Formate Dehydrogenase Mimics as Catalysts for Carbon Dioxide Reduction
Abstract
1. Introduction
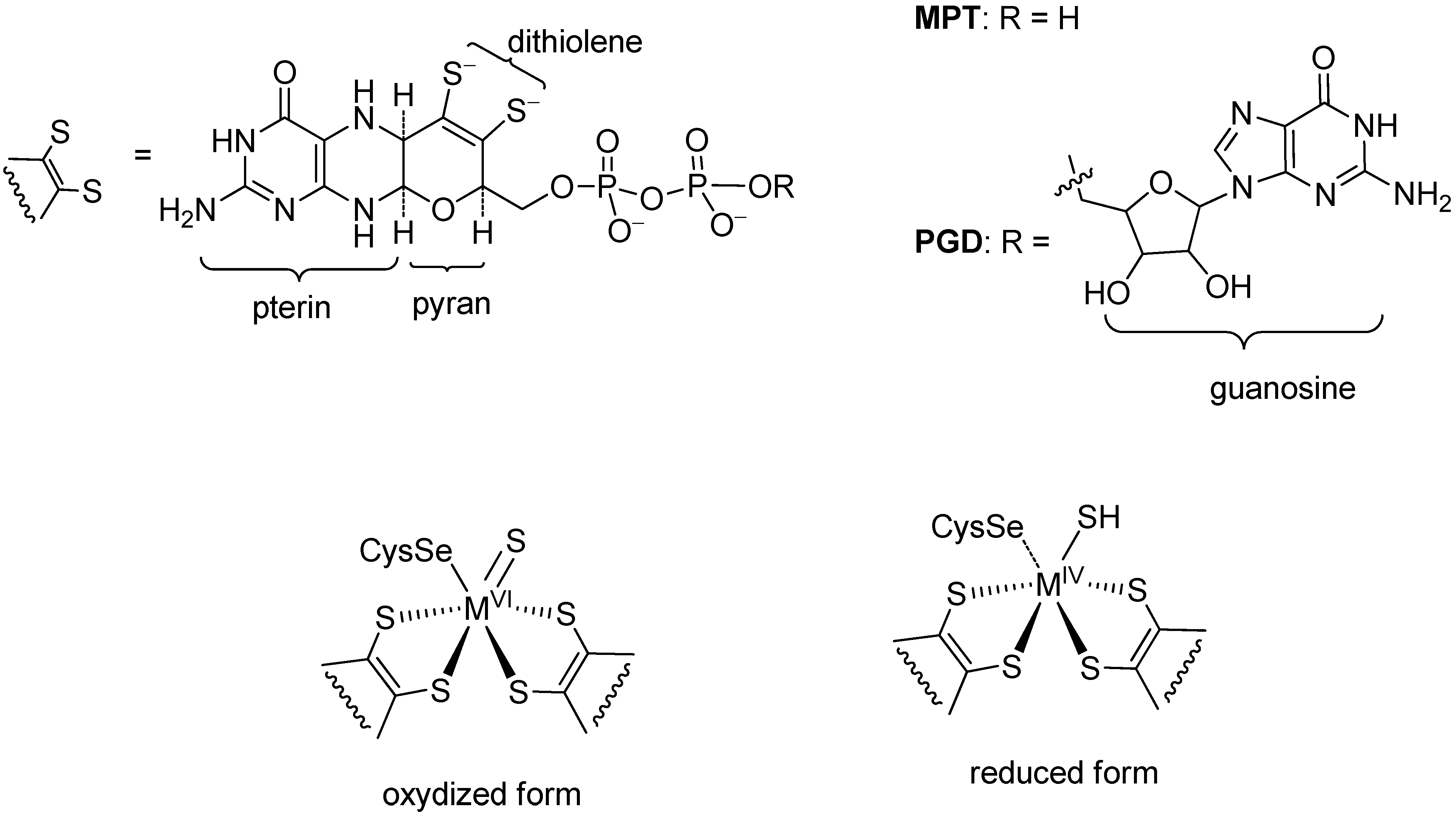
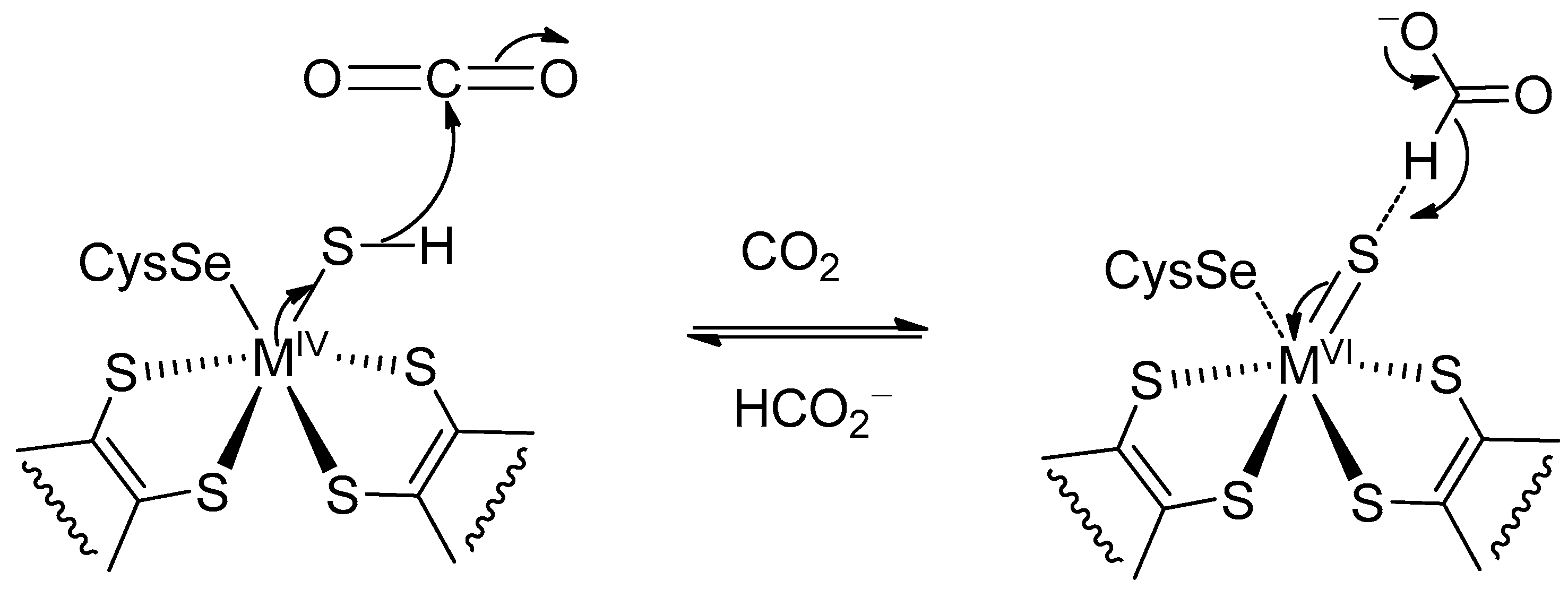
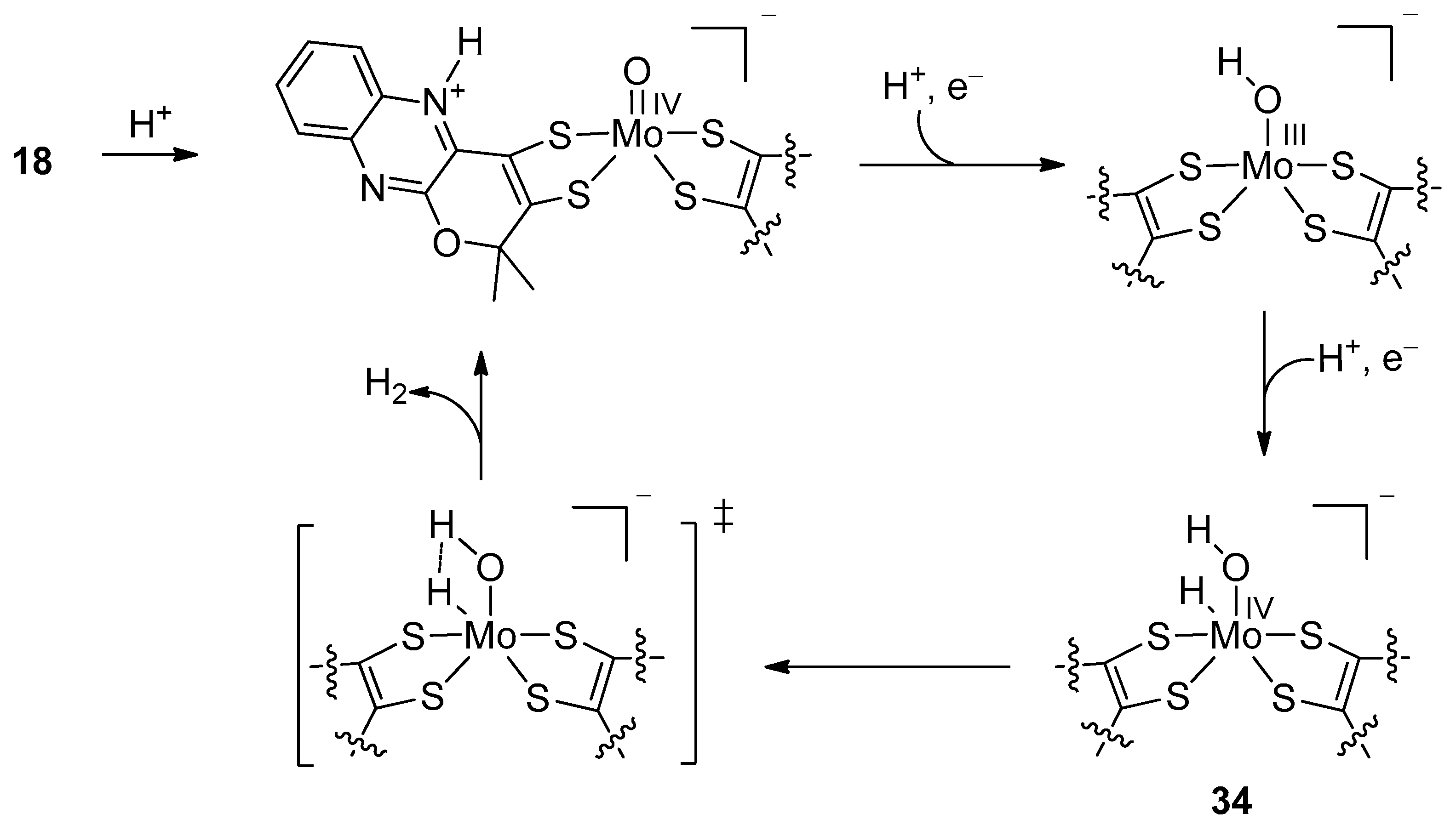
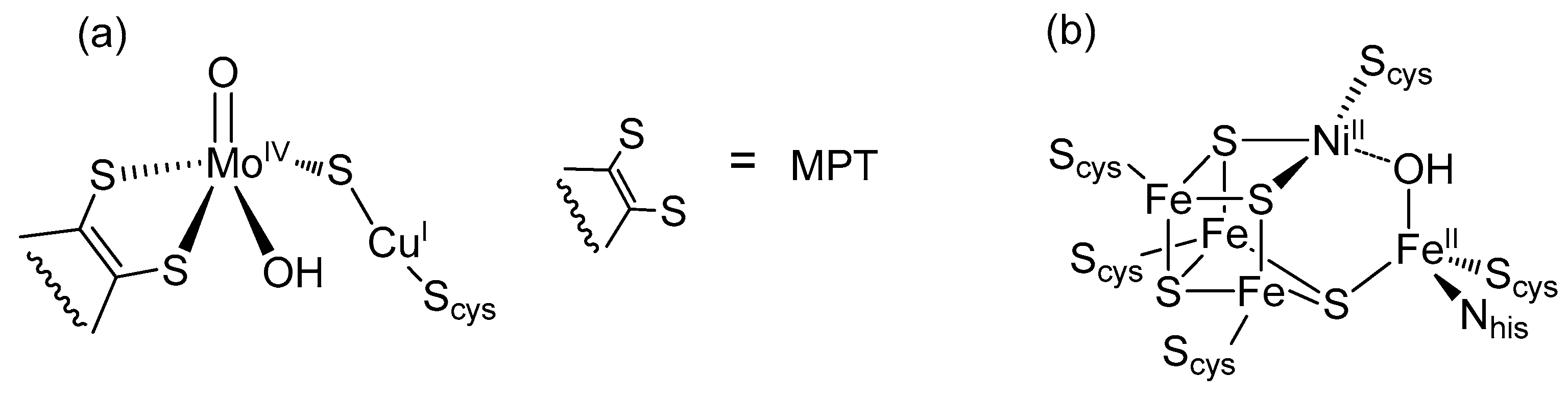
2. Formate Dehydrogenases (FDHs)
 (1)
(1)3. Structural Models of FDH
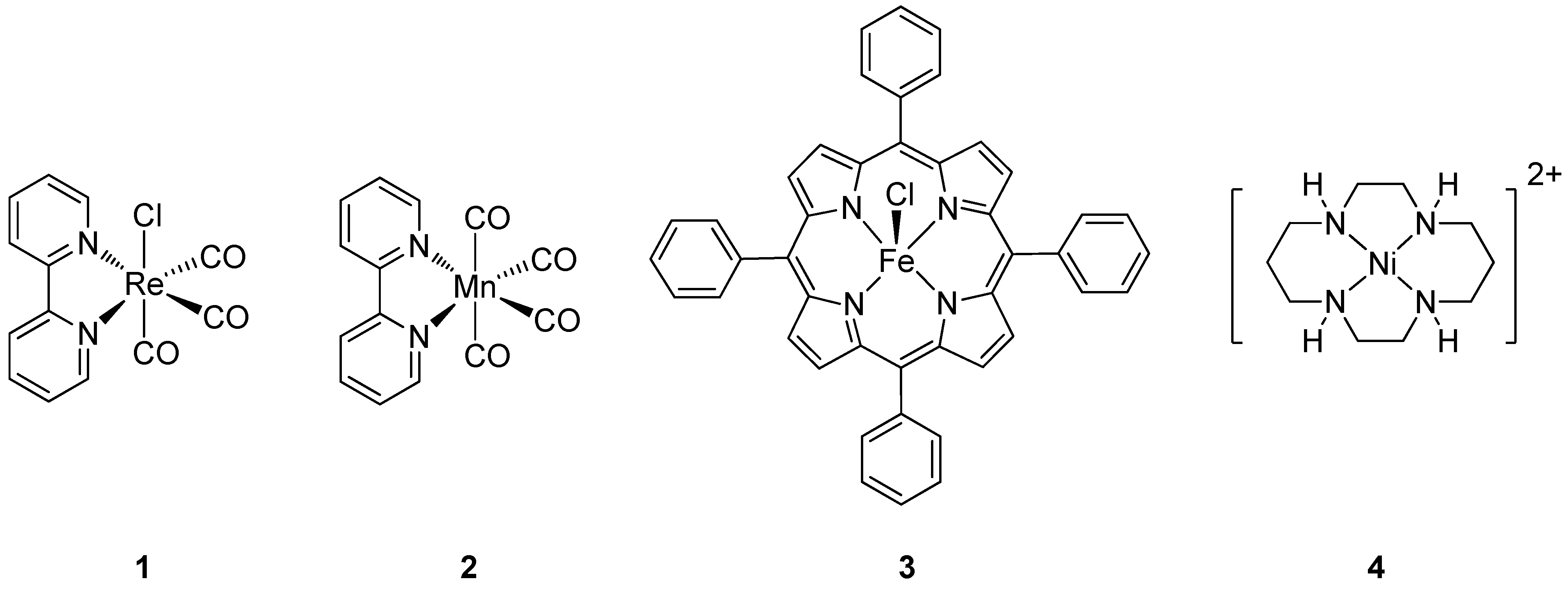
3.1. Variation of the Axial Ligands
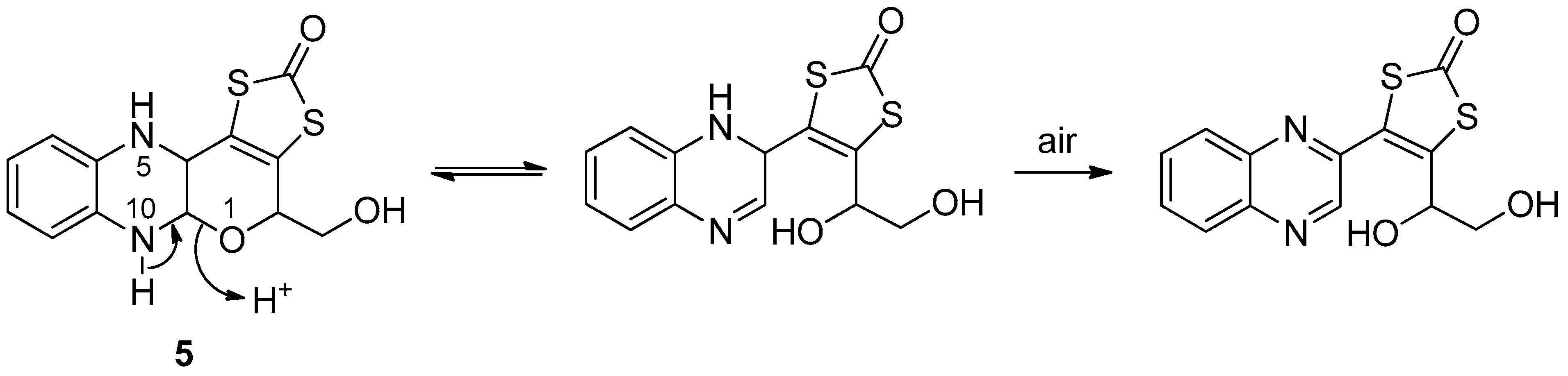
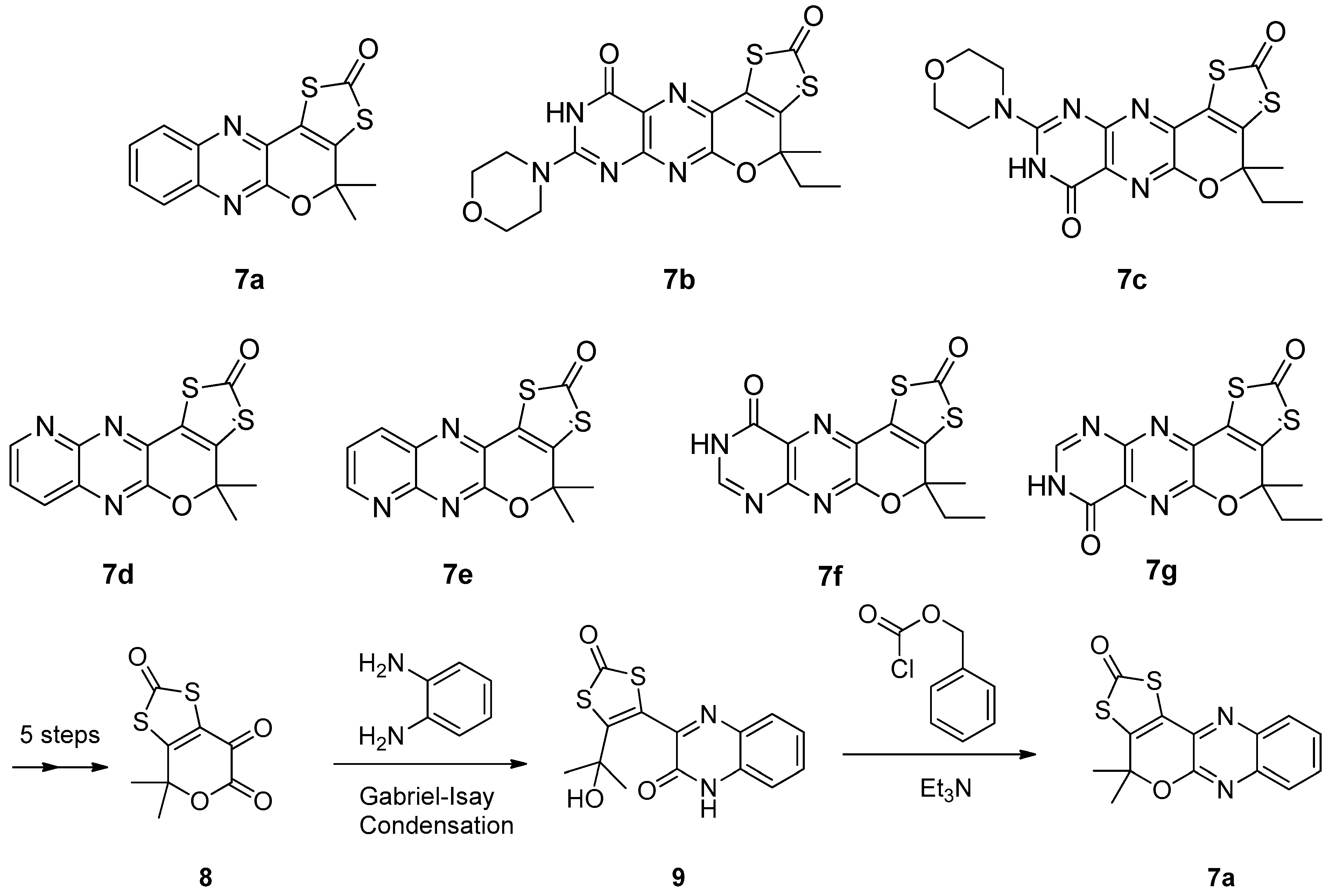
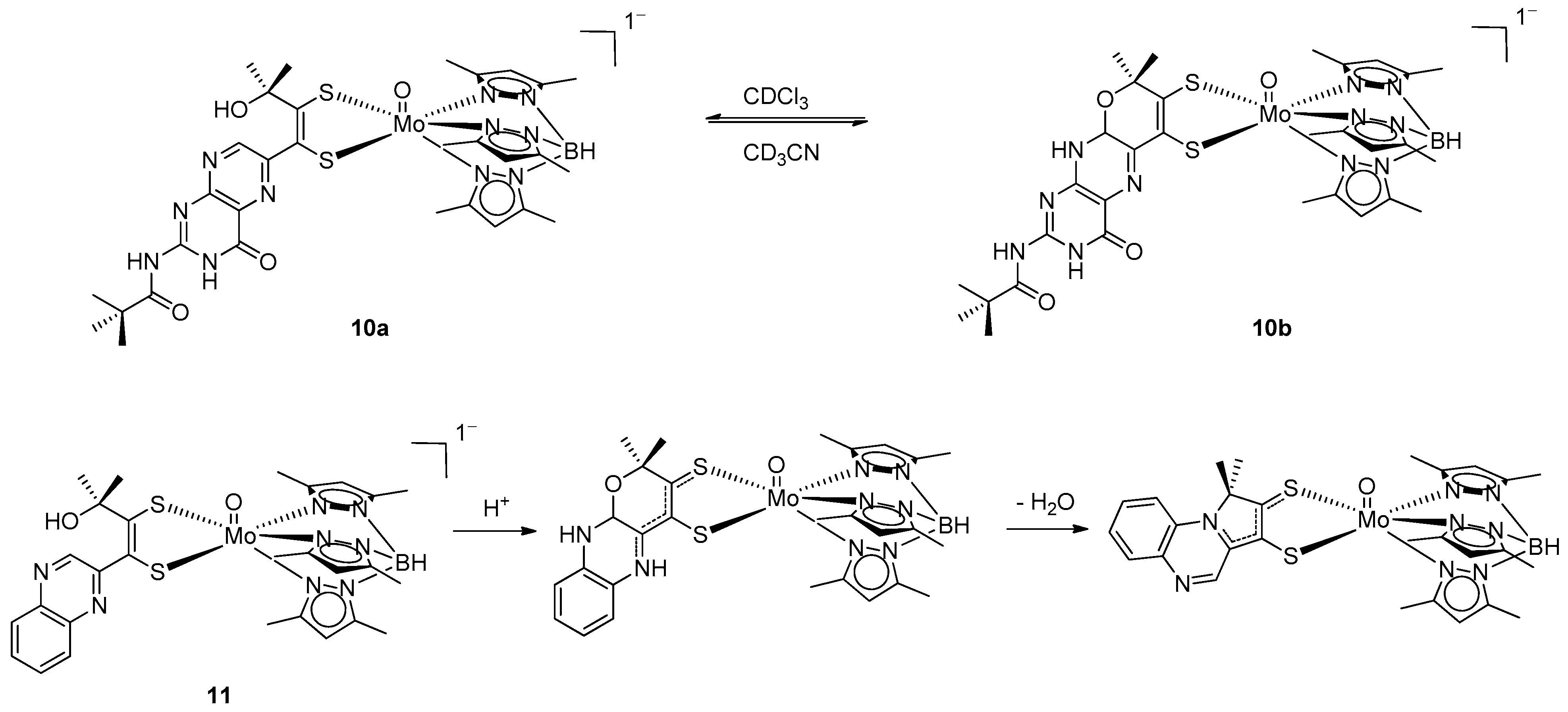
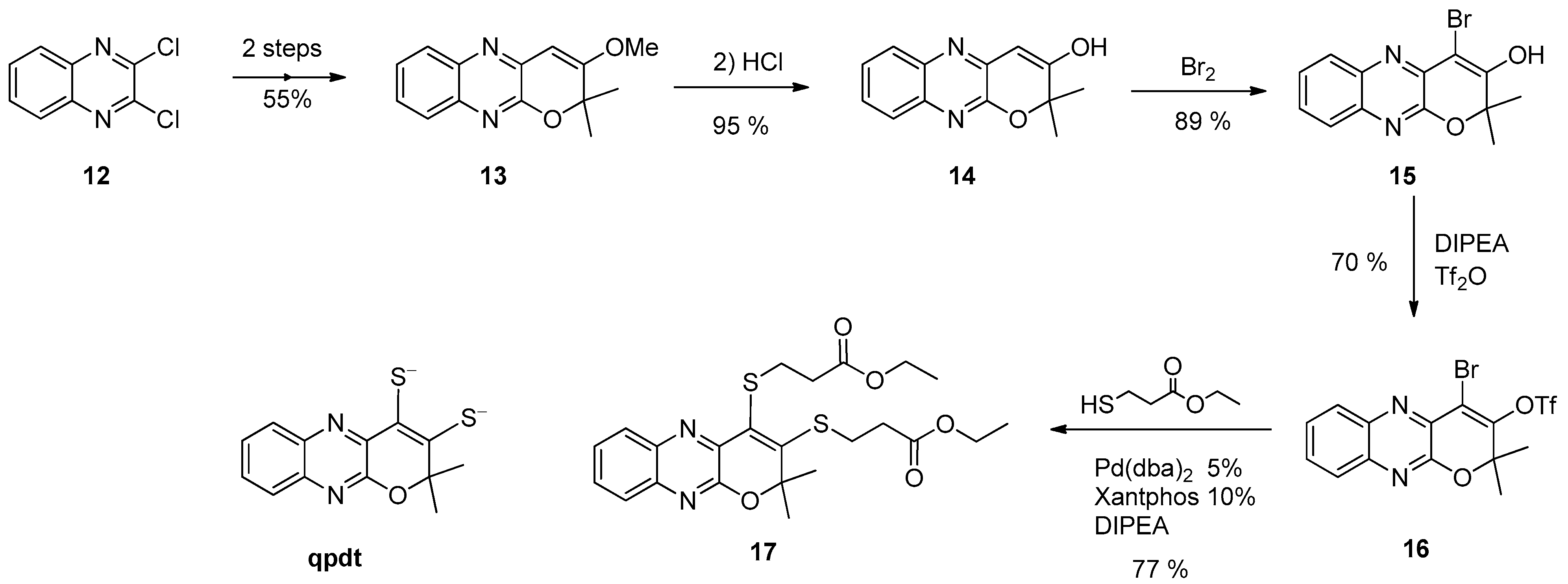
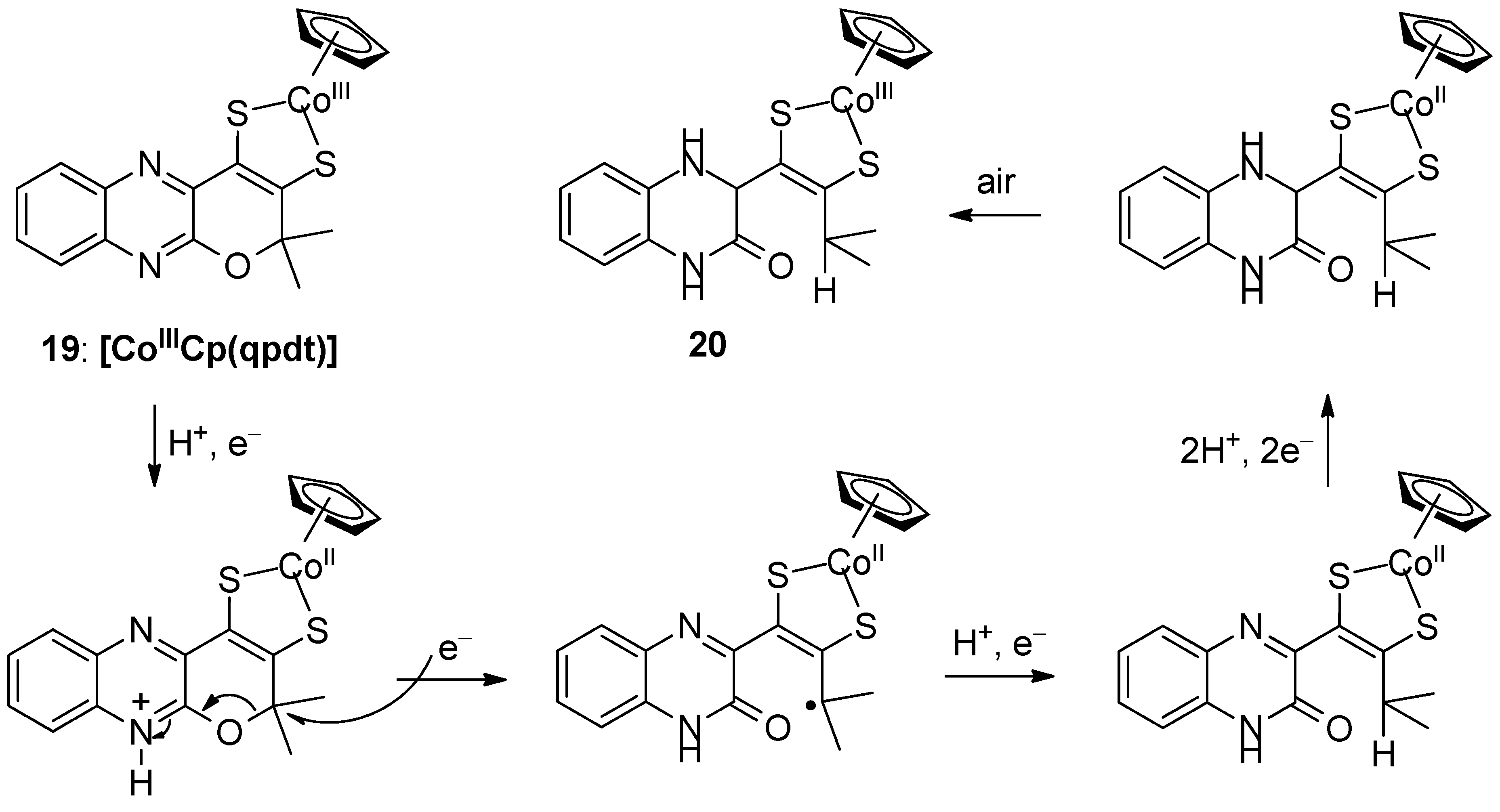
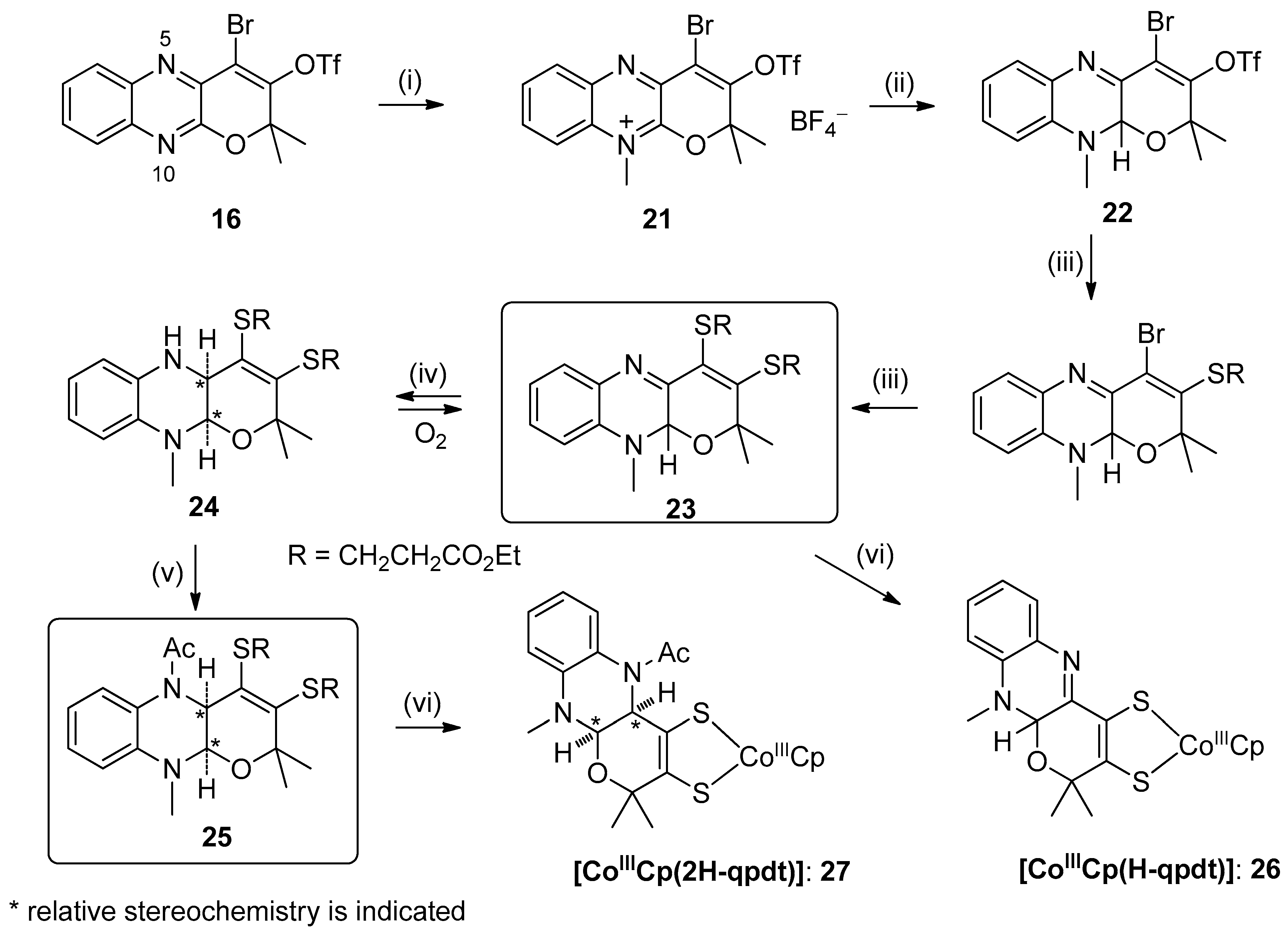
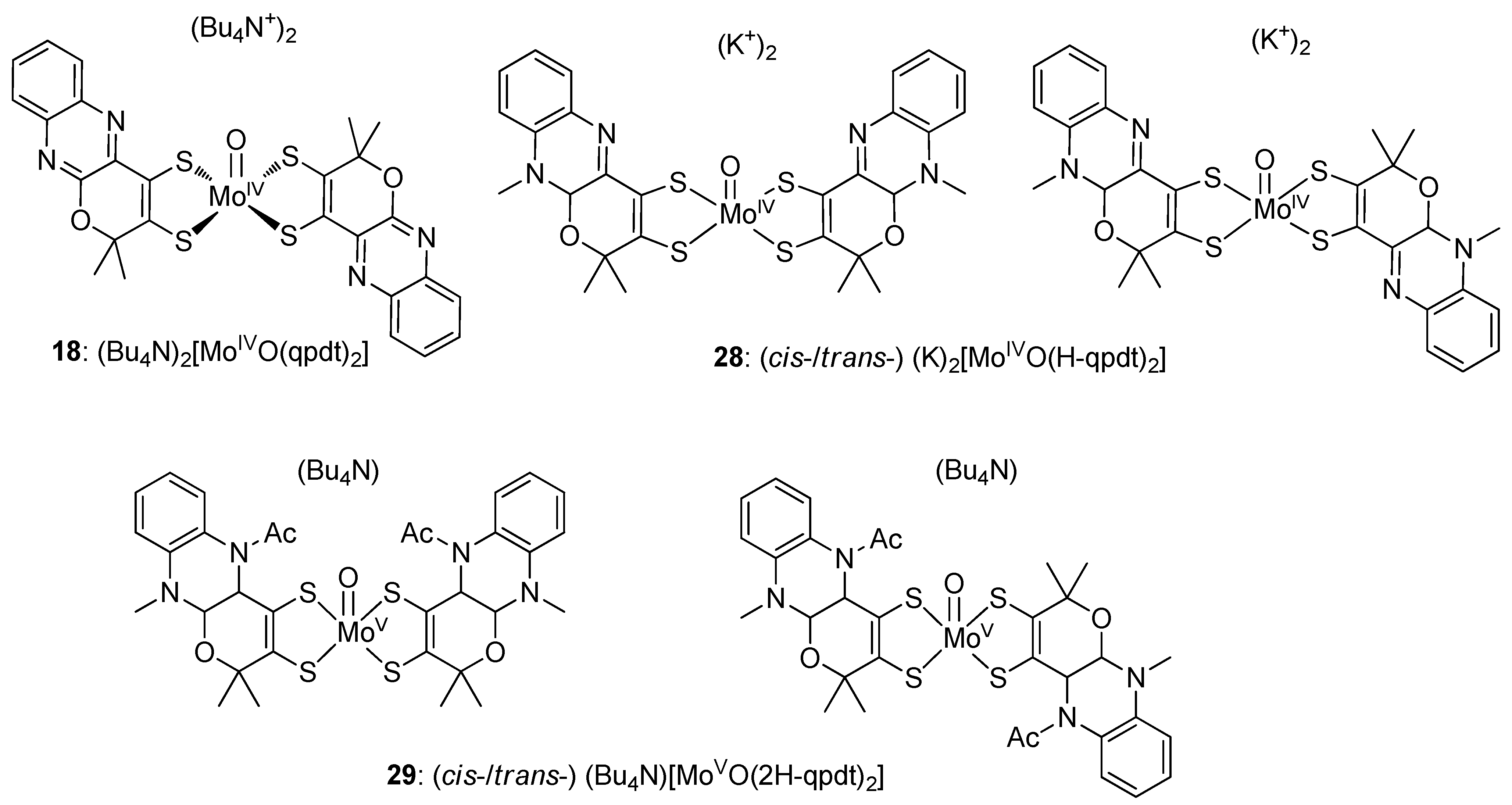
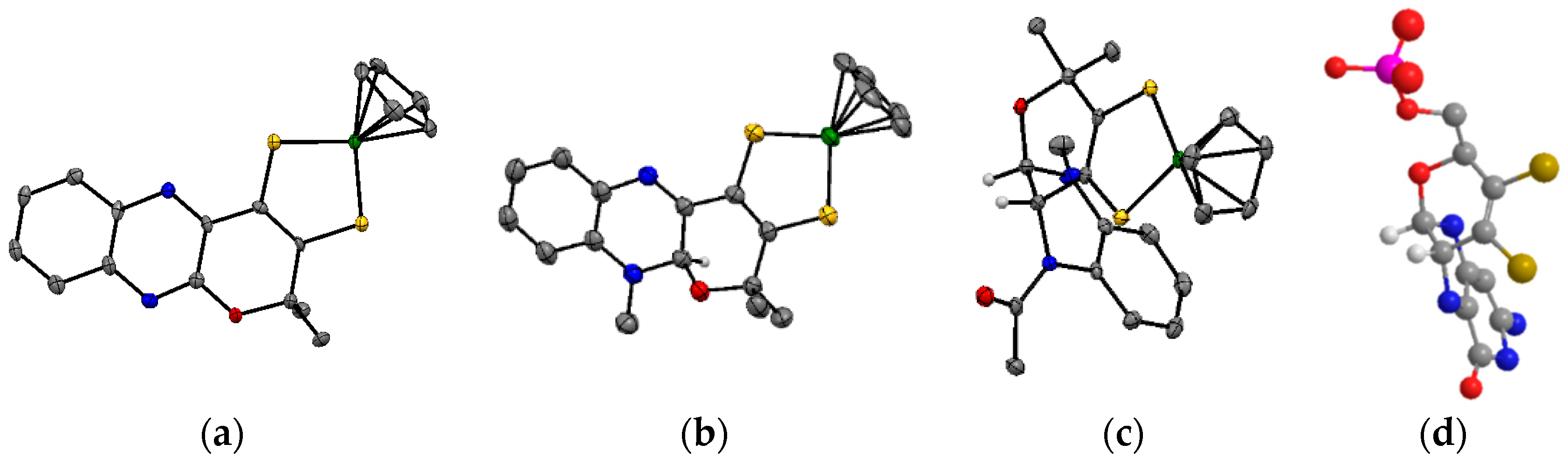
3.2. Pyrazine Containing Ligands
4. Functional Models
4.1. Mo/W-(Dithiolene)2 Complexes
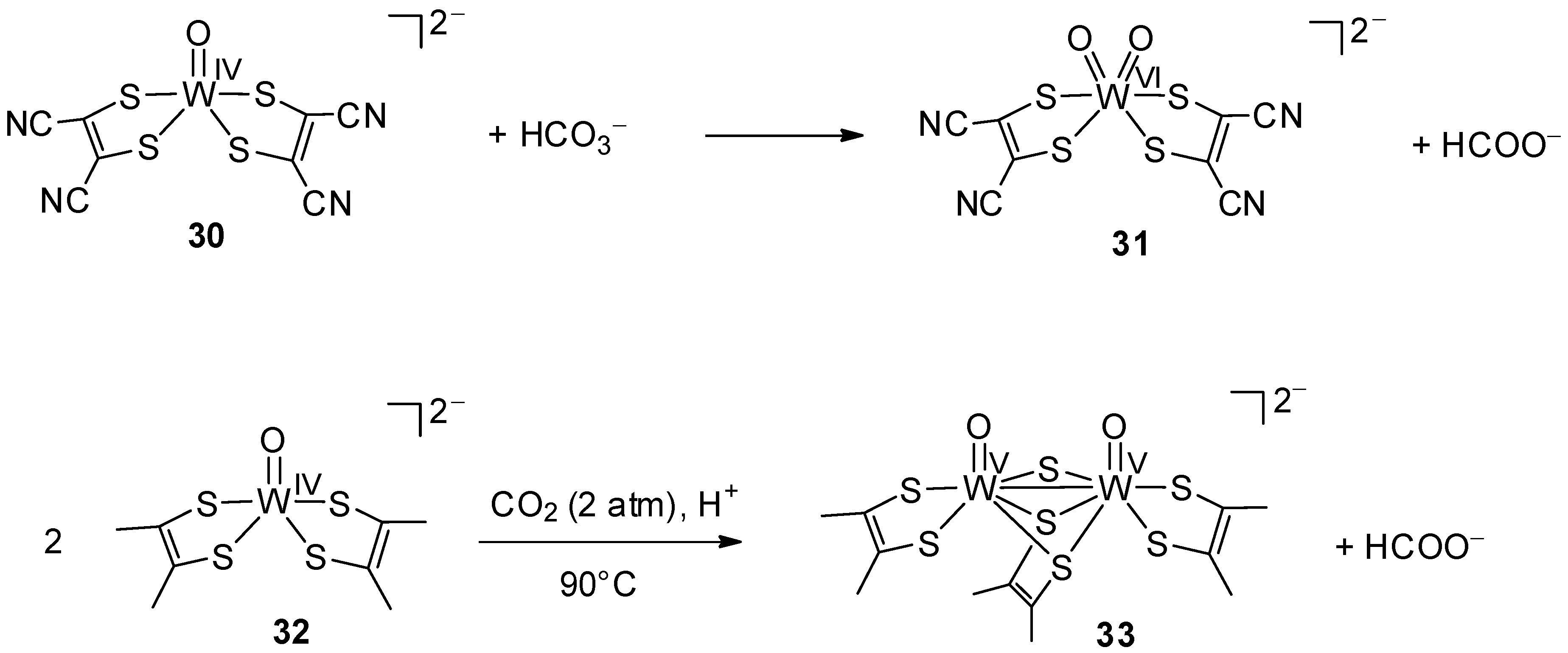
4.1.1. Equimolar Reaction with CO2
4.1.2. Catalytic Reduction of CO2
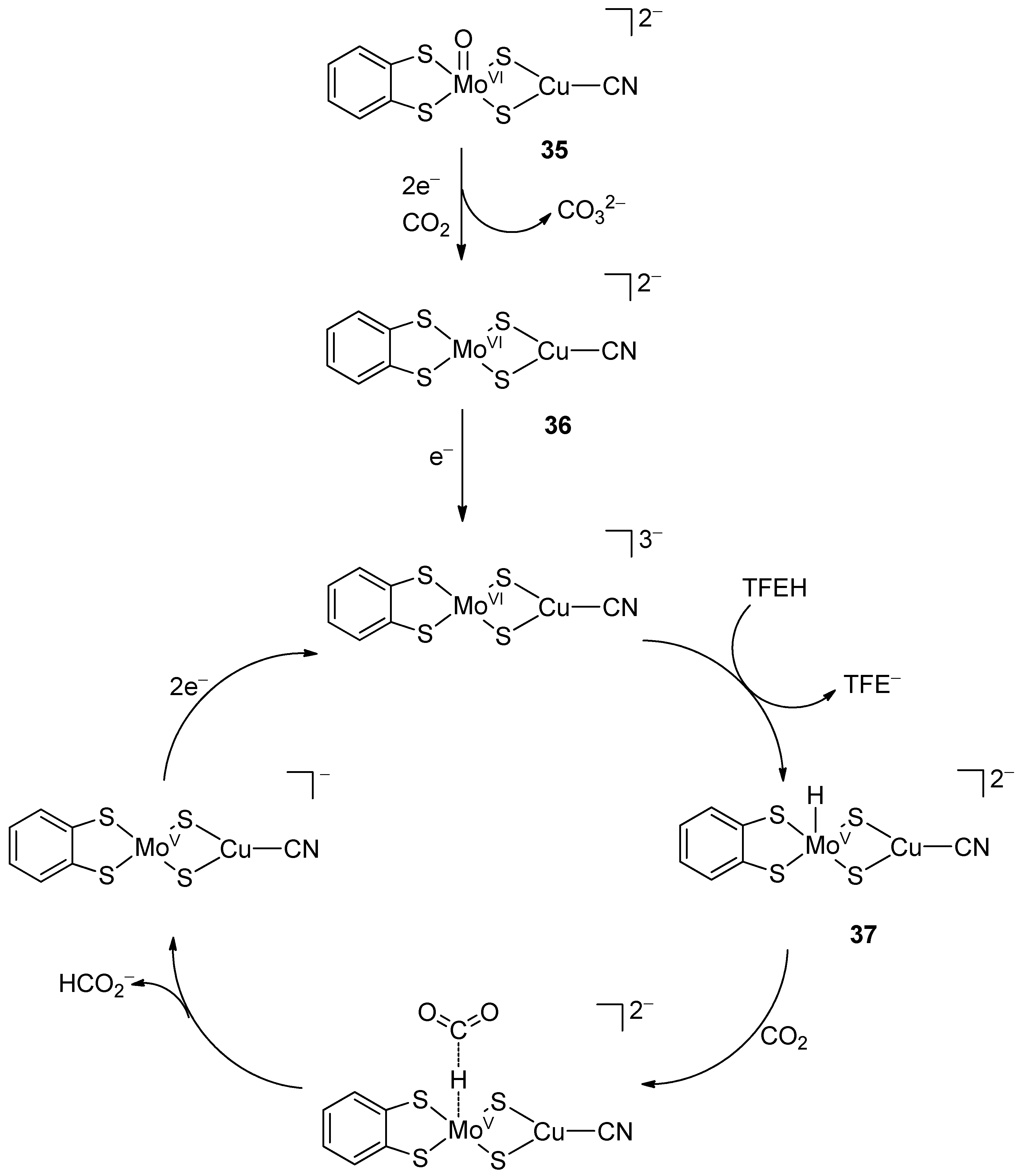
4.2. Mo/W-Cu Complexes, Models of CO-Dehydrogenases
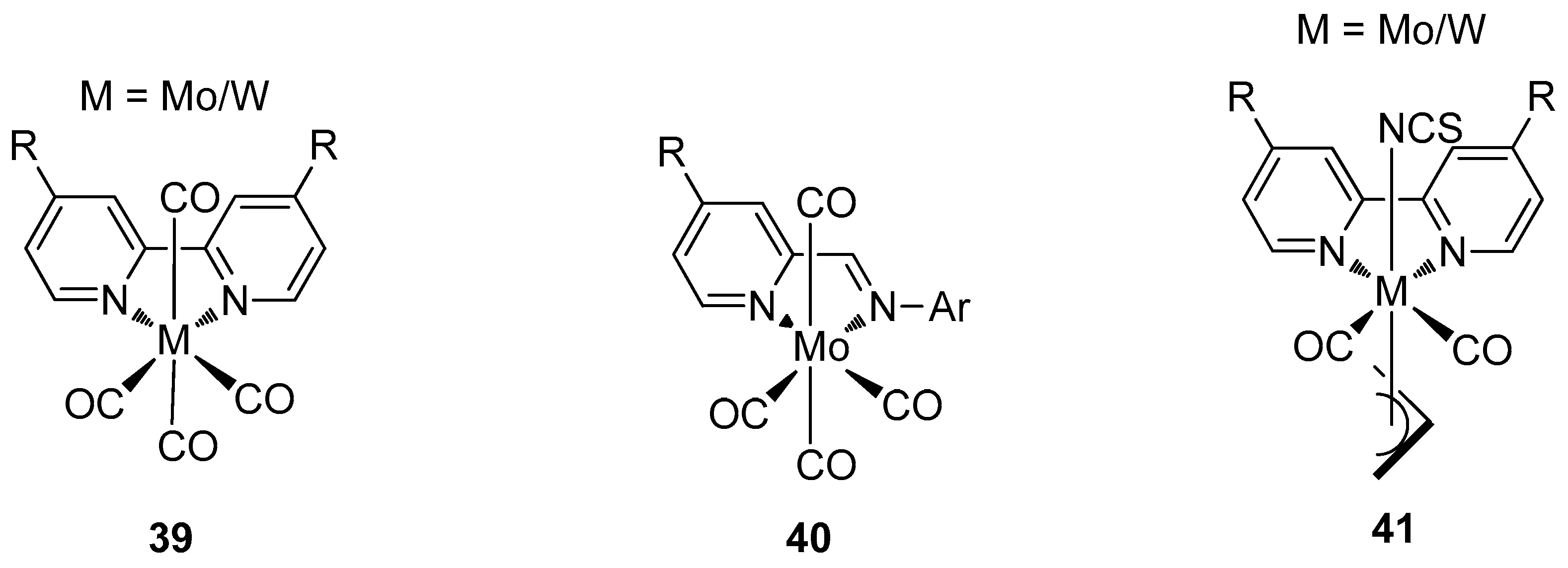
4.3. Other Mo/W Complexes
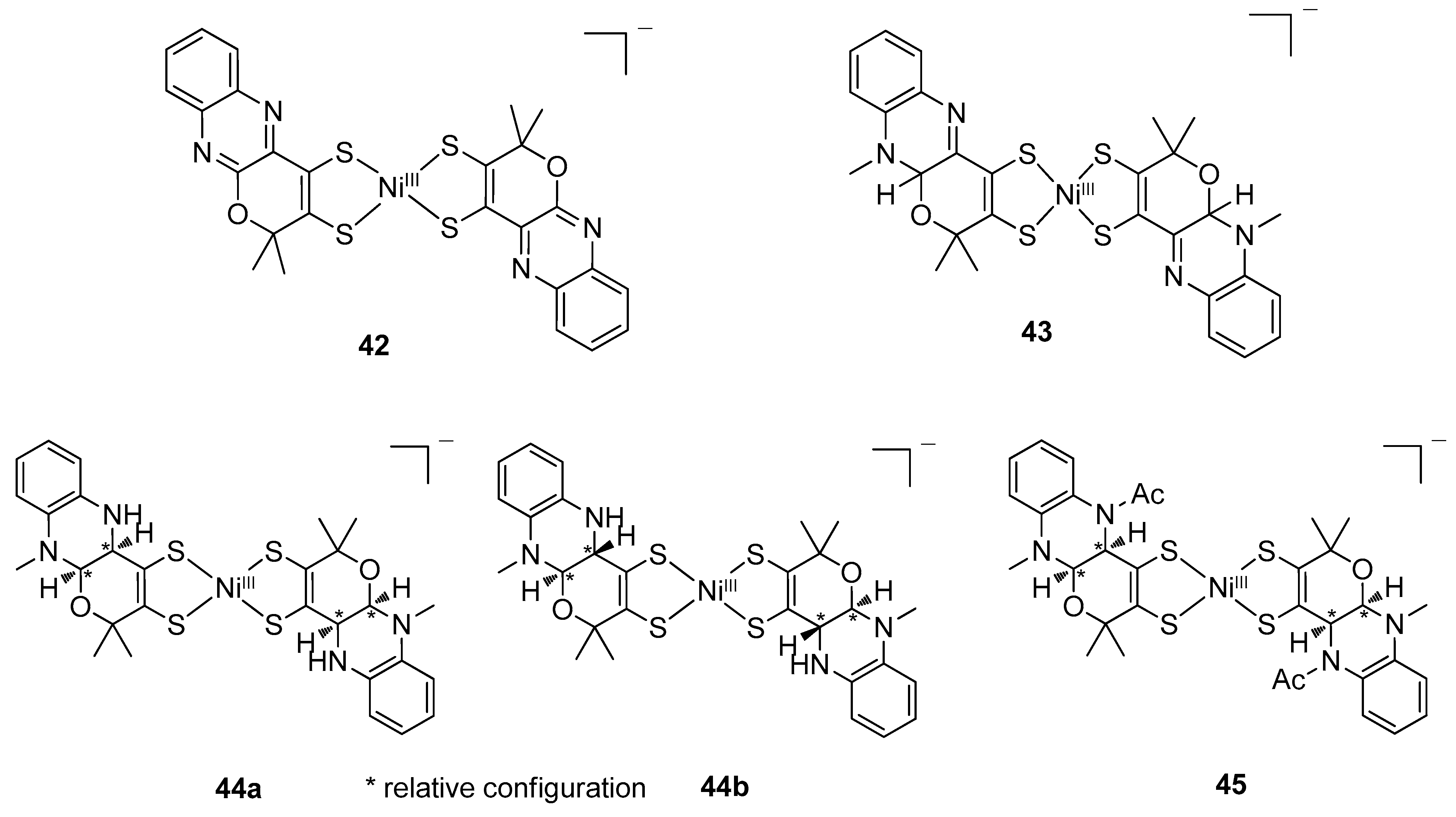
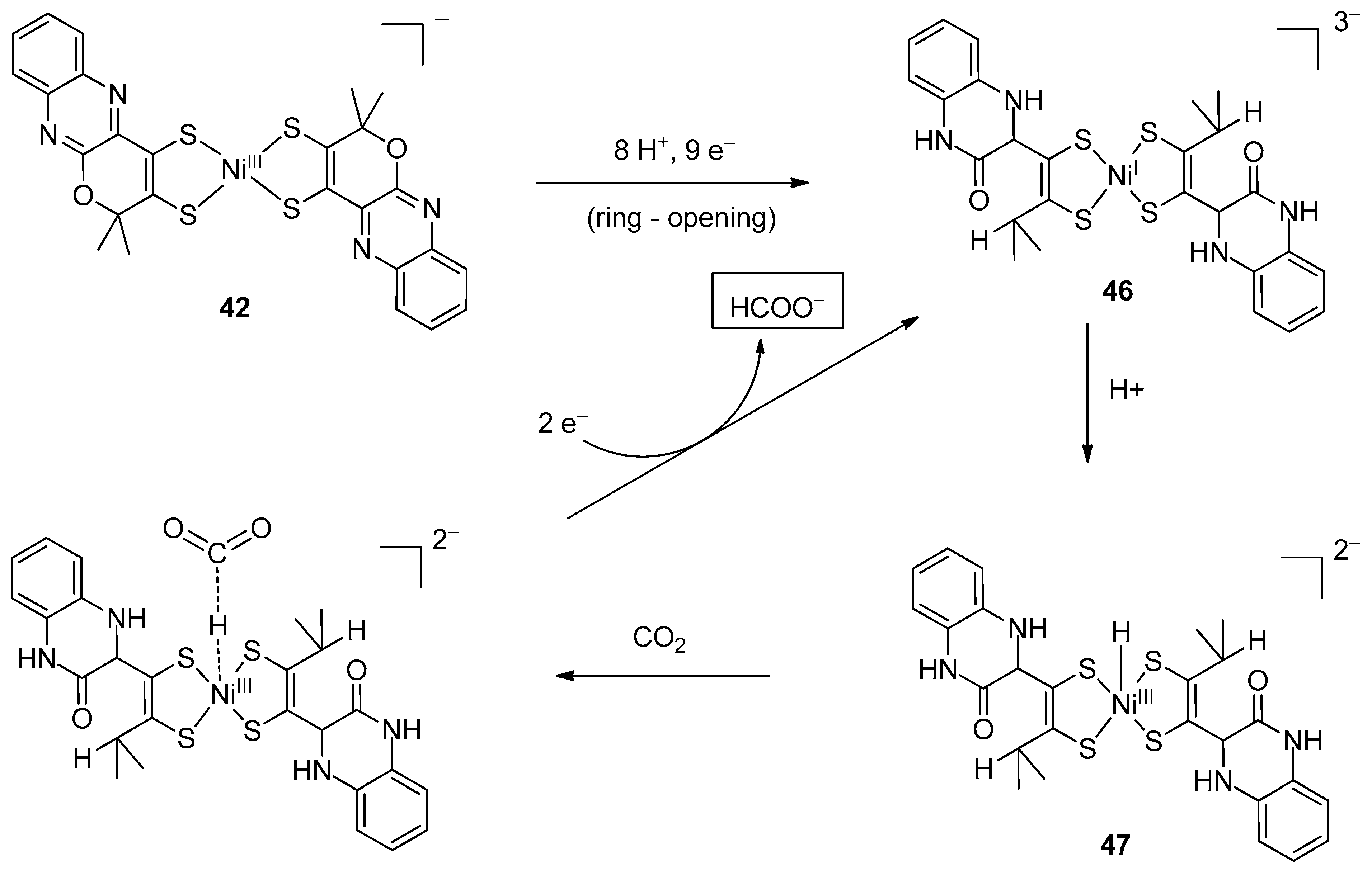
4.4. Ni(dithiolene)2 Complexes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, X.Y.; Yu, C.; Ren, Y.W.; Cui, S.; Li, W.B.; Qiu, J.S. Recent advances in innovative strategies for the CO2 electroreduction reaction. Energy Environ. Sci. 2021, 14, 765–780. [Google Scholar] [CrossRef]
- Grim, R.G.; Huang, Z.; Guarnieri, M.T.; Ferrell, J.R.; Tao, L.; Schaidle, J.A. Transforming the carbon economy: Challenges and opportunities in the convergence of low-cost electricity and reductive CO2 utilization. Energy Environ. Sci. 2020, 13, 472–494. [Google Scholar] [CrossRef]
- Kinzel, N.W.; Werle, C.; Leitner, W. Transition Metal Complexes as Catalysts for the Electroconversion of CO2: An Organometallic Perspective. Angew. Chem. Int. Ed. 2021, 60, 11628–11686. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Cometto, C.; Ishitani, O.; Robert, M. Electrons, Photons, Protons and Earth-Abundant Metal Complexes for Molecular Catalysis of CO2 Reduction. ACS Catal. 2017, 7, 70–88. [Google Scholar] [CrossRef]
