Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatment
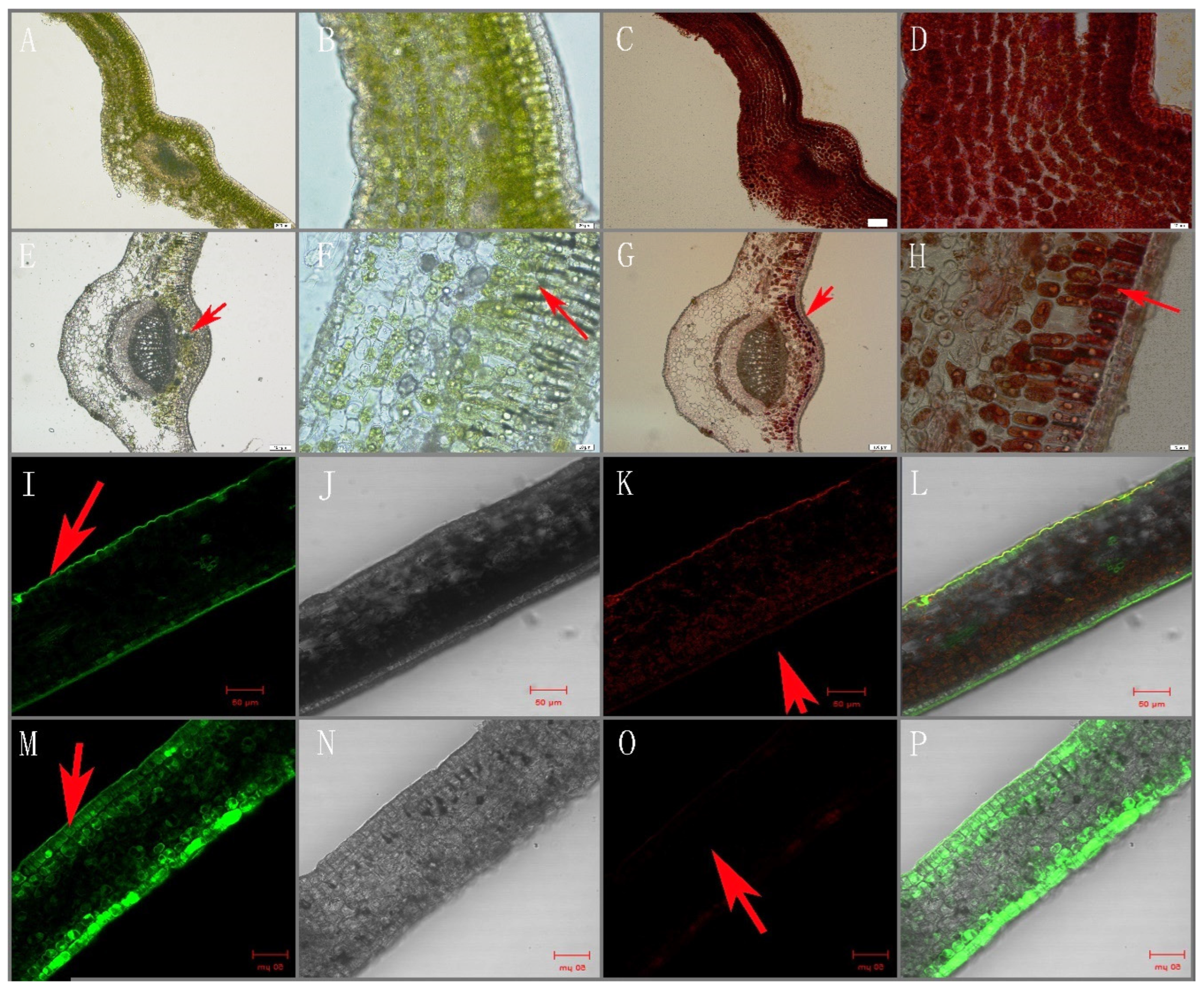
2.2. Electron Microscopy Analysis
2.3. Quantitative Determination of Catechin, Chlorophylls, Carotenoids, Total Flavonols and Total Polyphenols Content
2.4. Metabolome Analysis Using UPLC-Q-TOF/MS
2.5. Metabolome Analysis Using GC-TOF/MS
2.6. RNA Isolation, cDNA Library Construction, Illumina Deep Sequencing and Data Processing
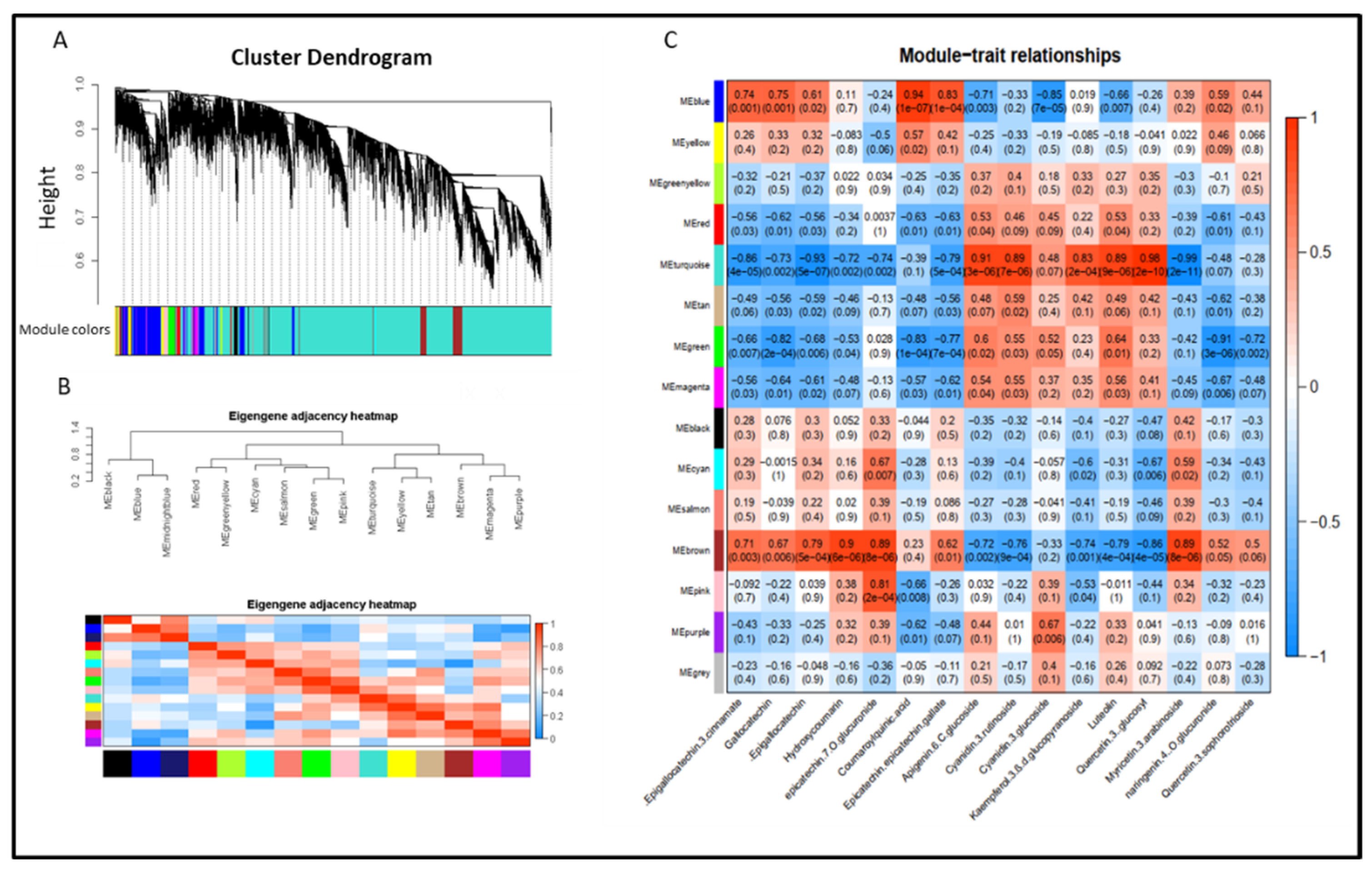
2.7. Gene Co-Expression Network, Module Identification and Gene Edge Number
2.8. Quantitative Reverse-Transcription PCR Analysis
2.9. Tissue Localization of Phenolic Compounds
3. Results
3.1. Phenotype and Ultra-Structure
3.2. Metabolic Changes of Catechins, Total Flavonol and Total Polyphenol Contents
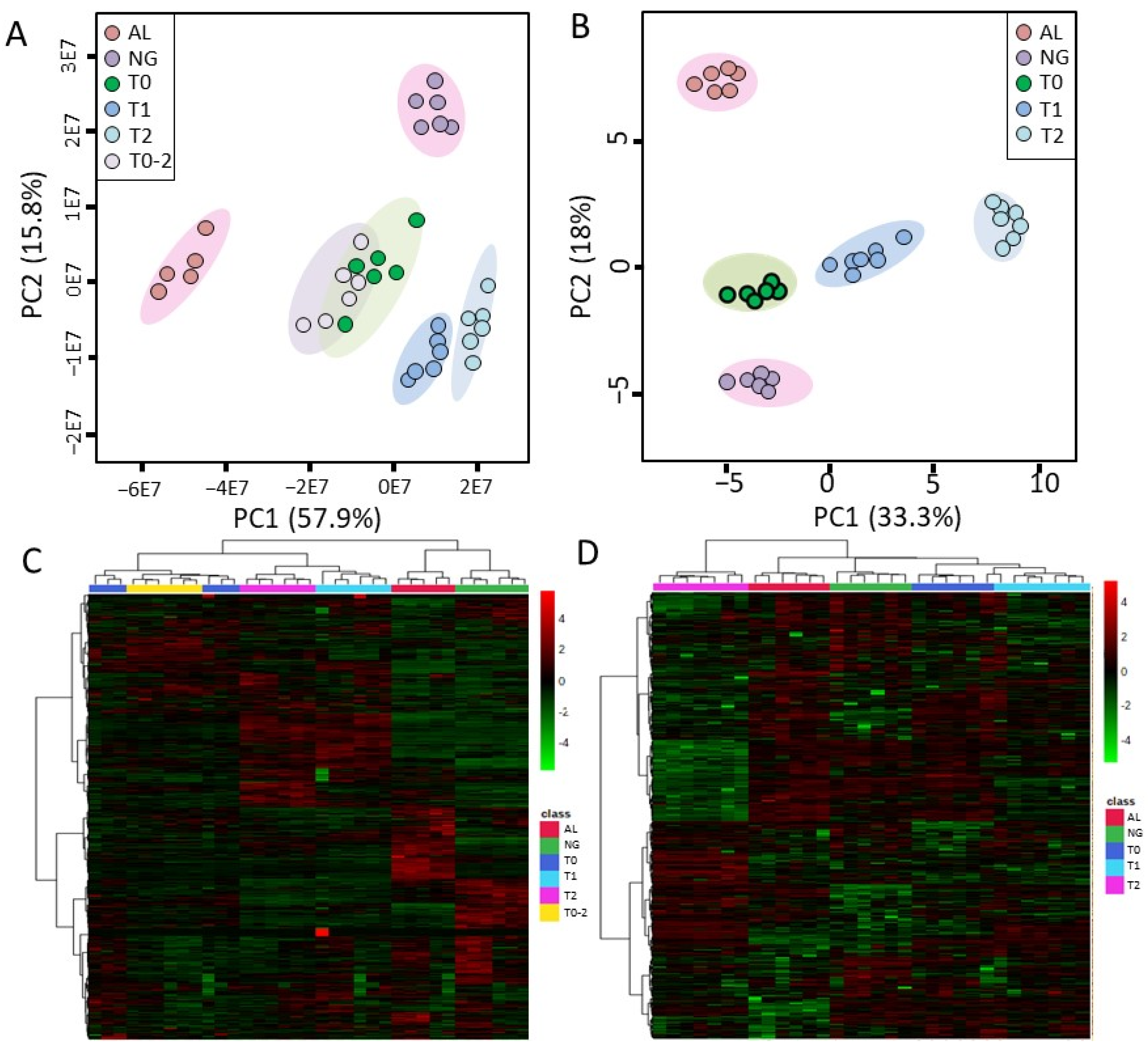
3.3. Overview of Metabolomic Profiling
3.4. Overview of Transcriptomic Analysis
3.5. Accumulation of Carbohydrates in Albino Shoots
3.6. Metabolic Changes of Flavanols in Albino Shoots
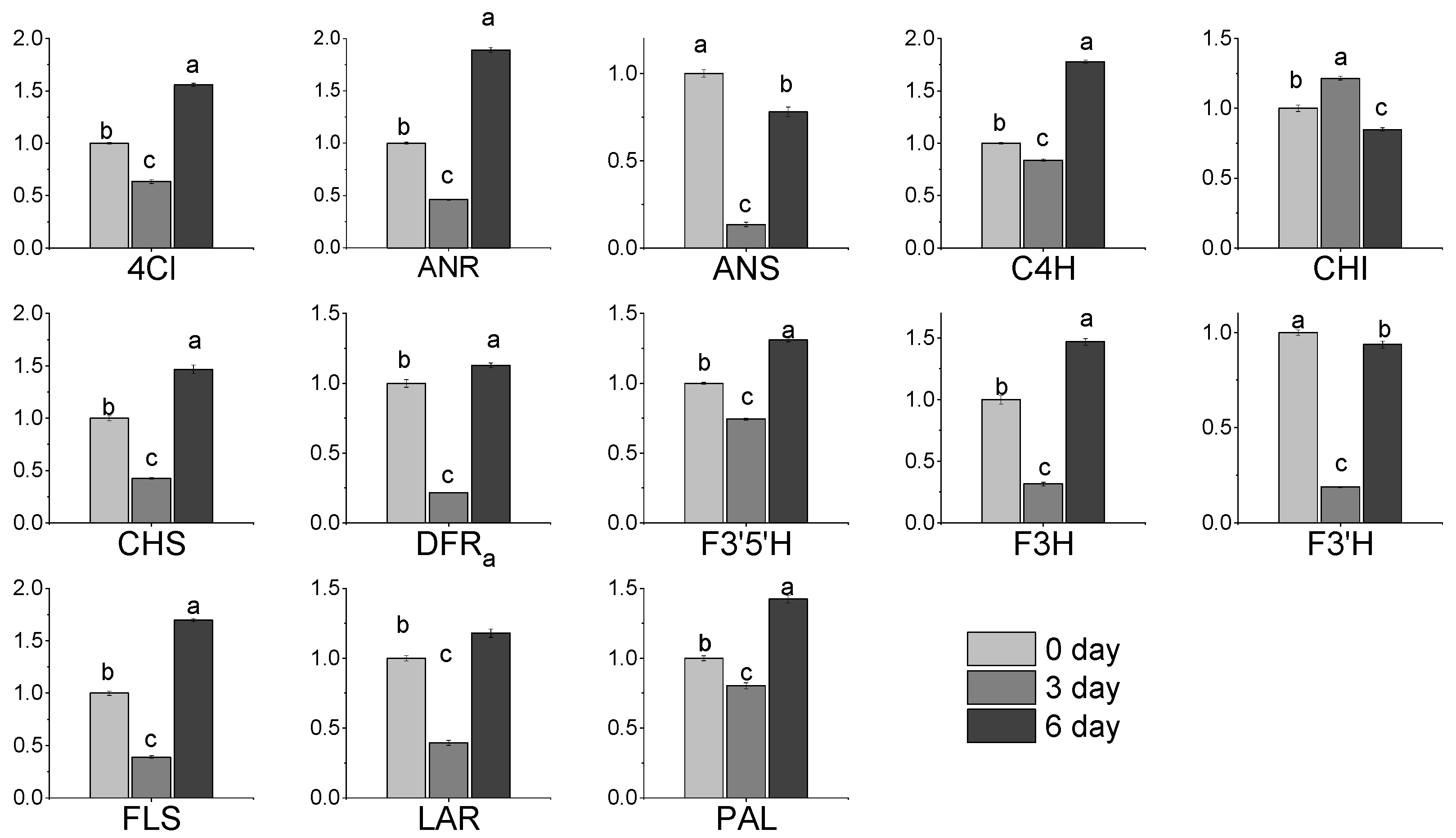
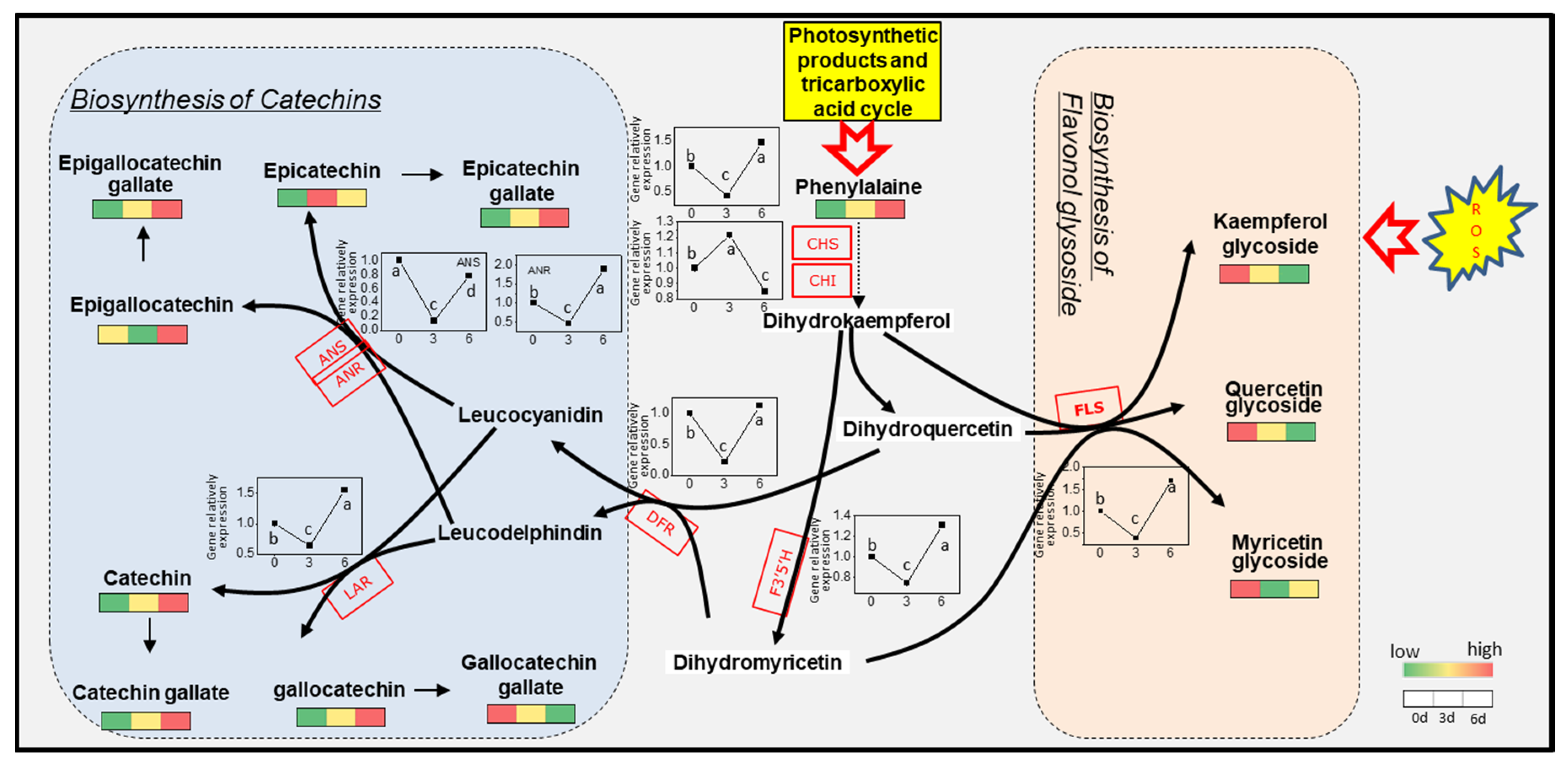
3.7. Expression of Gene Related to Flavonoids Metabolic in Albino Shoots
3.8. Intracellular Localization of Flavonoids
4. Discussion
4.1. Temperature-Stimulated Accumulation of Total Polyphenols and Catechins in Albino Mutant Tea Leaves Is Highly Induced by Enhanced Photosynthesis and Accumulation of Its Products
4.2. The Initial Accumulation and Temperature Inhibition of Flavonols in Albino Mutant Tea Leaves Are Associated with Metabolism Related to Oxidative Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.; Ruan, J. Tea: Analysis and Tasting. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 256–267. [Google Scholar]
- Tounekti, T.; Joubert, E.; Hernández, I.; Munné-Bosch, S. Improving the Polyphenol Content of Tea. Crit. Rev. Plant Sci. 2012, 32, 192–215. [Google Scholar] [CrossRef]
