |
| L + 3H+ ↔ LH33+ | 26.30 (4) |
| L + 4H+ ↔ LH44+ | 33.23 (5) |
| L + 5H+ ↔ LH55+ | 36.15 (5) |
| L + 6H+ ↔ LH66+ | 38.0 (1) |
| L + 2Cu2+ ↔ [Cu2L]4+ | 26.17 (7) |
| L + Cu2+ + 2H+ ↔ [CuLH2]4+ | 30.87 (4) |
| L + 2Cu2+ + OH− ↔ [Cu2L(OH)]3+ | 18.8 (1) |
| L + 2Cu2+ + 2OH− ↔ [Cu2L(OH)2]2+ | 10.6 (1) |
| Im− + H+ ↔ ImH | 14.50 (1) |
| Im− + 2H+ ↔ ImH2+ | 21.2 (1) |
| L + 2Cu2+ + Im− ↔ [Cu2L(Im)]3+ | 40.7 (1) |
| E. coli | S. aureus | C. albicans | ||
|---|---|---|---|---|
| Cu(CF3SO3)2 | MIC (mol/L) | 0.010 | 5.0 × 10−3 | >0.010 |
| MBC/MFC (mol/L) | 0.010 | 0.010 | >0.010 | |
| [Cu2(BPXD)](CF3SO3)4 | MIC (mol/L) | 4.0 × 10−4 | 3.0 × 10−4 | 7.0 × 10−4 |
| MBC/MFC (mol/L) | 0.010 | 1.25 × 10−3 | 2.5 × 10−3 |
| Microbicidal Effect (ME) | |||
|---|---|---|---|
| E. coli | S. aureus | C. albicans | |
| Contact time | |||
| 5 h | 3.8 (±0.8) | 2.9 (±0.8) | 1.2 (±0.7) |
| 24 h | >5 | 5 (±1) | >5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gazzola, V.; Grisoli, P.; Amendola, V.; Dacarro, G.; Mangano, C.; Pallavicini, P.; Poggi, A.; Rossi, S.; Vigani, B.; Taglietti, A. A Supramolecular Approach to Antimicrobial Surfaces. Molecules 2022, 27, 5731. https://doi.org/10.3390/molecules27175731
Gazzola V, Grisoli P, Amendola V, Dacarro G, Mangano C, Pallavicini P, Poggi A, Rossi S, Vigani B, Taglietti A. A Supramolecular Approach to Antimicrobial Surfaces. Molecules. 2022; 27(17):5731. https://doi.org/10.3390/molecules27175731
Chicago/Turabian StyleGazzola, Valentina, Pietro Grisoli, Valeria Amendola, Giacomo Dacarro, Carlo Mangano, Piersandro Pallavicini, Antonio Poggi, Silvia Rossi, Barbara Vigani, and Angelo Taglietti. 2022. "A Supramolecular Approach to Antimicrobial Surfaces" Molecules 27, no. 17: 5731. https://doi.org/10.3390/molecules27175731
APA StyleGazzola, V., Grisoli, P., Amendola, V., Dacarro, G., Mangano, C., Pallavicini, P., Poggi, A., Rossi, S., Vigani, B., & Taglietti, A. (2022). A Supramolecular Approach to Antimicrobial Surfaces. Molecules, 27(17), 5731. https://doi.org/10.3390/molecules27175731