- Romao, M.J. Molybdenum and tungsten enzymes: A crystallographic and mechanistic overview. Dalton Trans. 2009, 21, 4053–4068. [Google Scholar] [CrossRef]
- Niks, D.; Hille, R. Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism, and cofactor insertion. Protein Sci. 2019, 28, 111–122. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, I.; Moura, J.J.G. Molybdenum and tungsten-containing formate dehydrogenases: Aiming to inspire a catalyst for carbon dioxide utilization. Inorg. Chim. Acta 2017, 455, 350–363. [Google Scholar] [CrossRef]
- Maia, L.B.; Moura, J.J.G.; Moura, I. Molybdenum and tungsten-dependent formate dehydrogenases. J. Biol. Inorg. Chem. 2015, 20, 287–309. [Google Scholar] [CrossRef]
- Boyington, J.C.; Gladyshev, V.N.; Khangulov, S.V.; Stadtman, T.C.; Sun, P.D. Crystal structure of formate dehydrogenase H: Catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 1997, 275, 1305–1308. [Google Scholar] [CrossRef]
- Jormakka, M.; Richardson, D.; Byrne, B.; Iwata, S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 2004, 12, 95–104. [Google Scholar] [CrossRef]
- Raaijmakers, H.; Macieira, S.; Dias, J.M.; Teixeira, S.; Bursakov, S.; Huber, R.; Moura, J.J.G.; Moura, I.; Romao, M.J. Gene sequence and the 1.8 angstrom crystal structure of the tungsten-containing formate dehydrogenase from Desulfolvibrio gigas. Structure 2002, 10, 1261–1272. [Google Scholar] [CrossRef]
- Raaijmakers, H.C.A.; Romao, M.J. Formate-reduced E-coli formate dehydrogenase H: The reinterpretation of the crystal structure suggests a new reaction mechanism. J. Biol. Inorg. Chem. 2006, 11, 849–854. [Google Scholar] [CrossRef]
- Maia, L.B.; Fonseca, L.; Moura, I.; Moura, J.J.G. Reduction of Carbon Dioxide by a Molybdenum-Containing Formate Dehydrogenase: A Kinetic and Mechanistic Study. J. Am. Chem. Soc. 2016, 138, 8834–8846. [Google Scholar] [CrossRef] [PubMed]
- Enemark, J.H.; Cooney, J.J.A.; Wang, J.-J.; Holm, R.H. Synthetic Analogues and Reaction Systems Relevant to the Molybdenum and Tungsten Oxotransferases. Chem. Rev. 2004, 104, 1175–1200. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.; Burgmayer, S.J.N. Recent developments in the study of molybdoenzyme models. J. Biol. Inorg. Chem. 2015, 20, 373–383. [Google Scholar] [CrossRef]