- Jayasekera, S.; Kaur, L.; Molan, A.L.; Garg, M.L.; Moughan, P.J. Effects of season and plantation on phenolic content of unfermented and fermented Sri Lankan tea. Food Chem. 2014, 152, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Cheynier, V.; Comte, G.; Davies, K.M.; Lattanzio, V.; Martens, S. Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 2013, 72, 1–20. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, L.; Wang, Z.; Liu, Y.; Sun, M.; Yang, D.; Wei, C.; Shan, Y.; Xia, T. Light-induced expression of genes involved in phenylpropanoid biosynthetic pathways in callus of tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 133, 72–83. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, Y.; Ma, L.; Yi, X.; Ruan, J. Metabolomic Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole-Time of Flight Mass Spectrometry (UPLC-Q-TOF/MS) Uncovers the Effects of Light Intensity and Temperature under Shading Treatments on the Metabolites in Tea. PLoS ONE 2014, 9, e112572. [Google Scholar] [CrossRef]
- Punyasiri, P.A.N.; Abeysinghe, I.S.B.; Kumar, V.; Treutter, D.; Duy, D.; Gosch, C.; Martens, S.; Forkmann, G.; Fischer, T.C. Flavonoid biosynthesis in the tea plant Camellia sinensis: Properties of enzymes of the prominent epicatechin and catechin pathways. Arch. Biochem. Biophys. 2014, 431, 22–30. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, L.; Liu, L.; Yang, Q.; Lu, Z.; Nie, Z.; Wang, Y.; Xia, T. Purification and characterization of a novel galloyltransferase involved in catechin galloylation in the tea plant (Camellia sinensis). J. Biol. Chem. 2012, 287, 44406–44417. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Abeysinghe, I.S.B.; He, J.; He, X.; Huhman, D.; Mewan, K.M.; Sumner, L.W.; Yun, J.; Dixon, R.A. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013, 161, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Galardi, C.; Pinelli, P.; Massai, R.; Remorini, D.; Agati, G. Differential accumulation of flavonoids and hydroxycinnamates in leaves of Ligustrum vulgare under excess light and drought stress. New Phytol. 2004, 63, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Satou, M.; Enoki, H.; Oikawa, A.; Ohta, D.; Saito, K.; Hachiya, T.; Sakakibara, H.; Kusano, M.; Fukushima, A.; Saito, K.; et al. Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins. Plant Mol. Biol. 2014, 85, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Markham, K.R. Tautomerism of flavonol glucosides—Relevance to plant UV protection and flower colour. J. Photochem. Photobiol. A Chem. 1998, 118, 99–105. [Google Scholar] [CrossRef]