- Patsch, S.; Correia, J.V.; Elvers, B.J.; Steuer, M.; Schulzke, C. Inspired by Nature-Functional Analogues of Molybdenum and Tungsten-Dependent Oxidoreductases. Molecules 2022, 27, 3695. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, H.; Tsukube, H. Chemical analogues relevant to molybdenum and tungsten enzyme reaction centres toward structural dynamics and reaction diversity. Chem. Soc. Rev. 2008, 37, 2609–2619. [Google Scholar] [CrossRef]
- Majumdar, A.; Sarkar, S. Bioinorganic chemistry of molybdenum and tungsten enzymes: A structural–functional modeling approach. Coord. Chem. Rev. 2011, 255, 1039–1054. [Google Scholar] [CrossRef]
- Sung, K.-M.; Holm, R.H. Oxo Transfer Reactions Mediated by Bis(dithiolene)tungsten Analogues of the Active Sites of Molybdoenzymes in the DMSO Reductase Family: Comparative Reactivity of Tungsten and Molybdenum. J. Am. Chem. Soc. 2001, 123, 1931–1943. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.-M.; Holm, R.H. Synthesis and Structures of Bis(dithiolene)−Tungsten(IV) Complexes Related to the Active Sites of Tungstoenzymes. Inorg. Chem. 2000, 39, 1275–1281. [Google Scholar] [CrossRef]
- Lim, B.S.; Holm, R.H. Bis(dithiolene)molybdenum analogues relevant to the DMSO reductase enzyme family: Synthesis, structures, and oxygen atom transfer reactions and kinetics. J. Am. Chem. Soc. 2001, 123, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.S.; Donahue, J.P.; Holm, R.H. Synthesis and Structures of Bis(dithiolene)molybdenum Complexes Related to the Active Sites of the DMSO Reductase Enzyme Family. Inorg. Chem. 2000, 39, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Goddard, C.A.; Holm, R.H. Synthesis and Reactivity Aspects of the Bis(dithiolene) Chalcogenide Series [WIVQ(S2C2R2)2]2− (Q = O, S, Se). Inorg. Chem. 1999, 38, 5389–5398. [Google Scholar] [CrossRef]
- Elvers, B.J.; Schulzke, C.; Fischer, C. Photochemical Unmasking of 1,3-Dithiol-2-ones: An Alternative Route to Heteroleptic Dithiolene Complexes from Low-Valent Molybdenum and Tungsten Precursors. Eur. J. Inorg. Chem. 2019, 2019, 2796–2805. [Google Scholar] [CrossRef]
- Ryde, U.; Schulzke, C.; Starke, K. Which functional groups of the molybdopterin ligand should be considered when modeling the active sites of the molybdenum and tungsten cofactors? A density functional theory study. J. Biol. Inorg. Chem. 2009, 14, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Alphonse, F.A.; Karim, R.; Cano-Soumillac, C.; Hebray, M.; Collison, D.; Garner, C.D.; Joule, J.A. A bis(eta(5)-cyclopentadienyl)cobalt complex of a bis-dithiolene: A chemical analogue of the metal centres of the DMSO reductase family of molybdenum and tungsten enzymes, in particular ferredoxin aldehyde oxidoreductase. Tetrahedron 2005, 61, 11010–11019. [Google Scholar] [CrossRef]
- Bradshaw, B.; Collison, D.; Garner, C.D.; Joule, J.A. Stable pyrano[2,3-b]quinoxalines and pyrano[2,3-g]pteridines related to molybdopterin. Chem. Commun. 2001, 123–124. [Google Scholar] [CrossRef]
- Bradshaw, B.; Dinsmore, A.; Ajana, W.; Collison, D.; Garner, C.D.; Joule, J.A. Synthesis of the organic ligand of the molybdenum cofactor, in protected form. J. Chem. Soc. Perkin Trans. 1. 2001, 3239–3244. [Google Scholar]
- Bradshaw, B.; Dinsmore, A.; Collison, D.; Garner, C.D.; Joule, J.A. The synthesis of pyrano[2,3-b]quinoxalines related to molybdopterin. J. Chem. Soc. Perkin Trans. 1. 2001, 3232–3238. [Google Scholar]
- Bradshaw, B.; Dinsmore, A.; Garner, C.D.; Joule, J.A. Synthesis of a cobalt complex of a pyrano[2,3-b]quinoxaline-3,4-dithiolate related to molybdopterin. Chem. Commun. 1998, 417–418. [Google Scholar] [CrossRef]
- Bertero, M.G.; Rothery, R.A.; Palak, M.; Hou, C.; Lim, D.; Blasco, F.; Weiner, J.H.; Strynadka, N.C.J. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 2003, 10, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Kloer, D.P.; Hagel, C.; Heider, J.; Schulz, G.E. Crystal structure of ethylbenzene dehydrogenase from Aromatoleum aromaticum. Structure 2006, 14, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.; Burgmayer, S.J.N. Pterin chemistry and its relationship to the molybdenum cofactor. Coord. Chem. Rev. 2011, 255, 1016–1038. [Google Scholar] [CrossRef]