- Yamasaki, H.; Sakihama, Y.; Ikehara, N. Flavonoid-peroxidase reaction as a detoxification mechanism of plant cell against H2O2. Plant Physiol. 1997, 115, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.O.; Gould, K.S. Anthocyanins in leaves: Light attenuators or antioxidants? Funct. Plant Biol. 2003, 30, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Guidi, L.; Morassi, B.; Pinelli, P.; Remorini, D.; Degl’Innocenti, E.; Giordano, C.; Massai, R.; Agati, G. On the role of flavonoids in the integrated mechanisms of response of Ligustrum vulgare and Phillyrea latifolia to high solar radiation. New Phytol. 2005, 167, 457–470. [Google Scholar] [CrossRef]
- Ma, C.L.; Chen, L.; Wang, X.; Jin, J.; Ma, J.; Yao, M.; Wang, Z. Differential expression analysis of different albescent stages of ‘baiye 1′ (Camellia sinensis (L.) O. Kuntze) using cDNA microarray. Sci. Hortic. 2012, 148, 246–254. [Google Scholar] [CrossRef]
- Feng, L.; Gao, M.; Hou, R.; Hu, X.; Zhang, L.; Wan, X.; Wei, S. Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 2014, 155, 98–104. [Google Scholar] [CrossRef]
- Li, Q.; Huang, J.; Liu, S.; Li, J.; Yang, X.; Liu, Y.; Liu, Z. Proteomic analysis of young leaves at three developmental stages in an albino tea cultivar. Proteome Sci. 2011, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Li, J.; Li, Y.; Yuan, L.; Liu, S.; Huang, J.; Liu, Z. Dynamic changes in catechin levels and catechin biosynthesis-related gene expression in albino tea plants (Camellia sinensis L.). Plant Physiol. Biochem. 2013, 71, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, H.; Zhong, W.; Wu, L.; Ye, J.; Lin, C.; Zheng, X.; Lu, J.; Liang, Y. Effect of temperature on accumulation of chlorophylls and leaf ultrastructure of low temperature induced albino tea plant. Afr. J. Biotechnol. 2008, 7, 1881–1885. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Mumm, R.; Vos, R.C.; Ruan, J. Metabolomics reveals the within-plant spatial effects of shading on tea plants. Tree Physiol. 2021, 41, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; De Vos, C.H.R.; van Tunen, A.J.; Verhoeyen, M.E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Lu, J.; Zhang, L.; Wu, S.; Wu, Y. Estimation of black tea quality by analysis of chemical composition and colour difference of tea infusions. Food Chem. 2003, 80, 283–290. [Google Scholar] [CrossRef]
- Mokochinski, J.B.; Mokochinski, J.B.; Mazzafera, P.; Sawaya, A.C.H.F.; Mumm, R.; de Vos, R.C.H.; Hall, R.D. Metabolic responses of Eucalyptus species to different temperature regimes. J. Integr. Plant Biol. 2018, 60, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000, 72, 3573–3580. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Liu, F.; Chen, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Sun, K.; Wu, Y.; Chen, X.; et al. Transcriptome Analysis and Identification of Differentially Expressed Transcripts of Immune-Related Genes in Spleen of Gosling and Adult Goose. Int. J. Mol. Sci. 2015, 16, 22904–22926. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Li, A.; Horvath, S. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics 2007, 23, 222–231. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Piko, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods. 2012, 9, 1069. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, L.; Xia, T.; Zhao, L. Investigation of the site-specific accumulation of catechins in the tea plant (Camellia sinensis (L.) O. Kuntze) via vanillin-HCl staining. J. Agric. Food Chem. 2009, 57, 10371–10376. [Google Scholar] [CrossRef]
- Hutzler, P.; Fischbach, R.; Heller, W.; Jungblut, T.P.; Reuber, S.; Schmitz, R.; Veit, M.; Weissenbock, G.; Schnitzler, J.P. Tissue localization of phenolic compounds in plants by confocal laser scanning microscopy. J. Exp. Bot. 1998, 49, 953–965. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, D.; Liu, M.; Ruan, J. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2018, 231, 272–281. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and Metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef]
- Masanori, I.; Jun, H.; Amane, M.; Hiroyuki, I. Autophagy Contributes to Nighttime Energy Availability for Growth in Arabidopsis. Plant Physiol. 2013, 161, 1682–1693. [Google Scholar]