- Marbella, L.; Serli-Mitasev, B.; Basu, P. Development of a Fluorescent Pb2+ Sensor. Angew. Chem. Int. Edit. 2009, 48, 3996–3998. [Google Scholar] [CrossRef] [PubMed]
- Pimkov, I.V.; Serli-Mitasev, B.; Peterson, A.A.; Ratvasky, S.C.; Hammann, B.; Basu, P. Designing the Molybdopterin Core through Regioselective Coupling of Building Blocks. Chem. Eur. J. 2015, 21, 17057–17072. [Google Scholar] [CrossRef]
- Williams, B.R.; Gisewhite, D.; Kalinsky, A.; Esmail, A.; Burgmayer, S.J.N. Solvent-Dependent Pyranopterin Cyclization in Molybdenum Cofactor Model Complexes. Inorg. Chem. 2015, 54, 8214–8222. [Google Scholar] [CrossRef]
- Williams, B.R.; Fu, Y.; Yap, G.P.A.; Burgmayer, S.J.N. Structure and Reversible Pyran Formation in Molybdenum Pyranopterin Dithiolene Models of the Molybdenum Cofactor. J. Am. Chem. Soc. 2012, 134, 19584–19587. [Google Scholar] [CrossRef][Green Version]
- Gisewhite, D.R.; Nagelski, A.L.; Cummins, D.C.; Yap, G.P.A.; Burgmayer, S.J.N. Modeling Pyran Formation in the Molybdenum Cofactor: Protonation of Quinoxalyl–Dithiolene Promoting Pyran Cyclization. Inorg. Chem. 2019, 58, 5134–5144. [Google Scholar] [CrossRef]
- Porcher, J.P.; Fogeron, T.; Gomez-Mingot, M.; Chamoreau, L.M.; Li, Y.; Fontecave, M. Synthesis and Reactivity of a Bio-inspired Dithiolene Ligand and its Mo Oxo Complex. Chem. Eur. J. 2016, 22, 4447–4453. [Google Scholar] [CrossRef]
- Porcher, J.P.; Fogeron, T.; Gomez-Mingot, M.; Derat, E.; Chamoreau, L.M.; Li, Y.; Fontecave, M. A Bioinspired Molybdenum Complex as a Catalyst for the Photo- and Electroreduction of Protons. Angew. Chem. Int. Ed. 2015, 54, 14090–14093. [Google Scholar] [CrossRef]
- Ames, D.E.; Mitchell, J.C.; Takundwa, C.C. Preparation and Reactions of 2-Alkynyl-3-chloroquinoxaline. J. Chem. Res. Miniprint 1985, 1683–1696. [Google Scholar]
- Fogeron, T.; Retailleau, P.; Chamoreau, L.M.; Fontecave, M.; Li, Y. The unusual ring scission of a quinoxaline-pyran-fused dithiolene system related to molybdopterin. Dalton Trans. 2017, 46, 4161–4164. [Google Scholar] [CrossRef] [PubMed]