- Agati, G.; Galardi, C.; Gravano, E.; Romani, A.; Tattini, M. Flavonoid distribution in tissues of Phillyrea latifolia L. leaves as estimated by microspectrofluorometry and multispectral fluorescence microimaging. Photochem. Photobiol. 2002, 76, 350–360. [Google Scholar] [CrossRef]
- Yang, Z.; Kobayashi, E.; Katsuno, T.; Asanuma, T.; Fujimori, T.; Ishikawa, T.; Tomomura, M.; Mochizuki, K.; Watase, T.; Nakamura, Y.; et al. Characterisation of volatile and non-volatile metabolites in etiolated leaves of tea (Camellia sinensis) plants in the dark. Food Chem. 2012, 135, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Brunetti, M.; Ferdinando, F.; Ferrini, S. Pollastri and M. Tattini. Functional roles of flavonoids in photoprotection: New evidence, lessons from the past. Plant Physiol. Biochem. 2013, 72, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.R. Review: Molecular response of plant cells to UV-B stress. Funct. Plant Biol. 2002, 29, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.O.; Gould, K.S.; Kilmartin, P.A.; Mitchell, K.A.; Markham, K.R. Antioxidant capacities of green and cyanic leaves in the sun species, Quintinia serrata. Funct. Plant Biol. 2002, 29, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Matteini, P.; Goti, A.; Tattini, M. Chloroplast-located flavonoids can scavenge singlet oxygen. New Phytol. 2007, 174, 77–89. [Google Scholar] [CrossRef]
- Bernal, M.; Llorens, L.; Julkunen-Tiitto, R.; Badosa, J.; Verdaguer, D. Altitudinal and seasonal changes of phenolic compounds in Buxus sempervirens leaves and cuticles. Plant Physiol. Biochem. 2013, 70, 471. [Google Scholar] [CrossRef] [PubMed]

| NG | AM | T0 | T1 | T2 | |
|---|---|---|---|---|---|
| Catechins (µmol/mg dry weight) | |||||
| Gallocatechin | 10.82 ± 3.54 a | 4.33 ± 0.65 c | 4.57 ± 0.65 c | 6.86 ± 0.33 b | 11.75 ± 4.9 a |
| Epigallocatechin | 22.38 ± 2.92 b | 19.46 ± 3.51 b | 23.35 ± 3.43 b | 20.9 ± 0.16 b | 26.12 ± 3.43 b |
| Epigallocatechin gallate | 143.39 ± 2.42 b | 98.45 ± 6.47 c | 107.62 ± 8.01 c | 136.26 ± 1.38 b | 138.05 ± 1.18 b |
| Gallocatechin gallate | 0.69 ± 0.13 b | 0.43 ± 0.02 a | 0.62 ± 0.03 a | 0.55 ± 0.01 b | 0.52 ± 0.01 b |
| Catechin | 3.52 ± 0.32 a | 2.01 ± 0.23 c | 1.77 ± 0.06 c | 1.93 ± 0.04 b | 2.28 ± 0.2 a |
| Epicatechin | 17.84 ± 0.71 c | 14.13 ± 3.53 c | 16.58 ± 4.03 c | 21.2 ± 2.96 c | 18.48 ± 0.45 c |
| Epicatechin gallate | 23.47 ± 1.8 b | 18.34 ± 1.53 d | 20.29 ± 1.36 d | 25.33 ± 0.23 c | 27.35 ± 1.7 b |
| Catechin gallate | 1.39 ± 0.15 c | 0.67 ± 0.02 e | 0.81 ± 0.02 e | 1.13 ± 0.02 d | 1.27 ± 0.05 c |
| Gallocatechin | 12.82 ± 2.7 a | 4.02 ± 0.39 c | 4.57 ± 0.65 c | 6.86 ± 0.33 b | 11.75 ± 4.9 a |
| Chlorophylls and carotenoids (μg/g, fresh weight) | |||||
| Chlorophyll b | 240.21 ± 6.25 a | 94.24 ± 7.73 c | 104.36 ± 8.48 c | 196.62 ± 8.61 b | 255.70 ± 9.73 a |
| Chlorophyll a | 804.21 ± 26.81 b | 291.25 ± 14.53 d | 280.04 ± 19.92 d | 664.27 ± 46.77 c | 900.17 ± 40.80 a |
| Lutein | 223.07 ± 15.92 a | 88.87 ± 3.24 d | 79.63 ± 6.13 d | 143.91 ± 13.57 c | 183.87 ± 17.56 b |
| alpha-Carotene | 2.39 ± 0.05 d | 32.99 ± 2.45 a | 30.08 ± 2.35 a | 8.76 ± 0.55 b | 5.61 ± 0.16 c |
| beta-Carotene | 2.98 ± 0.15 d | 89.59 ± 6.92 a | 80.55 ± 7.02 a | 12.75 ± 0.33 b | 7.98 ± 0.79 c |
| Neoxanthin | 66.96 ± 3.21 b | 26.84 ± 1.19 c | 24.44 ± 5.39 c | 71.76 ± 8.47 a | 76.11 ± 9.04 a |
| Violaxanthin | 87.65 ± 3.56 a | 50.31 ± 1.88 c | 49.34 ± 3.08 c | 68.38 ± 7.66 b | 77.93 ± 10.08 a |
| zeaxanthin | 1.67 ± 0.04 c | 3.75 ± 0.32 a | 3.02 ± 0.22 a | 0.97 ± 0.46 d | 2.80 ± 0.55 b |
| Total polyphenols and total flavonol | |||||
| Total polyphenols (mg/g, dry weight) | 215.21 ± 9.20 a | 114.45 ± 11.58 c | 110.55 ± 14.46 c | 176.71 ± 10.06 b | 234.82 ± 14.85 a |
| Myricetin (μg/g, dry weight) | 2.51 ± 0.14 a | 2.55 ± 0.25 b | 2.68 ± 0.20 b | 2.18 ± 0.27 c | 2.64 ± 0.18 a |
| Quercetin (μg/g, dry weight) | 1.25 ± 0.08 c | 1.87 ± 0.05 a | 1.73 ± 0.05 a | 1.49 ± 0.04 b | 1.18 ± 0.06 c |
| Kaempferol (μg/g, dry weight) | 1.52 ± 0.10 b | 1.66 ± 0.04 a | 1.75 ± 0.11 a | 1.42 ± 0.07 b | 1.44 ± 0.07 b |
| AM/NG | T1/T0 | T2/T0 | T2/T1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | LOG2fold | VIP | p | LOG2fold | p | LOG2fold | VIP | p | LOG2fold | VIP | p | |
| Epiafzelechin 3-gallate | −1.11 | 1.40 | * | 1.33 | 1.76 | * | 1.42 | 1.69 | * | 0.08 | 0.54 | |
| Epigallocatechin 3-cinnamate | −2.08 | 1.41 | * | −0.37 | 0.89 | 0.88 | 1.52 | * | 1.26 | 1.75 | * | |