- Fogeron, T.; Retailleau, P.; Chamoreau, L.M.; Li, Y.; Fontecave, M. Pyranopterin Related Dithiolene Molybdenum Complexes as Homogeneous Catalysts for CO2 Photoreduction. Angew. Chem. Int. Ed. 2018, 57, 17033–17037. [Google Scholar] [CrossRef] [PubMed]
- McNamara, W.R.; Han, Z.; Alperin, P.J.; Brennessel, W.W.; Holland, P.L.; Eisenberg, R. A Cobalt–Dithiolene Complex for the Photocatalytic and Electrocatalytic Reduction of Protons. J. Am. Chem. Soc. 2011, 133, 15368–15371. [Google Scholar] [CrossRef]
- McNamara, W.R.; Han, Z.; Yin, C.-J.M.; Brennessel, W.W.; Holland, P.L.; Eisenberg, R. Cobalt-dithiolene complexes for the photocatalytic and electrocatalytic reduction of protons in aqueous solutions. Proc. Natl. Acad. Sci. USA 2012, 109, 15594–15599. [Google Scholar] [CrossRef]
- Eckenhoff, W.T.; Brennessel, W.W.; Eisenberg, R. Light-Driven Hydrogen Production from Aqueous Protons using Molybdenum Catalysts. Inorg. Chem. 2014, 53, 9860–9869. [Google Scholar] [CrossRef]
- Das, A.; Han, Z.; Brennessel, W.W.; Holland, P.L.; Eisenberg, R. Nickel Complexes for Robust Light-Driven and Electrocatalytic Hydrogen Production from Water. ACS Catal. 2015, 5, 1397–1406. [Google Scholar] [CrossRef]
- Gomez-Mingot, M.; Porcher, J.P.; Todorova, T.K.; Fogeron, T.; Mellot-Draznieks, C.; Li, Y.; Fontecave, M. Bioinspired Tungsten Dithiolene Catalysts for Hydrogen Evolution: A Combined Electrochemical, Photochemical, and Computational Study. J. Phys. Chem. B 2015, 119, 13524–13533. [Google Scholar] [CrossRef]
- Koutsouri, E.; Mitsopoulou, C.A. Photocatalytic Hydrogen Evolution by tris-dithiolene tungsten complexes. Open Chem. 2016, 14, 393–403. [Google Scholar] [CrossRef]
- Zarkadoulas, A.; Field, M.J.; Papatriantafyllopoulou, C.; Fize, J.; Artero, V.; Mitsopoulou, C.A. Experimental and Theoretical Insight into Electrocatalytic Hydrogen Evolution with Nickel Bis(aryldithiolene) Complexes as Catalysts. Inorg. Chem. 2016, 55, 432–444. [Google Scholar] [CrossRef]
- Zarkadoulas, A.; Field, M.J.; Artero, V.; Mitsopoulou, C.A. Proton-Reduction Reaction Catalyzed by Homoleptic Nickel-bis-1,2-dithiolate Complexes: Experimental and Theoretical Mechanistic Investigations. ChemCatChem 2017, 9, 2308–2317. [Google Scholar] [CrossRef]