| Epigallocatechin 3-coumaroate | −1.76 | 1.49 | * | 0.18 | 1.33 | * | 1.26 | 1.68 | * | 1.08 | 1.80 | * |
| 3 Hydroxycoumarin | −1.45 | 1.05 | * | 0.18 | 0.34 | 1.17 | 1.31 | * | 0.99 | 1.38 | * | |
| epicatechin 7-O-glucuronide | −3.11 | 1.45 | * | 0.43 | 1.08 | * | 0.96 | 1.49 | * | 0.53 | 1.40 | * |
| Apigenin glucoside arabinoside | −1.90 | 1.37 | * | 0.00 | 0.03 | 1.39 | 1.67 | * | 1.38 | 1.79 | * | |
| Cyanidin 3-rutinoside | −3.54 | 1.19 | * | 2.22 | 1.68 | * | 2.57 | 1.60 | * | 0.35 | 0.94 | |
| Delphinidin coumaroylglucoside) | −3.38 | 1.47 | * | 2.10 | 1.74 | * | 2.51 | 1.66 | * | 0.40 | 1.28 | * |
| Gladiatoside C2 | −6.79 | 1.48 | * | 2.39 | 1.54 | * | 2.90 | 1.59 | * | 0.50 | 1.01 | |
| Glycitin | −1.71 | 1.43 | * | 1.60 | 1.67 | * | 2.01 | 1.67 | * | 0.41 | 1.26 | * |
| Hyacinthin | −5.23 | 1.16 | * | 2.64 | 1.55 | * | 2.91 | 1.53 | * | 0.27 | 0.56 | |
| Isovitexin glucoside | −1.15 | 1.50 | * | 1.68 | 1.83 | * | 1.97 | 1.68 | * | 0.29 | 1.49 | * |
| Naringin | −2.39 | 1.50 | * | 0.17 | 0.98 | 0.53 | 1.58 | * | 0.36 | 1.70 | * | |
| Vanillic acid | −1.41 | 1.43 | * | 0.29 | 0.64 | 0.74 | 1.42 | * | 0.46 | 1.03 | ||
| Cyanidin 3-glucoside | 1.67 | 1.43 | * | −0.02 | 0.07 | −1.17 | 1.52 | * | −1.15 | 1.63 | * | |
| Kaempferol glucopyranoside | 1.67 | 1.46 | * | −0.60 | 1.57 | * | −1.22 | 1.61 | * | −0.62 | 1.69 | * |
| Rutin(Quercetin 3-Rutinoside) | 2.98 | 1.50 | * | −1.04 | 1.64 | * | −1.86 | 1.62 | * | −0.82 | 1.32 | * |
| Myricetin 3-arabinoside | 1.14 | 1.51 | * | −0.90 | 1.71 | * | −1.77 | 1.65 | * | −0.87 | 1.73 | * |
| Naringenin glucuronide | 2.22 | 1.43 | * | −0.67 | 1.57 | * | −1.84 | 1.63 | * | −1.17 | 1.83 | * |
| Quercetin 4′,7-diglucoside | 1.76 | 1.45 | * | −2.08 | 1.78 | * | −2.80 | 1.65 | * | −0.72 | 1.02 | * |
| AM/NG | T1/T0 | T2/T0 | T2/T1 | |
|---|---|---|---|---|
| Carbohydrate | ||||
| D-Arabinopyranose | −1.21 * | 0.53 * | 1.22 * | 0.69 ** |
| d-Galactose | −1.28 ** | 0.10 | 0.29 | 0.19 |
| D-Glucitol | −3.84 ** | 0.39 | 0.76 * | 0.37 |
| d-Glucose | −0.43 ** | 0.61 ** | 0.33 | 0.28 * |
| D-Psicose | −0.67 ** | −0.17 * | −0.24 * | −0.07 |
| Mannose | 0.26 | 0.33 | 0.71 ** | 0.38 * |