- Sarkar, S.; Das, S.K. CO2 fixation by [WIVO(S2C2(CN)2)2]2−: Functional model for the tungsten-formate dehydrogenase of Clostridium thermoaceticum. Proc. Indian Acad. Sci. (Chem. Sci.) 1992, 104, 533–534. [Google Scholar] [CrossRef]
- Seo, J.; Shearer, J.; Williard, P.G.; Kim, E. Reactivity of a biomimetic W(IV) bis-dithiolene complex with CO2 leading to formate production and structural rearrangement. Dalton Trans. 2019, 48, 17441–17444. [Google Scholar] [CrossRef] [PubMed]
- Shiekh, B.A.; Kaur, D.; Kumar, S. Bio-mimetic self-assembled computationally designed catalysts of Mo and W for hydrogenation of CO2/dehydrogenation of HCOOH inspired by the active site of formate dehydrogenase. Phys. Chem. Chem. Phys. 2019, 21, 21370–21380. [Google Scholar] [CrossRef]
- Dobbek, H.; Gremer, L.; Kiefersauer, R.; Huber, R.; Meyer, O. Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-angstrom resolution. Proc. Natl. Acad. Sci. USA 2002, 99, 15971–15976. [Google Scholar] [CrossRef]
- Jeoung, J.H.; Dobbek, H. Carbon dioxide activation at the Ni,Fe-cluster of anaerobic carbon monoxide dehydrogenase. Science 2007, 318, 1461–1464. [Google Scholar] [CrossRef] [PubMed]
- Mouchfiq, A.; Todorova, T.K.; Dey, S.; Fontecave, M.; Mougel, V. A bioinspired molybdenum–copper molecular catalyst for CO2 electroreduction. Chem. Sci. 2020, 11, 5503–5510. [Google Scholar] [CrossRef]
- Ghosh, D.; Sinhababu, S.; Santarsiero, B.D.; Mankad, N.P. A W/Cu Synthetic Model for the Mo/Cu Cofactor of Aerobic CODH Indicates That Biochemical CO Oxidation Requires a Frustrated Lewis Acid/Base Pair. J. Am. Chem. Soc. 2020, 142, 12635–12642. [Google Scholar] [CrossRef]
- Clark, M.L.; Grice, K.A.; Moore, C.E.; Rheingold, A.L.; Kubiak, C.P. Electrocatalytic CO2 reduction by M(bpy-R)(CO)4 (M = Mo, W.; R = H, tBu) complexes. Electrochemical, spectroscopic, and computational studies and comparison with group 7 catalysts. Chem. Sci. 2014, 5, 1894. [Google Scholar] [CrossRef]
- Tory, J.; Setterfield-Price, B.; Dryfe, R.A.W.; Hartl, F. [M(CO)4(2,2′-bipyridine)] (M=Cr, Mo, W) Complexes as Efficient Catalysts for Electrochemical Reduction of CO2 at a Gold Electrode. ChemElectroChem 2015, 2, 213–217. [Google Scholar] [CrossRef]
- Sieh, D.; Lacy, D.C.; Peters, J.C.; Kubiak, C.P. Reduction of CO2 by Pyridine Monoimine Molybdenum Carbonyl Complexes: Cooperative Metal-Ligand Binding of CO2. Chem. Eur. J. 2015, 21, 8497–8503. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.O.; Veenstra, F.L.P.; Chippindale, A.M.; Calhorda, M.J.; Hartl, F. Group 6 Metal Complexes as Electrocatalysts of CO2 Reduction: Strong Substituent Control of the Reduction Path of [Mo(η3-allyl)(CO)2(x,x′-dimethyl-2,2′-bipyridine)(NCS)] (x = 4–6). Organometallics 2019, 38, 1372–1390. [Google Scholar] [CrossRef]
- Grice, K.A.; Saucedo, C. Electrocatalytic Reduction of CO2 by Group 6 M(CO)6 Species without "Non-Innocent" Ligands. Inorg. Chem. 2016, 55, 6240–6246. [Google Scholar] [CrossRef] [PubMed]
- Fogeron, T.; Todorova, T.K.; Porcher, J.P.; Gomez-Mingot, M.; Chamoreau, L.M.; Mellot-Draznieks, C.; Li, Y.; Fontecave, M. A Bioinspired Nickel(bis-dithiolene) Complex as a Homogeneous Catalyst for Carbon Dioxide Electroreduction. ACS Catal. 2018, 8, 2030–2038. [Google Scholar] [CrossRef]
- Fogeron, T.; Retailleau, P.; Gomez-Mingot, M.; Li, Y.; Fontecave, M. Nickel Complexes Based on Molybdopterin-like Dithiolenes: Catalysts for CO2 Electroreduction. Organometallics 2019, 38, 1344–1350. [Google Scholar] [CrossRef]



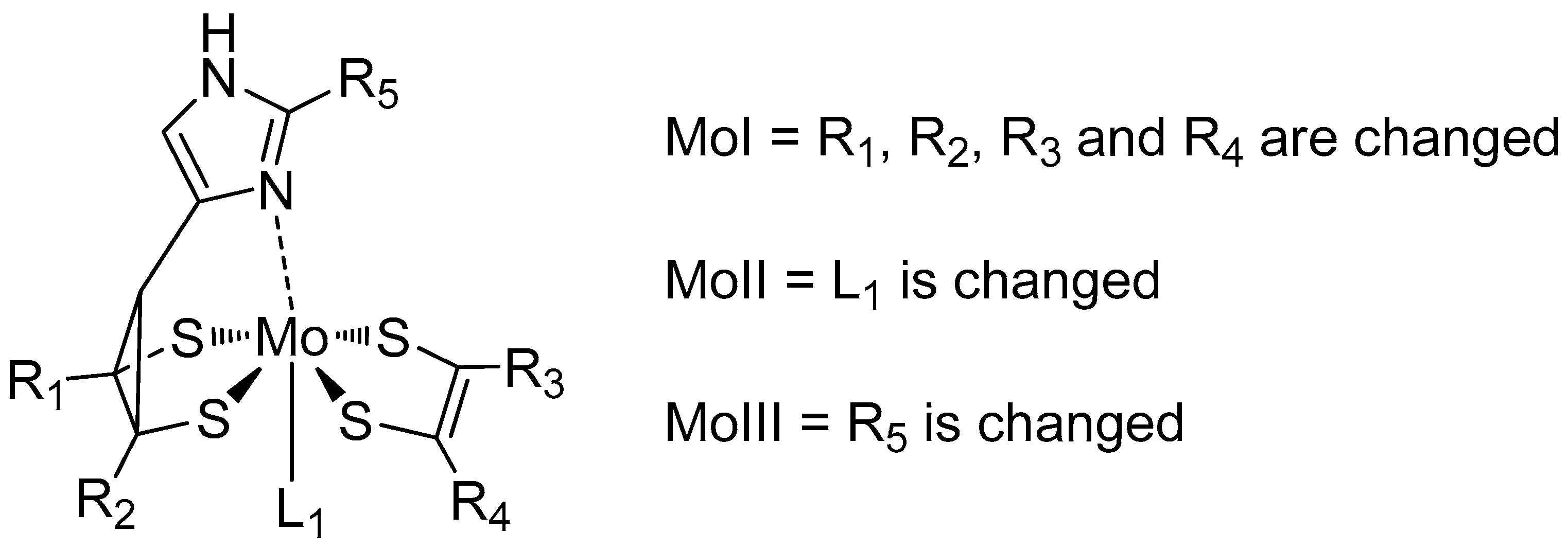

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fogeron, T.; Li, Y.; Fontecave, M. Formate Dehydrogenase Mimics as Catalysts for Carbon Dioxide Reduction. Molecules 2022, 27, 5989. https://doi.org/10.3390/molecules27185989
Fogeron T, Li Y, Fontecave M. Formate Dehydrogenase Mimics as Catalysts for Carbon Dioxide Reduction. Molecules. 2022; 27(18):5989. https://doi.org/10.3390/molecules27185989
Chicago/Turabian StyleFogeron, Thibault, Yun Li, and Marc Fontecave. 2022. "Formate Dehydrogenase Mimics as Catalysts for Carbon Dioxide Reduction" Molecules 27, no. 18: 5989. https://doi.org/10.3390/molecules27185989
APA StyleFogeron, T., Li, Y., & Fontecave, M. (2022). Formate Dehydrogenase Mimics as Catalysts for Carbon Dioxide Reduction. Molecules, 27(18), 5989. https://doi.org/10.3390/molecules27185989