| Inosose | −2.65 ** | 1.06 ** | 0.56 | −0.50 * |
| L-(-)-Sorbose | −0.59 ** | 0.18 | −0.17 | 0.00 |
| Amino acid | ||||
| l-Glutamine | 0.80 ** | −2.09 ** | −3.34 ** | −1.25 * |
| L-Proline | 1.53 ** | 0.08 | −2.24 ** | −2.32 * |
| L-Theanine | 1.53 ** | −0.23 * | −0.80 * | −0.57 * |
| L-Aspartic acid | 1.60 ** | −0.33 * | −1.22 ** | −0.89 * |
| L-Glutamic acid | 1.63 ** | −0.56 ** | −1.69 ** | −1.13 * |
| Serine | 1.94 ** | −0.56 * | −1.44 ** | −0.88 * |
| Asparagine | 0.56 | 1.46 ** | −0.18 | −1.64 * |
| Gene ID | Annotation | Class_Annotation |
|---|---|---|
| CSS0035261 | ADP glucose pyrophosphorylase | Starch and sucrose metabolism |
| CSS0026029 | photosystem II protein H (chloroplast) | Photosystem II reaction center protein |
| CSS0032826 | starch synthase 1, chloroplastic/amyloplastic-like | Starch and sucrose metabolism |
| CSS0004941 | UDP-Glycosyltransferase superfamily protein isoform 1 | Starch and sucrose metabolism |
| CSS0025971 | ferredoxin-3, chloroplastic | Photosynthesis |
| CSS0009596 | hypothetical protein PRUPE_ppa020609mg, partial | Defense mechanisms |
| CSS0040654 | NF-X1-type zinc finger protein NFXL1-like | response to stress | response to abiotic stimulus |
| CSS0019730 | peroxidase 12 | peroxidase activity | response to oxidative stress |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Li, C.; Jiao, Z.; Ruan, J.; Liu, M.-Y. Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves. Molecules 2022, 27, 5792. https://doi.org/10.3390/molecules27185792
Zhang Q, Li C, Jiao Z, Ruan J, Liu M-Y. Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves. Molecules. 2022; 27(18):5792. https://doi.org/10.3390/molecules27185792
Chicago/Turabian StyleZhang, Qunfeng, Chunlei Li, Zixin Jiao, Jianyun Ruan, and Mei-Ya Liu. 2022. "Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves" Molecules 27, no. 18: 5792. https://doi.org/10.3390/molecules27185792
APA StyleZhang, Q., Li, C., Jiao, Z., Ruan, J., & Liu, M.-Y. (2022). Integration of Metabolomics and Transcriptomics Reveal the Mechanism Underlying Accumulation of Flavonols in Albino Tea Leaves. Molecules, 27(18), 5792. https://doi.org/10.3390/molecules27185792









